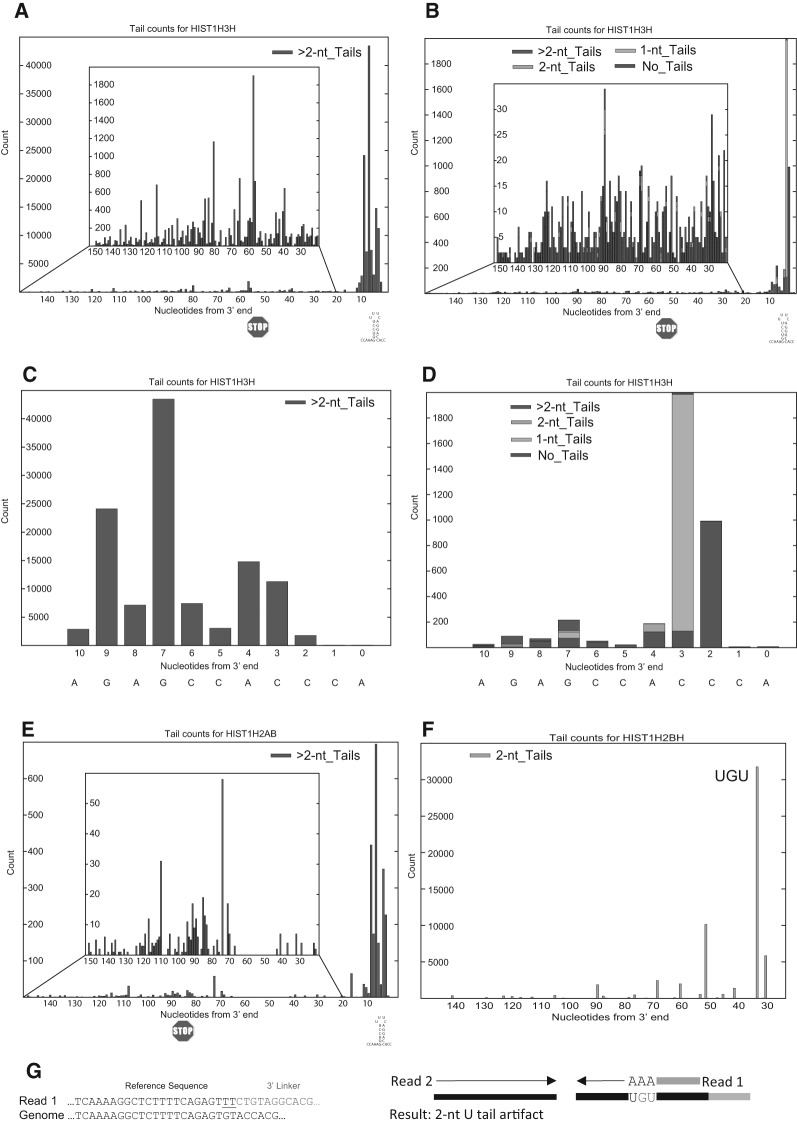
FIGURE 3.
Priming with a primer ending in three A's enhances sensitivity and enriches oligouridylated RNAs. (A–D) The profile of sequences obtained from the HIST1H3H mRNA from the same sample analyzed in Fig. 2F–I obtained from cDNA primed with the oligo(dA) primer (A,C) or the standard linker (B,D) are shown. In A,B, the sequences 5′ of the stem–loop are shown in the inset, and in C,D, the sequences from the 3′ side of the stem–loop are shown. Since the HIST1H3H mRNA is expressed at about a fivefold lower level than the highly expressed histone mRNAs, there were very few sequences in the body of the mRNA obtained with the standard primer. (E) The profile of sequences from the HIST1H2AB mRNA from the same sample obtained from cDNA primed with the oligo(dA) primer. This histone mRNA is expressed at a very low level, and there were not enough sequences obtained with the standard primer to analyze this mRNA. (F) Artifacts obtained with the oligo(dA) primer from the HIST1H2BH mRNA. The major peak resulted from mispriming at a UGU sequence, resulting in sequences containing three U's at the 3′ end, which were identified as a 2-nt tail since one U is encoded in the genome. (G) AppEnD analysis of sequences misprimed at a UGU sequence when cDNA is primed with the 3A-primer. We observed a number of these examples, but the AppEnD pipeline removes them as authentic tails, since the nontemplated tail is only 2 nt.