Abstract
X chromosome dosage compensation is required for male viability in Drosophila. Dosage compensation relative to autosomes is two-fold, but this is likely to be due to a combination of homeostatic gene-by-gene regulation and chromosome-wide regulation. We have baseline values for gene-by-gene dosage compensation on autosomes, but not for the X chromosome. Given the evolutionary history of sex chromosomes, these baseline values could differ. We used a series of deficiencies on the X and autosomes, along with mutations in the sex-determination gene transformer-2, to carefully measure the sex-independent X-chromosome response to gene dosage in adult heads by RNA sequencing. We observed modest and indistinguishable dosage compensation for both X chromosome and autosome genes, suggesting that the X chromosome is neither inherently more robust nor sensitive to dosage change.
Keywords: segmental aneuploidy, sex determination, dosage compensation, sex chromosomes, gene expression, genetics of sex
Like in many eukaryotes, reduction of gene dosage is deleterious in Drosophila melanogaster. Multi-locus deletions (deficiencies) are tolerated when the extent of the deletion is modest, but when >1% of genome is hemizygous, the result is reduced viability regardless of which part of the genome is deleted (Lindsley et al. 1972) . The X chromosome in males is an exception to this rule.
The sex chromosome dosage problem is inherent in the evolution of sex chromosomes, an event that has occurred many times during the course of evolution (Larsson and Meller 2006; Vicoso and Charlesworth 2006; Gurbich and Bachtrog 2008; Stenberg and Larsson 2011) . The initial signal for sex determination in Drosophila is the number of X chromosomes (Erickson and Quintero 2007), which results in males being hemizygous for ∼20% of the genome. X chromosome dosage compensation prevents male lethality by equalizing the expression level of genes on the X chromosome in males to that in females to maintain genic balance with the autosomes in both sexes (Parisi et al. 2003; Gupta et al. 2006; Sturgill et al. 2007; Deng et al. 2011). Chromosome-wide X-chromosome dosage compensation relative to autosomes requires the male-specific lethal (MSL) complex (Meller and Kuroda 2002; Hamada et al. 2005; Deng and Meller 2006; Zhang et al. 2010; Georgiev et al. 2011; Stenberg and Larsson 2011).
It is less clear how much of the X chromosome dosage compensation is due to chromosome-wide regulation and how much is due to gene-by-gene dosage responses. For example, there are X chromosome genes that do not show MSL association but that do show dosage compensation (Philip and Stenberg 2013), and some X chromosome genes are dosage compensated very early, prior to activation of the MSL system (Lott et al. 2011). Finally, removal of the MSL complex components does not result in halving of X chromosome gene expression relative to autosomes (Hamada et al. 2005; Deng and Meller 2006; Zhang et al. 2010), raising the possibility that other MSL-independent mechanisms may also contribute to dosage compensation in fly somatic cells.
A priori, an X-chromosome-wide mechanism does not need to result in a precise two-fold upregulation of gene expression relative to autosomes, because even autosomal genes can show some dosage compensation. In Drosophila S2 cells, the observed autosomal dosage compensation level was roughly equal to the approximately 1.4-fold “missing” compensation following MSL complex knockdown, suggesting that there may be an essentially additive effect of MSL and general dosage compensation mechanisms (Zhang et al. 2010). However, other Drosophila cell lines show a variety of global dosage compensation levels (Lee et al. 2014), raising the distinct possibility that this additive effect in S2 cells is coincidence.
Several studies have reported the transcriptional effect of systematically manipulating autosomal gene dosage (Gupta et al. 2006; Stenberg et al. 2009; Stenberg and Larsson 2011; Malone et al. 2012). These studies suggest that the same type of gene regulation that modulates levels in transcriptional networks results in gene-by-gene dosage compensation (Malone et al. 2012; Chen et al. 2014; Lee et al. 2014). This gene-by-gene response also highlights the need to use a matched sample for baseline measurements of dosage compensation. Simply comparing autosomes to X chromosomes is misleading when individual genes differ in dosage responses and when the population of genes residing on the X is highly biased due to the evolution of sex chromosomes (Vicoso and Charlesworth 2006; Gurbich and Bachtrog 2008). If X chromosomes were like autosomes, gene-by-gene responses at subsets of genes should not require specialized factors to achieve dosage compensation (dosage compensation of such genes would result in overexpression), and in other subsets where gene expression collapses in response to a 50% dosage reduction even a two-fold effect might not be sufficient (Malone et al. 2012). However, a host of studies have shown that the X chromosome is quite different than autosomes (Vicoso and Charlesworth 2006; Gurbich and Bachtrog 2008), showing a distinct chromatin signature even in females, for example (Zhang and Oliver 2010). As another example, there is also strong gene traffic off the X in the genus, which could drive dosage-sensitive genes off the X (Sturgill et al. 2007; Vibranovski et al. 2009). The gradual acquisition of dosage compensation during sex chromosome evolution could make the X either more or less sensitive to gene dosage changes, depending on which forces are dominant. For example, a long history of chromosome-wide regulation in males could reduce endogenous gene-by-gene dosage compensation by relaxing feedback in gene regulation networks, whereas in contrast a long history of pressure to increase expression of hemizygous genes on the X before acquisition of chromosome-wide dosage compensation mechanisms could boost these same gene-by-gene regulatory mechanisms. Thus, it is unclear if X chromosome gene-by-gene dosage responses are similar to autosomal gene-by-gene responses.
