Abstract
Chemerin, highly expressed in adipose and liver tissues, regulates glucose and lipid metabolism and immunity in these tissues in ruminants and mice. Our previous reports showed that chemerin is involved in adipogenesis and lipid metabolism in adipose tissue as an adipokine. The aim of the present study was to identify single nucleotide polymorphisms (SNPs) in the coding region of the chemerin gene and to analyze their effects on carcass traits and intramuscular fatty acid compositions in Japanese Black cattle. The SNPs in the bovine chemerin gene were detected in 232 Japanese Black steers (n = 161) and heifers (n = 71) using DNA sequencing. The results revealed five novel silent mutations: NM_001046020: c.12A>G (4aa), c.165G<A (55aa), c.276C>T (92aa), c.321 A>G (107aa), and c.396C>T (132aa). There was no association between 4 of the SNPs (c.12A>G [4aa], c.165G<A [55aa], c.321 A>G [107aa], and c.396C>T) and carcass traits or intramuscular fatty acid compositions. Regarding the remaining SNP, c.276C>T, we found that cattle with genotype CC had a higher beef marbling score than that of cattle with genotype CT, whereas cattle with genotype CT had a higher body condition score (p<0.10). Further, cattle with genotype CC had significantly higher C18:0 content in their intramuscular fat tissue than that of cattle with genotype CT (p<0.05). On the other hand, cattle with genotype CT had significantly higher C14:0 and C16:0 content in their intramuscular fat tissue (p<0.05). Moreover, the number of individuals carrying the minor allele of c.276C>T SNP is small. It is suggested that the c.276C>T SNP of the chemerin gene has potential in cattle breeding using modern methods, such as marker assisted selection. So, further functional and physiological research elucidating the impact of the chemerin gene on bovine lipid metabolism including fatty acid synthesis will help in understanding these results.
Keywords: Japanese Black Cattle, Chemerin, Single Nucleotide Polymorphism, Carcass Trait
INTRODUCTION
Chemerin, also known as retinoic acid receptor responder protein 2 or tazarotene-induced gene 2, plays a regulatory role not only in the immune system but also in energy metabolism. Chemerin is a secreted protein whose gene expression is observed in several tissues, especially in adipose tissue and the liver (Roh et al., 2007), which are essential organs for energy metabolism. Recent studies in mice and humans indicate that chemerin regulates glucose and lipid metabolism (Goralski et al., 2007; Sell et al., 2010) and is involved in metabolic disorders such as obesity, insulin resistance in skeletal muscle tissue, and non-alcoholic fatty liver disease (Bozaoglu et al., 2007; Sell et al., 2009). In our previous studies, chemerin was up-regulated during bovine adipocyte differentiation, and chemerin analog regulated insulin secretion related to glucose metabolism and the release of triglycerides in sheep in vivo (Roh et al., 2006; Song et al., 2010; Suzuki et al., 2012). In addition, in fully differentiated adipocytes, the treatment of tumor necrosis factor (TNF)-α and adiponectin up-regulated both chemerin and chemokine-like receptor 1 (CMKLR1) gene expression levels, although leptin did not show such effects (Suzuki et al., 2012). These results suggest that the expression of chemerin in bovine adipocytes might be regulated by chemerin itself and other adipokines, which indicates the possible role of chemerin in modulating the adipokine secretions in adipose tissues.
Japanese Black cattle are well known for their highly marbled beef and greater content of unsaturated fatty acids and they seem well adapted to accumulate oleic acid in their adipose tissue (Smith et al., 2006). Marbling abundance is an important component of the beef carcass quality grade, and a high concentration of oleic acid contributes to palatability and softness of beef (Yang et al., 1999). These traits are influenced by a large number of genes, and many genetic factors related to these traits have been investigated.
The bovine chemerin gene is located on chromosome 4 (NCBI reference sequence: NM_001046020.2) and consists of six exons, with exons 2, 3, 4, and 5 coding a protein with 162 amino acids. Single nucleotide polymorphism (SNP) A868G (c.12 A>G), located in exon 2 of the Bos taurus chemerin gene, was detected in six Chinese cattle breeds (Tian et al., 2011). A868G was a synonymous mutation, and it affected some carcass traits in Qinchuan heifers. Chemerin expression in subcutaneous adipose tissue from cross-bred heifers was correlated with body condition score (BCS) and yearling weight (Lindholm-Perry et al., 2012).
In this paper we report the discovery of new SNPs in the coding region of the bovine chemerin gene and attempt to show their significance for carcass traits and intramuscular fatty acids composition in Japanese Black cattle.
