Abstract
Hexachlorocyclohexane (HCH), a synthetic organochloride, was first used as a broad-acre insecticide in the 1940s, and many HCH-degrading bacterial strains have been isolated from around the globe during the last 20 years. To date, the same degradation pathway (the lin pathway) has been implicated in all strains characterized, although the pathway has only been characterized intensively in two strains and for only a single HCH isomer. To further elucidate the evolution of the lin pathway, we have biochemically and genetically characterized three HCH-degrading strains from the Czech Republic and compared the genomes of these and seven other HCH-degrading bacterial strains. The three new strains each yielded a distinct set of metabolites during their degradation of HCH isomers. Variable assembly of the pathway is a common feature across the 10 genomes, eight of which (including all three Czech strains) were either missing key lin genes or containing duplicate copies of upstream lin genes (linA-F). The analysis also confirmed the important role of horizontal transfer mediated by insertion sequence IS6100 in the acquisition of the pathway, with a stronger association of IS6100 to the lin genes in the new strains. In one strain, a linA variant was identified that likely caused a novel degradation phenotype involving a shift in isomer preference. This study identifies a number of strains that are in the early stages of lin pathway acquisition and shows that the state of the pathway can explain the degradation patterns observed.
Keywords: IS6100, evolution, hexachlorocyclohexane, lindane, transposon
Hexachlorocyclohexane (HCH) is a synthetic organochloride that saw widespread global use after the discovery of its insecticidal properties in the 1940s. However, the off-target toxicity of HCH has caused it to be phased out of use during the last two decades in many countries and it was added to the Stockholm Convention list of persistent organic pollutants in 2009 (Vijgen et al. 2011). The manufacture of HCH produces a number of different isomers that differ in the equatorial and axial arrangement of hydrogen and chlorine atoms. Only one isomer (γ-HCH, known commercially as lindane) has useful insecticidal properties, and this isomer makes up only a small proportion (8–15%) of the HCH produced (Breivik et al. 1999). The remaining isomers (predominantly α-HCH, 55–80%, β-HCH, 5–14%, and δ-HCH, 2–16%), most of which still show high mammalian toxicity (Breivik et al. 1999), are separated and dumped in large waste stockpiles around the world (Lal et al. 2010). The long half-life (e.g., 1.6 and 4.8 years for α- and β-HCH, respectively, in soil) of many HCH isomers means that they will remain an environmental hazard for many years (Wöhrnschimmel et al. 2012).
More than 60 HCH-degrading bacterial strains have been isolated from contaminated fields since the first such strain was reported in 1989 (Johri et al. 1998; Senoo and Wada 1989; Thomas et al. 1996). These strains are predominantly from the family Sphingomonadaceae; however, strains from distant genera such as Microbacterium and Bacillus also are represented (Lal et al. 2010). The most complete understanding of bacterial HCH degradation to date concerns the study of the insecticidal γ-HCH isomer. Interestingly, only one pathway (mediated by the lin genes) has currently been identified for its degradation, with examples found both inside and outside the Sphingomonadaceae. Many aspects of the pathway are still unclear; however, two recent reviews extensively cover our current understanding (Lal et al. 2010; Nagata et al. 2007). The lin pathway was originally identified in Sphingobium japonicum UT26 and is composed of an upstream [conversion of HCH to 2,5-dichlorohydroquinone (2,5-DCHQ)] and downstream component (conversion of 2,5-DCHQ into tricarboxylic acid cycle intermediates). The upstream pathway initially proceeds via two successive dehydrochlorinations (mediated by LinA) followed by two LinB-catalyzed hydrolytic dechlorinations and a dehydrogenation by LinC to produce 2,5-DCHQ (Nagata et al. 1993a,b; Nagata et al. 1994). The downstream pathway involves an initial reductive dechlorination by the glutathione S-transferase LinD, followed by ring cleavage and conversion to β-ketoadipate by LinE and LinF, respectively (Miyauchi et al. 1999, 1998). β-Ketoadipate is a common intermediate in aromatic metabolism and is converted into tricarboxylic acid cycle intermediates by LinGH and LinJ (Nagata et al. 2007).