In wild-type flies, sexual identity, sex-biased expression, and any special evolved features of the X chromosome are confounding factors if you are interested in what could be very subtle effects on dosage compensation. In this study, we separated these factors using autosomal and X-linked deficiencies in conjunction with sex transformation, and we further reduced confounding by restricting our analysis to heads, which show limited sex-biased expression. We performed next-generation RNA sequencing (RNA-seq) and determined expression of matched hemizygous and homozgyous genes to avoid comparing expression of X chromosomes to autosomal genes directly. We observed indistinguishable dosage responses of chromosomes X and 3L in this study, suggesting that X and autosomes have similar overall responses to dosage.
Materials and Methods
Samples and sequencing
We used congenic Drosophila strains from the European Drosophila deletion collection (DrosDel) project (Ryder et al. 2007; Cook et al. 2010) to assay expression changes due to gene dosage. In addition to the DrosDel lines, we examined expression in the w1118 DrosDel parental line and used a Dp(1;Y)BS; cn1 tra2B bw1/CyO and Df(2R)trix/CyO stocks for sex-transforming flies (Bloomington Drosophila Stock Center, Bloomington, IN). We also used OreR (gift from Elissa Lei, Laboratory of Cellular and Developmental Biology, NIDDK, NIH, Bethesda, Maryland). Flies were grown at 25° on San Diego Stock Center cornmeal media. We collected 30 5-d-old adult flies for each triplicate of 10 heads, which were dissected on dry ice and stored at −80°.
We homogenized frozen heads in 2 ml Axygen 96-well plates (Corning, Life Sciences, Union City, CA) preloaded with 200 µL 1-mm glass beads (Biospec Products Inc., Bartlesville, OK) and 200 µL RLT Buffer (Qiagen, Valencia, CA) and covered with Axygen sealing mats, in a Mini-BeadBeater 96 (Biospec Products Inc., Bartlesville, OK). We extracted total RNA with the RNeasy 96 Kit and QIAvac 96 vacuum manifold (Qiagen, Valencia, CA). We visually inspected RNA quality by gel electrophoresis detected with SYbrGold Nucleic Acid Gel Stain and we used a plate reader to quantify yield with Quant-iT RiboGreen (Life Technologies, Grand Island, NY). We used 100 ng of total RNA and the TruSeq RNA Sample Preparation V2 protocol (Illumina Inc., San Diego, CA) to make RNA-seq libraries from polyA+ RNA. We added 10 pg of pool 78A ERCC spike-in RNAs (Jiang et al. 2011) to the Elute, Prime, and Fragment Mix in the “Purify and fragment mRNA” step. We checked the library quality with the High-Sensitivity DNA Analysis Kit on a Bioanalyzer (Agilent, Santa Clara, CA) and concentration by Quant-iT PicoGreen (Life Technologies, Grand Island, NY), and performed 76nt single-end sequencing on an Illumina HiSequation 2000 running CASAVA-1.8.2/RTA 1.17.21.3/HiSeq Control Software 2.0.10.0 (Illumina Inc., San Diego, CA).
Sequence mapping and quantification
To measure expression genome-wide, we performed single-end RNA-Seq on poly-A+ RNA extracted from adult female and male heads in biological triplicate. We uniquely mapped reads to FlyBase genome release 5.57 (St. Pierre et al. 2014) and spike-in RNAs (Jiang et al. 2011) that we added to the libraries to empirically determine sampling variance in complex mixtures of RNA species. We used DESeq2 (Anders et al. 2015) normalized read counts and cufflinks (Trapnell et al. 2012) fragments per kilobase per million mapped reads (FPKM) to report gene-level steady-state expression.
We downloaded GFF3 gene annotation files from NCBI (http://www.ncbi.nlm.nih.gov/), extracted CDS and exon features on existing chromosomes (excluding chromosome U and Uextra), and converted to GTF format, which is compatible with all analysis tools we used, using a custom Perl script. Some peculiar gene features were incompatible with analysis tools. We “corrected” the boundaries of 10 CDS, which were beyond that of the parent genes and changed strand information for 9 CDS (11 exons) according to the annotation of the parent genes. We also added 96 ERCC spike-in annotations in the final GTF file. We mapped RNA reads with Tophat v2.0.10 (Kim et al. 2013). We used parameter −g 1 to retain only uniquely mapped reads and provided the GTF format annotation.
We measured the gene expression using cufflinks v2.1.1 (Trapnell et al. 2012) in fragments per kilobase of transcript per million mapped reads (FPKM). We measured gene expression using HT-Seq v0.5.4p1 (Anders et al. 2015), which reports in read counts. We normalized read counts using DESeq2 v1.6.3 (Love et al. 2014) in R v3.1.1. We used coefficient of variance for genes, intergenic space, and spike-in RNAs to derive a low expression cut-off value (see below).