MATERIALS AND METHODS
Animals and data collection
Japanese Black cattle (161 steers and 71 heifers) were raised and slaughtered in Miyagi Prefecture, Japan, and used to analyze the chemerin allelic frequencies and for the association study. The average slaughtering age and carcass weight (CW) of the cattle were 30.7±2.2 months and 459.9±67.3 kg, respectively. A cross-sectional portion of the intramuscular adipose tissue was collected from longissimus thoracis muscle at the 6th to 7th thoracic vertebrae, and a portion of skeletal muscle tissue was sampled from Japanese Black carcasses. The adipose and muscle tissues were stored at −20°C until analyses of fatty acid composition and genomic DNA extraction, respectively.
DNA extraction
Genomic DNA was extracted from approximately 50 mg of muscle tissue using DNA extraction apparatus QG-800 (Fujifilm, Tokyo, Japan) and QuickGene DNA tissue kit S (Fujifilm, Japan). The DNA concentration was measured using a NanoDrop ND-1000 Spectrophotometer V3.7.1 (Thermo Fisher Scientific, Wilmington, MA, USA).
Single nucleotide polymorphism identification and genotyping
According to the sequence of the bovine chemerin gene (GenBank accession no. NM_001046020), two pairs of primers were designed to amplify a coding region of the chemerin gene that included exons 2 to 4. The gene primer sequences were: E1-forward: 5′-GGCTGGGCTAAGGAACAGTG-3′, E1-reverse:5′-GGTCTCCAACCTCAGGCTTC-3′. E2-forward: 5′-CTGCAGGATAGTTCTGACTTTTG-3′, E2-reverse: 5′-GCTTTATTAGCTCAGGGGTCA-3′. Polymerase chain reaction (PCR) amplifications were performed in 30-μL reaction mixtures, each containing 10 to 30 ng DNA template, 5 μM of each primer, 3 μL 10×PCR Buffer for KOD-Plus-, 0.20 mM dNTPs, 1.67 mM MgSO4, and 0.5 U KOD-Plus- Polymerase (Toyobo, Osaka, Japan). The PCR protocol was 94°C for 9 min followed by 35 cycles of 30 s at 94°C, 30 s at 63°C or 60°C, and 1 min at 72°C and a final extension at 72°C for 5 min. Next, the PCR products were electrophoresed through 1.5% agarose gel and single band was confirmed. Then, the products were purified using Fast Gel/PCR Extraction Kit (Nippon Genetics Co, Ltd, Tokyo, Japan). Finally, the PCR products were sequenced to detect the presence of SNPs. Bi-directional DNA sequencing of PCR amplicons was carried out using an ABI PRISM 3730 sequencer (Applied Biosystems, Foster, CA, USA). Sequencing variants were detected by visual examination of the sequencing map followed by alignment using CLUSTAL (Higgins and Sharp, 1988).
Fatty acids composition
To analyze fatty acid compositions, we performed gas chromatography analysis as described in a previous report (Yokota et al., 2012). Briefly, total lipid was extracted by adding 1 mL of n-hexane to approximately 10 mg of intramuscular adipose tissue of longissimus thoracis muscle. The mixture was vortexed and incubated at room temperature for 30 min. Two hundred microliters of 2 N sodium hydrate/methanol solution (1:1 v/v) and 600 μL of hydrogen chloride-methanol reagent (5% to 10%) were added to saponify and methylate the mixture (Tokyo Chemical Industry, Tokyo, Japan). The methylated lipid was concentrated by flowing in N2 gas. The concentrated lipid was then diluted with 500 μL of hexane before proceeding to GC analysis. The fatty acid was analyzed by gas chromatography (6850 Network GC System; Agilent Technologies, Palo Alto, CA, USA). The inlet temperature was 230°C, and the oven temperature was programmed to gradually increase from 100°C to 170°C at 10°C/min and from 170°C to 224°C at 3°C/min. The detector sensor temperature was set at 230°C. We used helium as the carrier gas and a capillary column (DB-23, 0.25 mm ID×30 m, df = 0.25 μm; Agilent Technologies, USA) and a flame ionization detector for detection. The data for fatty acid composition comprised percentages of C14:0 (myristic acid), C16:0 (palmitic acid), C16:1 (palmitoleic acid), C18:0 (stearic acid), C18:1 (oleic acid), C18:2 (linoleic acid), saturated fatty acid (SFA) (C14:0+C16:0+C18:0), and monounsatureated fatty acid (MUFA) (C16:1 + C18:1).