The catabolic pathway for the major isomers other than γ-HCH has not been fully elucidated; however, lin genes are known to play a role. The initial steps in the degradation of these other isomers can be catalyzed by either LinA or LinB, depending on the axial and equatorial arrangement of chlorine atoms in each isomer. β-HCH lacks the 1,2-biaxial HCl pair required by the reaction mechanism of LinA, and is instead initially metabolized by LinB (Trantirek et al. 2001). α- and δ-HCH are each capable of degradation by LinA in vitro, but in bacterial cell cultures δ-HCH appears to be initially degraded by LinB (Geueke et al. 2013; Kumar et al. 2005; Suar et al. 2005; Wu et al. 2007). The expression ratio of LinA to LinB is known to affect the initial metabolism of HCH isomers and is the likely explanation for why δ-HCH is degraded via LinB (Geueke et al. 2013). The subsequent steps in degradation of these three isomers are as yet unknown. A summary of the degradation pathway for each isomer relevant to this work can be found in Supporting Information, Figure S1.
As a recently introduced synthetic compound, HCH also provides a model system for examining how bacteria evolve new catabolic pathways. Although they are chromosomally located in S. japonicum UT26, most upstream components of the lin pathway are found in unique regions of the UT26 genome associated with the insertion sequence IS6100, suggesting that they have been acquired through horizontal transfer (Lal et al. 2006; Nagata et al. 2011). IS6100 is a member of the IS6 family, which can mobilize genes between two directly repeated IS6100 elements through replicon fusion and subsequent resolution by homologous recombination (Mahillon and Chandler 1998). This process results in an increase in the number of copies of IS6100 in a genome, with a copy of the element left at the original site.
The horizontal transfer of lin genes is observed in other strains, such as the recently sequenced Sphingomonas sp. MM-1, in which all the upstream lin genes are found on plasmids (Tabata et al. 2011; Tabata et al. 2013). A number of other genomes of HCH-degrading Sphingobium strains isolated from India and Japan have been sequenced recently, although their genomic organization of lin genes is yet to be fully described (Kohli et al. 2013; Kumar Singh et al. 2013; Mukherjee et al. 2013). In addition to these isolated strains, the metagenomes of soil samples have been sequenced to assess the population dynamics of lin genes in contaminated sites. These studies revealed that the prevalence of upstream lin genes is correlated with the level of HCH contamination in the soil, suggesting selective pressure drives acquisition of the transmissible lin genes in HCH contaminated soil (Sangwan et al. 2012).
Although most characterized strains have a full complement of lin genes, there is a growing body of evidence from newly sequenced strains that key components of the pathway frequently are missing (Dogra et al. 2004; Kohli et al. 2013; Kumar Singh et al. 2013; Mukherjee et al. 2013). Given the apparent nature of pathway acquisition through a series of horizontal transfer events, these missing genes are likely to reflect early stages in the acquisition of the pathway, with some species yet to acquire some genes. With an incomplete pathway, these strains are unlikely to be able to completely mineralize HCH, instead converting it cometabolically. Many HCH-degrading strains have been poorly characterized in this regard, however, leaving open the possibility that there may be alternate steps in HCH degradation yet to be discovered. An additional limitation with the currently sequenced species is that, apart from S. japonicum UT26, the others have all been isolated from Indian soil samples, with many being isolated from the same field. Any insights gained from these strains might therefore be specific to the particular environmental conditions they encounter and restrict our ability to draw broader conclusions about the evolution of the lin pathway.