Results
Datasets
We generated 264 samples for RNA-Seq profiling with a median of ∼8 million uniquely mapped reads per sample (Table 1). We failed 15 samples during library or sequencing quality checks. We also ensured that the lines still bore the originally generated deletions. Previously, we found that when a Df/+ line did not alter gene expression in the hemizygous segment, there was no Df in the source stock as measured by DNA-Seq (Malone et al. 2012). Therefore, we scanned the gene expression ratios of each Df/+ sample relative to +/+ controls across chromosomes 3L and X. We suspected that Df(3L)4685 was not a deficiency based on this criterion and cytological examination also failed to show evidence of a deletion (K. Cook, personal communication). We removed nine “Df(3L)4685” samples as well as five samples without replicates from the analysis. These data are still useful for other purposes. Data from all 249 high-quality samples are available at the Gene Expression Omnibus (GEO) (Barrett and Edgar 2006) under accession GSE60571. Triplicates showed excellent linear correlations (Figure 1A), giving us confidence in measuring subtle differences in gene expression.
Table 1. Summary of samples used.
| Genotypes | XX | XY | XX; tra2B/Dfa | XY; tra2B/Df | No.b |
|---|---|---|---|---|---|
| Df(1)ED13478/+ | 3 | NAc | 3 | NA | 6 |
| Df(1)ED14021/+ | 3 | NA | 3 | NA | 6 |
| Df(1)ED409/+ | 3 | NA | 3 | NA | 6 |
| Df(1)ED6443/+ | 3 | NA | 3 | NA | 6 |
| Df(1)ED6630/+ | 3 | NA | 3 | NA | 6 |
| Df(1)ED6712/+ | 3 | NA | 3 | NA | 6 |
| Df(1)ED6727/+ | 3 | NA | 3 | NA | 6 |
| Df(1)ED6829/+ | 3 | NA | 3 | NA | 6 |
| Df(1)ED6878/+ | 3 | NA | 3 | NA | 6 |
| Df(1)ED6906/+ | 3 | NA | 3 | NA | 6 |
| Df(1)ED6957/+ | 3 | NA | 3 | NA | 6 |
| Df(1)ED6989/+ | 3 | NA | 3 | NA | 6 |
| Df(1)ED7147/+ | 0 | NA | 3 | NA | 3d |
| Df(1)ED7153/+ | 3 | NA | 3 | NA | 6 |
| Df(1)ED7161/+ | 3 | NA | 3 | NA | 6 |
| Df(1)ED7170/+ | 3 | NA | 3 | NA | 6 |
| Df(1)ED7225/+ | 1 | NA | 0 | NA | 1e |
| Df(1)ED7261/+ | 3 | NA | 3 | NA | 6 |
| Df(1)ED7289/+ | 3 | NA | 3 | NA | 6 |
| Df(1)ED7331/+ | 2 | NA | 3 | NA | 5 |
| Df(1)ED7374/+ | 1 | NA | 0 | NA | 1e |
| Df(1)ED7635/+ | 3 | NA | 3 | NA | 6 |
| Df(3L)ED210/+ | 3 | 3 | 3 | 0 | 9 |
| Df(3L)ED211/+ | 3 | 3 | 3 | 0 | 9 |
| Df(3L)ED217/+ | 3 | 3 | 3 | 0 | 9 |
| Df(3L)ED225/+ | 3 | 3 | 3 | 0 | 9 |
| Df(3L)ED230/+ | 3 | 3 | 3 | 0 | 9 |
| Df(3L)ED4287/+ | 3 | 3 | 3 | 0 | 9 |
| Df(3L)ED4421/+ | 3 | 3 | 3 | 0 | 9 |
| Df(3L)ED4457/+ | 3 | 3 | 3 | 0 | 9 |
| Df(3L)ED4475/+ | 3 | 3 | 3 | 0 | 9 |
| Df(3L)ED4543/+ | 3 | 3 | 3 | 0 | 9 |
| Df(3L)ED4685/+ | 3 | 3 | 3 | 0 | 9f |
| Df(3L)ED4978/+ | 3 | 3 | 3 | 0 | 9 |
| w1118/ w1118 or Y | 3 | 3 | 3 | 3 | 12 |
| w1118 /w+ | 2 | NA | 3 | NA | 5 |
| OreR | 3 | 3 | 0 | 0 | 6 |
| No. | 102 | 42 | 102 | 3 | 249 |
Df(2R)trix.
Number of independent RNA-Seq profiles.
Not applicable.
Present in GEO, but removed from this analysis due to lack of a matching nonsex-transformed sample.
Present in GEO, but removed from this analysis due to lack of replicates.
Present in GEO, but removed from this analysis because we observed no dosage effect in the region of the annotated Df, and because no Df was detected in the source stock. See text.