Statistical analysis
Gene frequencies were determined by direct counting. The effects of chemerin genotypes on carcass traits and intramuscular fatty acid compositions in Japanese Black cattle were analyzed by analysis of variance using the general linear model procedure of SAS (SAS Institute, Cary, NC, USA). The statistical model included fixed effects (sex, sampling year, and genetic information of the chemerin gene) and the covariate effect of slaughter age. Differences were considered significant at p<0.05, and differences at p<0.10 were considered to indicate a tendency. The statistical model of the genotype of c.276 of the chemerin gene was as follows:
yijk: the trait measured on the individual i
μ: the overall mean for the trait
sexj: the fixed effect of sex j (2 classes: male and female)
yeark: the fixed effect of sampling year k (3 classes: years 20093 classes: years 2010, and 2011)
b: the covariate coefficient with xijk
xijk: the random effect of slaughter age ijk
Chemerin276: the fixed effect of the c.276 genotype in the chemerin gene (2 classes: CC and CT)
eijk: the residual effect
RESULTS
Single nucleotide polymorphisms identified and genotypes
Eight hundred fifteen and 823 bp fragments of the chemerin gene were amplified and sequenced with two different primers, respectively. As a result, five silent mutations, c.12A>G (4aa), c.165G>A (55aa), c.276C>T (92aa), c.321A>G (107aa), and c.396C>T (132aa), were detected within the coding region of the chemerin gene (Figure 1).
Figure 1.
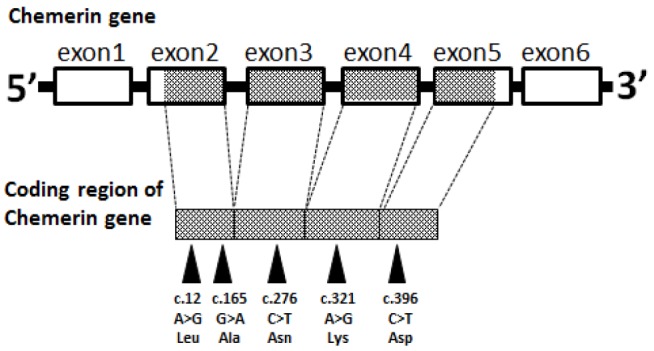
Single nucleotide polymorphisms present in the coding region of the bovine chemerin gene.
Table 1 shows the genotype distribution for each SNP site. The highest frequencies at c.12A>G, c.165G>A, c.276C>T, c.321A>G, and c.396C>T were for genotypes AA, GG, CC, AA, and CC, respectively. Most of the alleles at c.276 were allele C, and frequency of allele T was low. Because the frequency of genotypes with the minor allele T was very low (<0.03), we analyzed the effects of individual SNPs only.
Table 1.
Genotype frequencies (%) and allelic frequencies (%) of the chemerin gene determined by gene sequencing
| SNPs | Genotype frequencies | Allele frequencies | |||
|---|---|---|---|---|---|
|
|
|
||||
| c.12A>G | AA (80) | AG (112) | GG (40) | A | G |
| 34.5 | 48.3 | 17.2 | 58.6 | 41.4 | |
| c.165G>A | AA (1) | AG (5) | GG (226) | A | G |
| 0.4 | 2.2 | 97.4 | 1.5 | 98.5 | |
| c.276C>T | CC (214) | CT (18) | C | T | |
| 92.2 | 7.8 | 96.1 | 3.9 | ||
| c.321A>G | AA (109) | AG (98) | GG (25) | A | G |
| 47.0 | 42.2 | 10.8 | 68.1 | 31.9 | |
| c.396C>T | CC (231) | CT (1) | C | T | |
| 99.6 | 0.4 | 99.8 | 0.2 | ||
SNPs, single nucleotide polymorphisms.