Here, we present an analysis of three HCH-degrading strains (Sphingobium czechense LL01, Novosphingobium barchaimii LL02, and Sphingobium baderi LL03) isolated from soil samples at a Czech lindane-manufacturing site (Kaur et al. 2013; Niharika et al. 2013a,b). Strain LL01 has been reported to degrade α-, β-, γ-, and δ-HCH and LL03 to degrade only α-, γ-, and δ-HCH. Strain LL02 has been reported to degrade technical-HCH (a mix of HCH isomers); however, it is unknown which pure isomers it is capable of degrading. No information has yet been reported on how the HCH degradation in these strains compares with other, well-characterized, strains or on the relative rates of isomer degradation in each strain. Both of these features are known to be affected by the lin gene complement of a strain (Sharma et al. 2011; Wu et al. 2007)
We first compared the degradation patterns and metabolites of α-, β-, γ-, and δ-HCH in each strain with the well-characterized strain S. japonicum UT26, which degrades all four isomers. We were able to confirm the previously published abilities of LL01 and LL03 and show that N. barchaimii LL02 degrades α-, γ-, and δ-HCH but with a shift in isomer preference, degrading δ-HCH much faster and γ-HCH much slower than the other strains. We then sequenced the genomes of the three Czech strains to compare them with the published genomes of UT26 (isolated in Japan) and six other strains isolated in India. The comparative genomic analysis presented here finds partial lin pathways in half of the studied strains, with variable organization of those genes that are present. It also highlights the key role of IS6100 elements in lin gene transmission, showing a variety of insertion sites for IS6100 elements around the lin genes, and the first association of IS6100 with the linGHIJ cluster. Finally, we identify a LinA variant in N. barchaimii LL02 that is presumed to be responsible for the shift in isomer preference.
Methods
Strains and media
Bacterial strains used in this study were obtained from the DSMZ (www.dsmz.de) under the accession numbers DSM-16413 (S. japonicum UT26), DSM-25410 (S. czechense LL01), DSM-25411 (N. barchaimii LL02), and DSM-25433 (S. baderi LL03). Quarter-strength Luria Bertani medium (QSLB; 2.5 g of tryptone, 1.25 g of yeast extract, 2.5 g of NaCl, 20 g of sodium succinate, and 20 mL of glycerol per liter) was used for the growth of these strains at 30°.
Degradation assays
Seed cultures of each strain were prepared by inoculating glycerol stocks into QSLB and incubating at 30° overnight with shaking (180 rpm). The seed cultures were then centrifuged at 8000g for 10 min at 4° and the pellet resuspended in QSLB. After standardizing to an OD600 of 0.3, the cells were added (1% v/v) into a 500-mL flask with 20 mL of QSLB containing either α-, β-, γ-, or δ-HCH at a concentration of 0.5 ppm. Uninoculated QSLB containing HCH was used as a negative control, and all treatments were tested in triplicate cultures.
Samples of growth media from each flask (500 µL) were taken at 0, 0.5, 1, 4, 8, 24, 32, and 52 hr after inoculation. Because of the slow degradation of γ-HCH by strain LL02, an additional sample was taken at 96 hr after inoculation. Residual HCH and any metabolites were extracted with equal volumes of hexane and analyzed by gas chromatography using an Agilent 7890A gas chromatograph with an electron capture detector and a BPX-50 column (length: 30 m, internal diameter: 0.32 mm, film thickness: 0.25 µm). The temperature conditions for the analysis were held isothermally at 100° for 5 min, followed by an increase at a rate of 10° per minute to 200°, which was held for a further 5 min. Quantitation of HCH isomers was performed with standard curves produced from serial dilutions of the control flask media. A qualitative assessment of the metabolites produced by each strain was made by comparison to the retention time of 1,2,3-TCB, 1,2,4-TCB, and 1,3,5-TCB as authentic standards (Sigma-Aldrich) or to the pentachlorocyclohexane (PCCH) or pentachlorocyclohexanol metabolites produced with purified LinA and LinB respectively from the relevant HCH isomer (Trantirek et al. 2001).