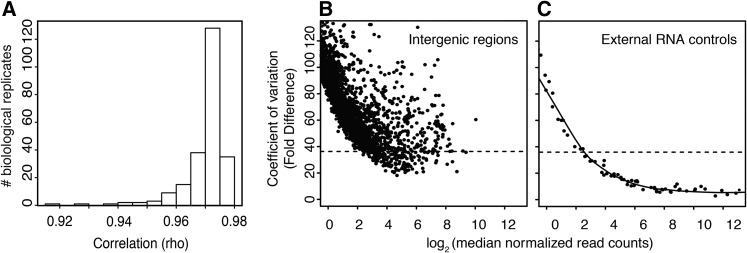
Figure 1.
RNA-Seq libraries and expression cut-offs. (A) Correlations among biological replicate RNA-seq experiments on adult heads. (B, C) Coefficients of variance (CV) for reads mapping to intergenic space (B) and spike-in RNA controls (C). Ninety-five percent of intergenic regions have a CV above the dotted line.
Although RNA-seq gene expression measurements show excellent correlations, understanding dosage responses and compensation requires precision at less than two-fold differential expression. Low abundance transcripts are difficult to accurately quantify, because noise and sampling errors are more pronounced for genes with low expression (Jiang et al. 2011; Malone and Oliver 2011). These factors can confound dosage analysis by suggesting dosage compensation where none exists (taken to an extreme, a gene that is not expressed could be said to be perfectly dosage compensated).
To rigorously avoid overcalling compensation due to low expression, we directly modeled error profiles by comparing intergenic gene expression to genic expression. Although there may be unannotated genes in intergenic space, signal from these regions may also represent biological noise and/or DNA contamination in RNA preparations (Zhang et al. 2010). We also analyzed quantification variance for the synthetic spike-in RNAs with known input concentrations as determined by mass spectrophotometry (Jiang et al. 2011). For intergenic regions (Figure 1B), we found that 95% of intergenic regions had a coefficient of variance (CV) >36 for fold differences. At this CV value, the normalized read counts of external RNA standards fitted with a local polynomial regression was six normalized read counts (Figure 1C). Sixty-four percent of all genes were expressed at more than six, and 69% showed CVs <36. Therefore, we used six normalized read counts as a low-expression cut-off for ratiometric analysis.
Gene expression in wild-type and sex-transformed heads
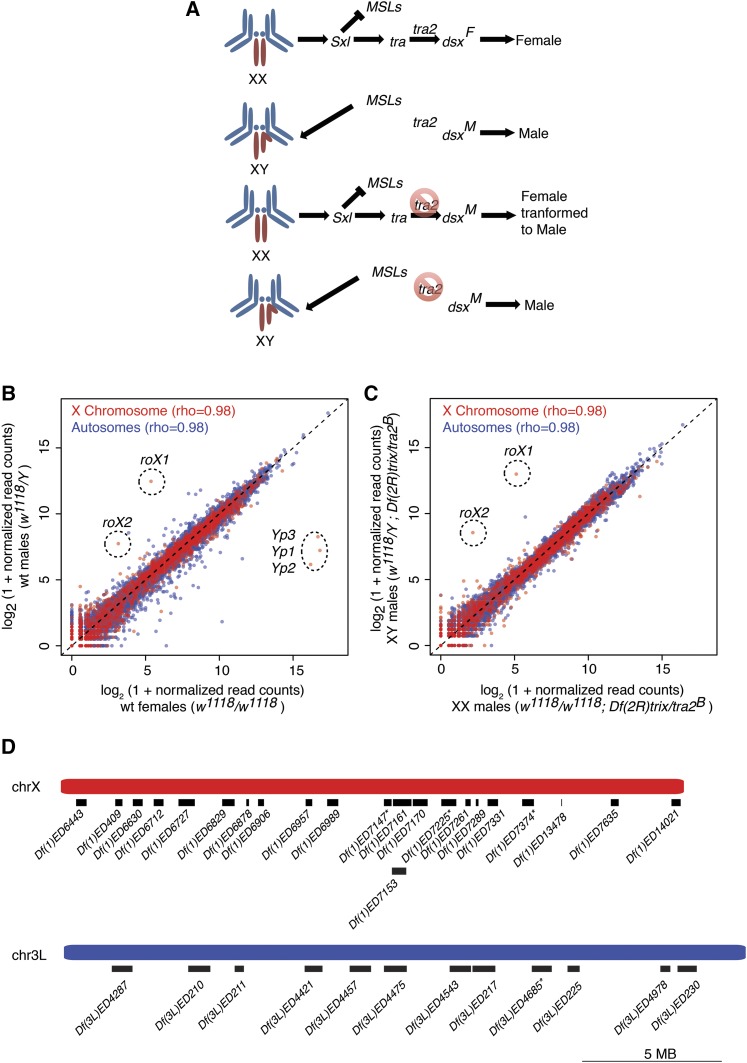
Sex-biased expression complicates the analysis of the subtle changes in gene expression between XX and XY flies. Therefore, we restricted our expression analysis to heads, because previous RNA-seq experiments revealed relatively limited sex-biased expression in this body part (Sturgill et al. 2013). As an additional measure to reduce sex-biased expression, we transformed XX females into somatic males (Figure 2A) using transformer-2 alleles (tra2Df/tra2B) (Mattox and Baker 1991). When we compared the expression profiles of wild-type female (w1118/w1118) and male flies (w1118/Y) (Figure 2B), we observed a tight overall correlation between the head expression profiles. However, we did observe 670 genes with significantly female-biased and 602 with male-biased expression (FDR-corrected P < 0.05) in wild-type heads (Figure 2B, Supporting Information, File S1). When we compared gene expression in XX females transformed into males (w1118/w1118; tra2Df/tra2B) relative to XY males with tra2 mutations (w1118/Y; tra2Df/tra2B), we only observed 78 genes with significant XX-biased and 34 with XY-biased expression (Figure 2C, File S1). As expected, sex differences regulated upstream of tra2, such as expression of the dosage compensation transcripts encoded by roX1 and roX2, remained XY-biased, whereas genes known to be regulated downstream of tra2 (Salz and Erickson 2010), such as the three Yolk protein genes (Clough et al. 2014), were lowered to male levels in XX; tra2Df/tra2B flies (Figure 2, B and C).