Single nucleotide polymorphism and carcass trait associations
In Table 2, the genotypes of 232 individuals were compared with the phenotypic data for 7 carcass traits and 6 intramuscular fatty acid compositions. The carcasses were evaluated by official graders according to the guidelines of the Japanese Meat Grading Association. The recorded traits, including carcass weight, rib eye area, rib thickness, subcutaneous fat thickness, and yield estimate, were measured or calculated. There were no associations between 4 of the SNPs (c.12A>G [4aa], c.165G<A [55aa], c.321 A>G [107aa], and c.396C>T [132aa]) and carcass traits or intramuscular fatty acid compositions. In the case of the SNP marker of c.276C>T in the exon 3 region, however, cattle with the genotype CC tended to have a higher beef marbling score (BMS), and cattle with genotype CT tended to have a higher BCS (p<0.10) (Table 2). In addition, cattle with genotype CC had a significantly higher percentage of C18:1 (p<0.05) and tended to have a higher percentage of MUFA (p<0.10) (Table 2). In contrast, cattle with genotype CT had significantly higher percentages of C14:0 and C16:0 (p<0.05) (Table 2). These findings indicate that a larger proportion of these two saturated fatty acids led to the high proportion of SFA in cattle with genotype CT (p<0.10). These results suggested that the c.276C>T SNP of the chemerin gene may affect carcass and meat quality traits in Japanese Black cattle.
Table 2.
Effects of c.276C>T of the chemerin gene on carcass traits and intramuscular fatty acids composition in Japanese Black cattle (n = 232)
| Traits | c.276C>T | |
|---|---|---|
|
| ||
| CC | CT | |
| Carcass trait | ||
| CW (kg) | 462.1±4.6 | 433.4±16.2 |
| REA (cm2) | 61.1±0.7 | 58.4±2.5 |
| RT (cm) | 8.0±0.1 | 7.8±0.2 |
| SFT (cm) | 2.5±0.1 | 2.7±0.2 |
| YE (%) | 74.7±0.1 | 74.4±0.4 |
| BMS No. | 6.5±0.2** | 5.1±0.4 |
| BCS No. | 3.8±0.0 | 4.1±0.1** |
| Intramuscular fatty acid compositions | ||
| C14:0 | 2.6±0.0 | 2.7±0.1* |
| C16:0 | 26.3±0.1 | 26.9±0.4* |
| C16:1 | 4.0±0.1 | 4.2±0.2 |
| C18:0 | 12.0±0.1 | 12.1±0.6 |
| C18:1 | 52.9±0.2* | 51.9±0.7 |
| C18:2 | 2.2±0.0 | 2.2±0.1 |
| SFA | 40.9±0.2 | 41.7±0.8** |
| MUFA | 56.9±0.2** | 56.1±0.8 |
CW, carcass weight; REA, rib eye area; RT, rib thickness; SFT, subcutaneous fat thickness; YE, yield estimate; BMS No., beef marbling standard number; BCS No., beef color standard number; SFA, saturated fatty acids; MUFA, monounsaturated fatty acids.
p<0.1,
p<0.05.
DISCUSSION
Our study is the first to demonstrate that the polymorphic loci identified within the bovine chemerin gene might be associated with marbling score and intramuscular fatty acid compositions in Japanese Black cattle. Previously, several genetic variations of the fatty acid synthase, stearoyl-CoA desaturase, and GH (growth hormone) genes associated with bovine carcass traits had been reported (Taniguchi et al., 2004; Abe et al., 2009; Matsuhashi et al., 2011; Yokota et al., 2012). Our previous studies showed that SNPs in the GH gene of Japanese Black cattle were associated with some carcass traits, fatty acid composition, the expression of lipogenic genes, and the regulation of plasma insulin level in Japanese Black heifers (Ardiyanti et al., 2009; Ardiyanti et al., 2012). The c.12 A>G SNP that we detected in the coding region of the chemerin gene in Japanese Black cattle was identical to the A868G SNP detected in six Chinese cattle breeds (Tian et al., 2011). In agreement with their report, genotype AG of this SNP was the dominant genotype in Japanese Black cattle. However, there was no association between carcass traits and AA/AG/GG genotypes in the Japanese Black cattle used in our study. Thus, these results suggest that the effect of the c.12A>G SNP might differ among breeds.
A human study revealed that the rs17173608 SNP located on intron 3 of the chemerin gene was associated with visceral adipose tissue mass (Mussig et al., 2009). Another report showed that this SNP increased the risk of metabolic syndrome in an Iranian population (Hashemi et al., 2012). These results suggest that chemerin gene polymorphisms located in noncoding regions could affect the regional fat deposition in humans. Intramuscular fat deposition (marbling) measured as BMS is one of the economically important traits in Japanese Black cattle, with a high level of BMS associated with higher intramuscular fat deposition. To date, there are no reports on the direct mechanism underlying chemerin’s regulation of the intramuscular fat deposition of cattle, to the best of our knowledge. However, increasing lipid content induced higher chemerin gene expression in bovine differentiated adipocytes (Song et al., 2010).