Genome sequencing and analysis
Genomic DNA for S. czechense LL01, N. barchaimii LL02 and S. baderi LL03 was prepared using the QIAGEN Genomic Tip kit (QIAGEN) following the manufacturer’s protocol. DNA was sequenced on an Illumina HiSeq2000 by Macrogen (Seoul, Korea). Both a short (500 bp) and long (5 kb) insert library was prepared and 100-bp paired reads were obtained from each sample. The initial reads were quality filtered (minimum average quality = 3) and trimmed (minimum quality = 20) using PrinSeq (Schmieder and Edwards 2011) and then downsampled to 100−120× coverage based on predicted genome size. FastQC was used to inspect the reads for quality issues before proceeding with the assembly.
Initial assemblies were constructed using the ABySS assembler (Simpson et al. 2009) with a range of k-mer lengths and the best assembly based on the contig N50 metric for each strain was selected for scaffolding using the matepair reads. Gaps in the final assembly were closed with GapFiller (Nadalin et al. 2012) and annotation of the final assemblies was performed using PGAAP [National Center for Biotechnology Information (NCBI)].
Concurrent with this work, the genome of S. baderi LL03 has been independently sequenced using the 454 GS FLX Titanium technology. Comparison with the assembly produced in this study from Illumina sequencing indicates that the two have near identical sequences, with 98.2% of the 454 assembly aligning to the Illumina assembly with greater than 99.8% identity. All analysis described has been carried out on the Illumina assembly. The Whole Genome Shotgun sequences for S. czechense LL01, N. barchaimii LL02, and S. baderi LL03 have been deposited at DDBJ/EMBL/GenBank under the accession numbers JACT00000000, JACU00000000, and JACV00000000 respectively. The versions described in this paper are JACT01000000, JACU01000000, and JACV01000000.
In addition to the aforementioned sequences, genome sequences for S. japonicum UT26 (GCF_000091125), S. indicum B90A (AJXQ01000000), Sphingomonas sp. MM-1 (GCF_000347675), Sphingobium sp. HDIP04 (ATDO01000000), S. chinhatense IP26 (AUDA01000000), S. quisquiliarum P25 (ATHO01000000), S. ummariense RL-3 (AUWY01000000), Sphingobium sp. SYK-6 (GCF_000283515), Sphingomonas wittichii RW-1 (GCF_000016765), and Novosphingobium aromaticivorans DSM12444 (GCF_000013325) and the linB containing plasmid pLB1 (NC_008330) were used in our analysis. Average nucleotide identities (ANIs) of all pairwise comparisons were calculated with JSpecies (Richter and Rosselló-Móra 2009). Whole-genome alignments were performed against UT26, LL01, LL02 and LL03 using MUMmer (Delcher et al. 2002) and regions of horizontal transfer in each target genome were predicted with the Genomic Island Suite of Tools (GIST), which combines the results of several common horizontal transfer prediction tools to produce a single, improved estimate (Hasan et al. 2012). This information was visualized using Circos (Krzywinski et al. 2009).
The lin genes of UT26 (linA; SJA_C1-18560, linB; SJA_C1-19590, linC; SJA_C1-00590, linD; SJA_P1-01390; linE; SJA_P1-01430, linR; SJA_P1-01440; linF; SJA_C2-04820, linG; SJA_C2-05210, linH; SJA_C2-05220, linI; SJA_C2-05200, linJ; SJA_C2-05190, linK; SJA_C1-00210, linL; SJA_C1-00220, linM; SJA_C1-00230, linN; SJA_C1-00240,) were used as queries for TBLASTX searches in the newly sequenced strains. The top hits of each search were then aligned to the non-redundant NCBI database to confirm their assignment. The genomic regions containing lin genes were aligned pairwise with MEGABLAST and displayed with genoPlotR (Guy et al. 2010) to examine the conserved regions between the studied sequences.