Figure 2.
Gene expression in wild-type and sex-transformed heads. (A) Cartoon of sex determination (from top) in wild-type females with two X chromosomes (XX), wild-type males (XY), females transformed into males (XX), and control males for tra2 (XY). Left panel shows sex chromosomes (red) and autosomes (blue). Major components of the sex determination hierarchy are Sex-lethal (Sxl), the MSL complex, transformer (tra), transformer2 (tra2), and doublesex (dsx). Dsx encodes female (F) and male (M) transcripts due to alternative splicing mediated by tra and tra2. Mutations in tra2 are indicted (red circle/slash). Gene expression scatterplots of wild-type XY male vs. XX female heads (B), or tra2 mutant heads from XY males vs. XX females transformed into males (C), with genes and global correlations for X (red) and autosome (blue) indicated. Genes with the most dramatic gene expression differences are shown (dotted circles). (D) Cartoon of 22 X chromosome (red) and 12 autosomal (blue) Dfs used in this study, including three X chromosome and one autosomal Dfs (asterisk) excluded from analysis (see Table 1). Scale bar is as shown.
Gene expression by chromosome and gene dosage
If the X chromosome is inherently less or more sensitive to gene dosage reduction than autosomes, then we should observe differences between the dosage responses of chromosome X and 3L in XX females and in XX females transformed into males. To generate these hemizygous segments on the X and autosomes, we chose 12 deficiencies (Dfs) on chromosome 3L as controls and 22 Dfs on the X chromosome (Figure 2D). To avoid any potential bias associated with a particular chromosomal location, we selected a distribution along the length of the chromosome arms. We also avoided deletions reducing the dosage of genes known to be involved in dosage compensation or sex determination, because these could also confound our baseline measurements. Although it is possible that relatively small deletions show different dosage responses than whole chromosomes, we observed no correlation between Df extent and compensation level (not shown), as previously reported for the chromosome arm 2L (Malone et al. 2012).
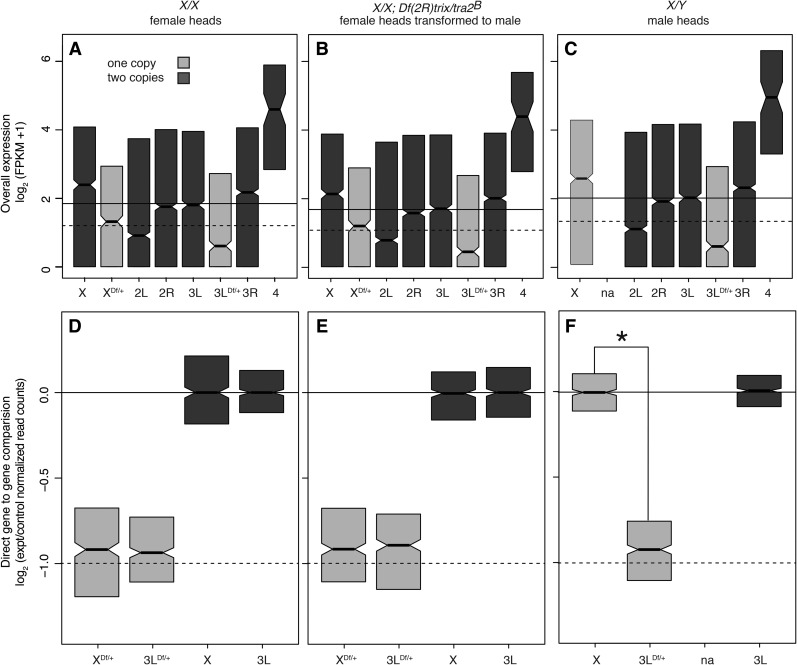
We first looked at expression genome-wide in the samples parsed by chromosome arm with expression of hemizygous genes from Df/+ chromosome regions binned separately (Figure 3, A–C). This analysis showed a strong bias for higher expression levels for chromosomes X and 4 genes in the regions of the genome with wild-type gene content. Chromosome 2L showed reduced overall expression in heads. This gene-content uncorrected analysis also showed essentially no compensation for X chromosome genes and a collapse in the expression of hemizygous genes on chromosome 3L. At face value, these observations show clear differences in baseline compensation between the X chromosome and the autosomes.