The difference in fatty acid composition between genotypes CC and CT of the c.276C>T SNP suggests that this SNP might affect the differentiation of adipocytes and intramuscular fatty acid compositions by changing the expression of chemerin. The ‘silent’ polymorphism could change the substrate specificity by altering its conformations (Kimchi-Sarfaty et al., 2007). Moreover, exonic cis-elements are important for correct splice-site identification and are distinct from the classical splicing signals (Cartegni et al., 2002). Chemerin regulates the differentiation of adipocytes and myoblast cells (Yang et al., 2012). Therefore, we need to evaluate the effects of c.276C>T SNP on chemerin gene expression and reveal the mechanisms that affect intramuscular fatty acid compositions.
In this study, we found 5 SNPs in the coding region of the bovine chemerin gene in Japanese Black cattle, and the c.276C>T SNP was associated with marbling and fatty acids compositions. However, further studies are required to elucidate the biological relevance and mechanism by which the chemerin gene polymorphism affects these traits in Japanese Black cattle.
ACKNOWLEDGMENTS
This study was partly supported by a grant-in-aid from JSPS.
Footnotes
CONFLICT OF INTEREST
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
REFERENCES
- Abe T, Saburi J, Hasebe H, Nakagawa T, Misumi S, Nade T, Nakajima H, Shoji N, Kobayashi M, Kobayashi E. Novel mutations of the FASN gene and their effect on fatty acid composition in Japanese Black beef. Biochem Genet. 2009;47:397–411. doi: 10.1007/s10528-009-9235-5. [DOI] [PubMed] [Google Scholar]
- Ardiyanti A, Abe F, Kobashikawa H, Hirayama T, Sugino T, Suzuki K, Katoh K. Plasma hormone and metabolite concentrations involved in the somatotropic axis of Japanese Black heifers in association with growth hormone gene polymorphism. Domest Anim Endocrinol. 2009;37:243–249. doi: 10.1016/j.domaniend.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Ardiyanti A, Abe T, Tameoka N, Kobayashi E, Shoji N, Ohtani Y, Suzuki K, Roh SG, Katoh K. Effects of growth hormone gene polymorphism on lipogenic gene expression levels in diaphragm tissues of Japanese Black Heifers. Asian Australas J Anim Sci. 2012;25:1055–1062. doi: 10.5713/ajas.2012.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozaoglu K, Bolton K, McMillan J, Zimmet P, Jowett J, Collier G, Walder K, Segal D. Chemerin is a novel adipokine associated with obesity and metabolic syndrome. Endocrinology. 2007;148:4687–4694. doi: 10.1210/en.2007-0175. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Goralski KB, McCarthy TC, Hanniman EA, Zabel BA, Butcher EC, Parlee SD, Muruganandan S, Sinal CJ. Chemerin, a novel adipokine that regulates adipogenesis and adipocyte metabolism. J Biol Chem. 2007;282:28175–28188. doi: 10.1074/jbc.M700793200. [DOI] [PubMed] [Google Scholar]
- Hashemi M, Rezaei H, Eskandari-Nasab E, Kaykhaei MA, Zakeri Z, Taheri M. Association between chemerin rs17173608 and vaspin rs2236242 gene polymorphisms and the metabolic syndrome, a preliminary report. Gene. 2012;510:113–117. doi: 10.1016/j.gene.2012.08.048. [DOI] [PubMed] [Google Scholar]
- Higgins DG, Sharp PM. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Lindholm-Perry AK, Kuehn LA, Rempel LA, Smith TPL, Cushman RA, McDaneld TG, Wheeler TL, Shackelford SD, King DA, Freetly HC. Evaluation of bovine chemerin (RARRES2) gene variation on beef cattle production traits. Front Genet. 2012;3:39. doi: 10.3389/fgene.2012.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuhashi T, Maruyama S, Uemoto Y, Kobayashi N, Mannen H, Abe T, Sakaguchi S, Kobayashi E. Effects of bovine fatty acid synthase, stearoyl-coenzyme A desaturase, sterol regulatory element-binding protein 1, and growth hormone gene polymorphisms on fatty acid composition and carcass traits in Japanese Black cattle. J Anim Sci. 2011;89:12–22. doi: 10.2527/jas.2010-3121. [DOI] [PubMed] [Google Scholar]