Figure 3.
Dosage response boxplots. (A–C) Head gene expression values by chromosome arm, including a separate category for Df/+ segments. (D–F) Ratiometric plots comparing the expression of genes in Df/+ segments to the same genes in +/+ (light). The controls (dark) were selected based on sampling the same number of genes as found in the experimental set. The +/+ genes from Df/+ flies were compared to the same +/+ genes from +/+ flies, or +/Y in the case of X-linked genes in XY males. (A, D) Females. (B, E) Females transformed into males. (C, F) Males. Plots show interquartile range (IQR; box), medians (bar), and confidence interval (±1.57 × IQR/√N).
However, expression variance by linkage and by sampled region could also be due to differing chromosome and/or regional gene content as a confounding factor for analyzing dosage effects. For example, when we simply examined expression levels, median chromosome 2L expression was lower than expression hemizygous genes in Df/+ regions on the X chromosome. These overall expression levels were essentially the reciprocal of the gene expression bias in reproductive tissues (Parisi et al. 2003; Sturgill et al. 2007), suggesting that different gene content, not dosage responses per se, are responsible. Specifically, there is under-representation of genes with male-biased expression in reproductive tissues on the X chromosome and fourth chromosome, and we show over-representation of genes expressed in heads on these chromosomes. Given that chromosome 4 is derived from an ancient X (Vicoso and Bachtrog 2013), these data suggest selection favors X linkage for genes expressed at high levels in heads. This is reminiscent of mammals where genes highly expressed in the brain are preferentially X chromosome–linked (Vallender and Lahn 2004). Reciprocally, there is an over-representation of genes with male-biased gene expression on chromosome 2L, and we show depletion for genes with high expression in heads on 2L. For our purpose here, sex-biased content is a confounding factor that hinders comparison of X chromosome expression levels with those of the autosomes.
Comparing the expression of the same genes when present in different copy number eliminates the effect of gene content differences (Figure 3, D–F). When we examined expression of a given gene when hemizygous, compared to the median expression of that gene in all lines (by bootstrapping), we observed a much more consistent dosage response, characterized by a strong decrease in median expression, with a median compensation level of ∼1.1-fold, in the case of both Dfs of chromosomes X and 3L. In both XX females and XX females transformed into males, the expression of hemizygous X chromosome and autosome genes was not significantly different, and there was not even evidence of a trend (Figure 3E). In XY males, X chromosome compensation was not significantly different from perfect, whereas compensation of autosomal hemizygosity was similar to that observed in XX fly heads (Figure 3F). These data indicate that the X chromosome has a generic response to decreased gene dosage that is similar to that of the autosomes.
Discussion
Drosophila melanogaster sex chromosomes are thought to have arisen from an autosome pair and diverged to the current state by the gradual loss of genes on the nonrecombining Y chromosome (Vicoso and Charlesworth 2006). With the loss of more and more genes from Y chromosome, the chromosome-wide dosage compensation mechanism evolved as an effective machine for balancing X chromosome gene expression in flies with a highly degenerate Y chromosome. According to theory, the initial compensation for the one-dose genes on proto-X should be the same as the autosomal ancestor, which we suggest is the ∼1.1-fold upregulation we observed in this study. If the X chromosome became hemizygous little by little rather than all at once (Marin et al. 2000), then this might have provided time for selection to strengthened transcriptional regulatory interactions either before or during the emergence of the chromosome-wide compensation system. Additionally, the evolution of X chromosomes with a gradually degenerating Y chromosome should result in selection against chromosome-wide dosage compensation when the Y is young due to deleterious overexpression of genes with functional alleles on both the X and Y chromosomes. This shifting selection during the course of X chromosome evolution might result in strengthened compensation or selection for gene content that favors X-linkage for genes with more robust gene regulation. We found very little evidence for inherent differences between X and autosome compensation, suggesting that the extant X has the same inherent dosage response as an autosome. This is somewhat surprising given that the X chromosome shows a specialized chromatin structure in females and males (Zhang and Oliver 2010; Lee et al. 2014) and responds differently to gene dosage balance in trans (Sun et al. 2013), in addition to the evolutionary history outlined above.
The generic response observed for the X and autosomes is consistent with gene regulation playing a role in the response to copy number. For any given gene, we observed a significant correlation in compensation values among female, male, and sex-transformed flies bearing the same Dfs (not shown), supporting the idea that inherent gene-specific compensation levels contribute to the general dosage response (Malone et al. 2012). This includes evidence for additive effects of generic and chromosome-wide regulation (genes with the best compensation in Df/X flies tend to be overcompensated in XY flies), as previously reported in S2 cells (Zhang et al. 2010). We found median compensation (1.1-fold) at the low end of the previously reported range (1.1- to 1.8-fold) for partial compensation in Drosophila adults and cell lines (Johansson et al. 2007; Stenberg et al. 2009; McAnally and Yampolsky 2010; Zhang et al. 2010; Lundberg et al. 2012; Malone et al. 2012). Many of the differences in compensation values in these studies are due to low expression cutoff decisions. Increasing the cutoff values in the previous datasets also results in lower compensation values and decreasing the low expression cutoff values increased median compensation values in the dataset reported here (not shown). Thus, either there is better compensation of genes with low expression (Stenberg et al. 2009; Malone et al. 2012) or there is data compression due to expression noise and nonlinearity at low expression levels (Malone and Oliver 2011; Wang et al. 2014), or both. Additionally, reanalysis of cell line RNA-Seq datasets using more conservative low expression cutoffs, newer short-read aligners, and copy number callers (Boeva et al. 2012; Langmead and Salzberg 2012; Kim et al. 2013) showed consistent partial compensation ≤1.2-fold in most cell lines (H. Lee and B. Oliver, unpublished data). Human HeLa cells also show ∼1.2-fold compensation (Landry et al. 2013). However, even within a study with common methods and data handling, there are unexplained outliers (Lee et al. 2014). This highlights the importance of using carefully matched experimental and control biological samples and careful consideration of data handling in the analysis of dosage compensation. Given that X chromosome dosage compensation is very close to two-fold, whereas the effect of MSL mutations and gain-of-function are not, and given that the X does not appear to show boosted gene-by-gene compensation, then a substantial amount of X chromosome dosage compensation remains unexplained.
Supplementary Material
Acknowledgments
We thank Harold Smith for sequencing, Hangnoh Lee for reanalysis of published data, Kevin Cook for Df cytology, Helix (http://helix.nih.gov) and Biowulf (http://biowulf.nih.gov) for providing high-performance computational capabilities, FlyBase for files, and our lab for stimulating discussions. We also thank the Bloomington Stock Center and Elissa Lei for fly lines. The Intramural Research Program of the National Institutes of Health, NIDDK (ZIA-DK015600), supported this research.
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.017632/-/DC1
Data accessions: GEO GSE60571.
Communicating editor: J. A. Birchler
Literature Cited
- Anders S., Pyl P. T., Huber W., 2015. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T., Edgar R., 2006. Gene expression omnibus: microarray data storage, submission, retrieval, and analysis. Methods Enzymol. 411: 352–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeva V., Popova T., Bleakley K., Chiche P., Cappo J., et al. , 2012. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 28: 423–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. X., Golovnina K., Sultana H., Kumar S., Oliver B., 2014. Transcriptional effects of gene dose reduction. Biol Sex Differ 5: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough E., Jimenez E., Kim Y. A., Whitworth C., Neville M. C., et al. , 2014. Sex- and tissue-specific functions of Drosophila doublesex transcription factor target genes. Dev. Cell 31: 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook K. R., Parks A. L., Jacobus L. M., Kaufman T. C., Matthews K. A., 2010. New research resources at the Bloomington Drosophila Stock Center. Fly (Austin) 4: 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Meller V. H., 2006. roX RNAs are required for increased expression of X-linked genes in Drosophila melanogaster males. Genetics 174: 1859–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Hiatt J. B., Nguyen D. K., Ercan S., Sturgill D., et al. , 2011. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat. Genet. 43: 1179–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson J. W., Quintero J. J., 2007. Indirect effects of ploidy suggest X chromosome dose, not the X:A ratio, signals sex in Drosophila. PLoS Biol. 5: e332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev P., Chlamydas S., Akhtar A., 2011. Drosophila dosage compensation: males are from Mars, females are from Venus. Fly (Austin) 5: 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V., Parisi M., Sturgill D., Nuttall R., Doctolero M., et al. , 2006. Global analysis of X-chromosome dosage compensation. J. Biol. 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurbich T. A., Bachtrog D., 2008. Gene content evolution on the X chromosome. Curr. Opin. Genet. Dev. 18: 493–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada F. N., Park P. J., Gordadze P. R., Kuroda M. I., 2005. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev. 19: 2289–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Schlesinger F., Davis C. A., Zhang Y., Li R., et al. , 2011. Synthetic spike-in standards for RNA-seq experiments. Genome Res. 21: 1543–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A. M., Stenberg P., Bernhardsson C., Larsson J., 2007. Painting of fourth and chromosome-wide regulation of the 4th chromosome in Drosophila melanogaster. EMBO J. 26: 2307–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., et al. , 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry, J. J., P. T. Pyl, T. Rausch, T. Zichner, M. M. Tekkedil et al., 2013 The genomic and transcriptomic landscape of a HeLa cell line. G3 (Bethesda) 3: 1213–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J., Meller V. H., 2006. Dosage compensation, the origin and the afterlife of sex chromosomes. Chromosome Res. 14: 417–431. [DOI] [PubMed] [Google Scholar]