- Mussig K, Staiger H, Machicao F, Thamer C, Machann J, Schick F, Claussen CD, Stefan N, Fritsche A, Haring HU. RARRES2, encoding the novel adipokine chemerin, is a genetic determinant of disproportionate regional body fat distribution: a comparative magnetic resonance imaging study. Metabolism. 2009;58:519–524. doi: 10.1016/j.metabol.2008.11.011. [DOI] [PubMed] [Google Scholar]
- Roh SG, Hishikawa D, Hong YH, Sasaki S. Control of adipogenesis in ruminants. Anim Sci J. 2006;77:472–477. [Google Scholar]
- Roh SG, Song SH, Choi KC, Katoh K, Wittamer V, Parmentier M, Sasaki S. Chemerin—A new adipokine that modulates adipogenesis via its own receptor. Biochem Biophys Res Commun. 2007;362:1013–1018. doi: 10.1016/j.bbrc.2007.08.104. [DOI] [PubMed] [Google Scholar]
- Sell H, Divoux A, Poitou C, Basdevant A, Bouillot JL, Bedossa P, Tordjman J, Eckel J, Clement K. Chemerin correlates with markers for fatty liver in morbidly obese patients and strongly decreases after weight loss induced by bariatric surgery. J Clin Endocrinol Metabol. 2010;95:2892–2896. doi: 10.1210/jc.2009-2374. [DOI] [PubMed] [Google Scholar]
- Sell H, Laurencikiene J, Taube A, Eckardt K, Cramer A, Horrighs A, Arner P, Eckel J. Chemerin is a novel adipocyte-derived factor inducing insulin resistance in primary human skeletal muscle cells. Diabetes. 2009;58:2731–2740. doi: 10.2337/db09-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SB, Lunt DK, Chung KY, Choi CB, Tume RK, Zembayashi M. Adiposity, fatty acid composition, and delta-9 desaturase activity during growth in beef cattle. Anim Sci J. 2006;77:478–486. [Google Scholar]
- Song SH, Fukui K, Nakajima K, Kozakai T, Sasaki S, Roh SG, Katoh K. Cloning, expression analysis, and regulatory mechanisms of bovine chemerin and chemerin receptor. Domest Anim Endonol. 2010;39:97–105. doi: 10.1016/j.domaniend.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Hong YH, Song SH, Ardiyanti A, Kato D, So KH, Katoh K, Roh SG. The regulation of chemerin and CMKLR1 genes expression by TNF-α, adiponectin, and chemerin analog in bovine differentiated adipocytes. Asian Australas J Anim Sci. 2012;25:1316–1321. doi: 10.5713/ajas.2012.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Song SH, Sato K, So KH, Ardiyanti A, Kitayama S, Hong YH, Lee SD, Choi KC, Hagino A, Katoh K, Roh SG. Chemerin analog regulates energy metabolism in sheep. Anim Sci J. 2012;83:263–267. doi: 10.1111/j.1740-0929.2011.01002.x. [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Utsugi T, Oyama K, Mannen H, Kobayashi M, Tanabe Y, Ogino A, Tsuji S. Genotype of stearoyl-coA desaturase is associated with fatty acid composition in Japanese Black cattle. Mamm Genome. 2004;15:142–148. doi: 10.1007/s00335-003-2286-8. [DOI] [PubMed] [Google Scholar]
- Tian WQ, Wang HC, Song FB, Zan LS, Wang H, Wang HB, Xin YP, Ujan JA. Association between a single nucleotide polymorphism in the bovine chemerin gene and carcass traits in Qinchuan cattle. Genet Mol Res. 2011;10:2833–2840. doi: 10.4238/2011.November.17.1. [DOI] [PubMed] [Google Scholar]
- Yang A, Larsen TW, Powell VH, Tume RK. A comparison of fat composition of Japanese and long-term grain-fed Australian steers. Meat Sci. 1999;51:1–9. doi: 10.1016/s0309-1740(98)00065-5. [DOI] [PubMed] [Google Scholar]
- Yang H, Li F, Kong X, Yuan X, Wang W, Huang R, Li T, Geng M, Wu G, Yin Y. Chemerin regulates proliferation and differentiation of myoblast cells via ERK1/2 and mTOR signaling pathways. Cytokine. 2012;60:646–652. doi: 10.1016/j.cyto.2012.07.033. [DOI] [PubMed] [Google Scholar]
- Yokota S, Sugita H, Ardiyanti A, Shoji N, Nakajima H, Hosono M, Otomo Y, Suda Y, Katoh K, Suzuki K. Contributions of FASN and SCD gene polymorphisms on fatty acid composition in muscle from Japanese Black cattle. Anim Genet. 2012;43:790–792. doi: 10.1111/j.1365-2052.2012.02331.x. [DOI] [PubMed] [Google Scholar]