- Lee H., McManus C. J., Cho D. Y., Eaton M., Renda F., et al. , 2014. DNA copy number evolution in Drosophila cell lines. Genome Biol. 15: R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley D. L., Sandler L., Baker B. S., Carpenter A. T., Denell R. E., et al. , 1972. Segmental aneuploidy and the genetic gross structure of the Drosophila genome. Genetics 71: 157–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott S. E., Villalta J. E., Schroth G. P., Luo S., Tonkin L. A., et al. , 2011. Noncanonical compensation of zygotic X transcription in early Drosophila melanogaster development revealed through single-embryo RNA-seq. PLoS Biol. 9: e1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg L. E., Figueiredo M. L., Stenberg P., Larsson J., 2012. Buffering and proteolysis are induced by segmental monosomy in Drosophila melanogaster. Nucleic Acids Res. 40: 5926–5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone J. H., Cho D. Y., Mattiuzzo N. R., Artieri C. G., Jiang L., et al. , 2012. Mediation of Drosophila autosomal dosage effects and compensation by network interactions. Genome Biol. 13: r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone J. H., Oliver B., 2011. Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biol. 9: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin I., Siegal M. L., Baker B. S., 2000. The evolution of dosage-compensation mechanisms. BioEssays 22: 1106–1114. [DOI] [PubMed] [Google Scholar]
- Mattox W., Baker B. S., 1991. Autoregulation of the splicing of transcripts from the transformer-2 gene of Drosophila. Genes Dev. 5: 786–796. [DOI] [PubMed] [Google Scholar]
- McAnally A. A., Yampolsky L. Y., 2010. Widespread transcriptional autosomal dosage compensation in Drosophila correlates with gene expression level. Genome Biol. Evol. 2: 44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller V. H., Kuroda M. I., 2002. Sex and the single chromosome. Adv. Genet. 46: 1–24. [DOI] [PubMed] [Google Scholar]
- Parisi M., Nuttall R., Naiman D., Bouffard G., Malley J., et al. , 2003. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 299: 697–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip P., Stenberg P., 2013. Male X-linked genes in Drosophila melanogaster are compensated independently of the male-specific lethal complex. Epigenetics Chromatin 6: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder E., Ashburner M., Bautista-Llacer R., Drummond J., Webster J., et al. , 2007. The DrosDel deletion collection: a Drosophila genomewide chromosomal deficiency resource. Genetics 177: 615–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salz H. K., Erickson J. W., 2010. Sex determination in Drosophila: The view from the top. Fly (Austin) 4: 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Pierre, S. E., Ponting L. Stefancsik R. McQuilton P., 2014. FlyBase 102–advanced approaches to interrogating FlyBase. Nucleic Acids Res. 42: D780–D788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg P., Larsson J., 2011. Buffering and the evolution of chromosome-wide gene regulation. Chromosoma 120: 213–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg P., Lundberg L. E., Johansson A. M., Ryden P., Svensson M. J., et al. , 2009. Buffering of segmental and chromosomal aneuploidies in Drosophila melanogaster. PLoS Genet. 5: e1000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill D., Zhang Y., Parisi M., Oliver B., 2007. Demasculinization of X chromosomes in the Drosophila genus. Nature 450: 238–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill D., Malone J. H., Sun X., Smith H. E., Rabinow L., et al. , 2013. Design of RNA splicing analysis null models for post hoc filtering of Drosophila head RNA-Seq data with the splicing analysis kit (Spanki). BMC Bioinformatics 14: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Johnson A. F., Li J., Lambdin A. S., Cheng J., et al. , 2013. Differential effect of aneuploidy on the X chromosome and genes with sex-biased expression in Drosophila. Proc. Natl. Acad. Sci. USA 110: 16514–16519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., et al. , 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallender E. J., Lahn B. T., 2004. How mammalian sex chromosomes acquired their peculiar gene content. BioEssays 26: 159–169. [DOI] [PubMed] [Google Scholar]
- Vibranovski M. D., Zhang Y., Long M., 2009. General gene movement off the X chromosome in the Drosophila genus. Genome Res. 19: 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B., Charlesworth B., 2006. Evolution on the X chromosome: unusual patterns and processes. Nat. Rev. Genet. 7: 645–653. [DOI] [PubMed] [Google Scholar]
- Vicoso B., Bachtrog D., 2013. Reversal of an ancient sex chromosome to an autosome in Drosophila. Nature 499: 332–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Gong B., Bushel P. R., Thierry-Mieg J., Thierry-Mieg D., et al. , 2014. The concordance between RNA-seq and microarray data depends on chemical treatment and transcript abundance. Nat. Biotechnol. 32: 926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Oliver B., 2010. An evolutionary consequence of dosage compensation on Drosophila melanogaster female X-chromatin structure? BMC Genomics 11: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Malone J. H., Powell S. K., Periwal V., Spana E., et al. , 2010. Expression in aneuploid Drosophila S2 cells. PLoS Biol. 8: e1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.