Results
Degradation of HCH
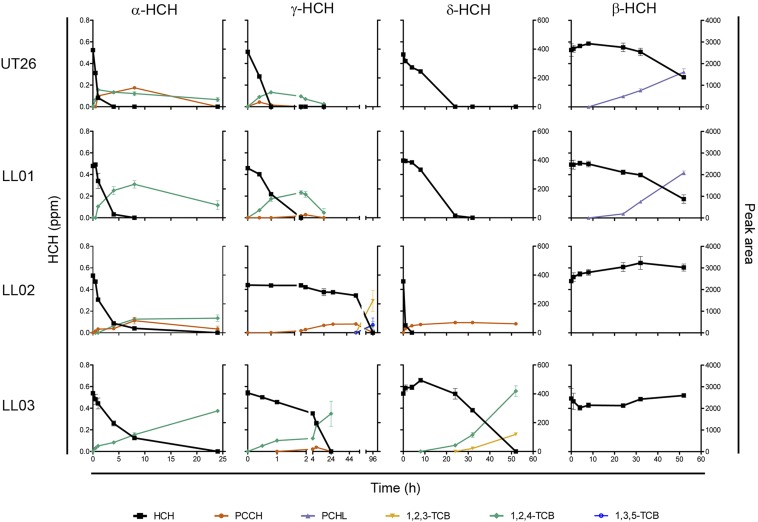
Growth assays with four HCH isomers allowed us to compare the abilities of LL01, LL02, and LL03 to degrade the various HCH isomers with those of the well characterized strain UT26 (Figure 1). We were able to confirm the ability of LL01 to degrade all four isomers tested (relative rates: α = γ > δ >> β), and of LL03 to degrade α-, γ-, and δ-HCH (α > γ >> δ) (Kaur et al. 2013; Niharika et al. 2013a). LL02 was found to degrade only α-, γ-, and δ-HCH (δ > α >> γ). Compared with UT26, LL01 exhibited a similar degradation pattern for each isomer, whereas LL03 was much slower than UT26 for the three isomers it could degrade. LL02 had a novel degradation pattern, with slower γ-HCH and faster δ-HCH degradation than the other strains. Although LinA variants are known to affect the α-HCH enantiomer specificity or the α/γ isomer preference (Sharma et al. 2011; Suar et al. 2005), this is the first strain reported to degrade δ-HCH faster than all other isomers.
Figure 1.
Degradation assays for HCH isomers. Degradation of α-HCH, β-HCH, γ-HCH, and δ-HCH (left-hand axis) by UT26, LL01, LL02, and LL03 and relative quantification of metabolites by peak area (right-hand axis). Time postinoculation is shown on the bottom axis and the timescale for the γ-HCH assay has been adjusted to observe both the very fast (less than 1 hr) and very slow (96 hr) degradation of γ-HCH by different strains. Values are the mean of three biological replicates, with standard deviations. Identity of metabolites was confirmed by comparison to authentic standards or to metabolites produced with purified UT26 LinA or LinB enzymes. HCH, hexachlorocyclohexane; PCCH, pentachlorocyclohexene; PCHL, pentachlorocyclohexanol; TCB, trichlorobenzene.
Metabolites produced in the growth assays were identified by comparison of their retention time to known standards. Strain LL01 produced the same metabolite profile for each isomer as UT26, with degradation of β-HCH−producing pentachlorocyclohexanol and the remaining isomers producing pentachlorocyclohexane (PCCH), and trichlorobenzene (TCB). β-PCCH was not directly observed in the degradation of α-HCH by strain LL01 but is inferred from the observed TCB metabolite. As the bulk of the δ-HCH degradation occurred between the 8- and 24-hr samples, no metabolites for δ-HCH degradation were observed in UT26 and LL01.
Strain LL03 produced the same metabolites as UT26 for α-HCH and γ-HCH but produced 1,2,4-trichlorobenzene (1,2,4-TCB) and 1,2,3-trichlorobenzene (1,2,3-TCB) when incubated with δ-HCH and could not degrade β-HCH at all. The δ-HCH metabolites produced by LL03 have been observed when δ-HCH is incubated with purified UT26 LinA enzyme but not in cell cultures of various strains, as δ-HCH is preferentially degraded by LinB (Geueke et al. 2013). These metabolites, combined with the lack of degradation of β-HCH (a LinB-initiated degradation), are consistent with the claim that LL03 lacks a linB gene, leading to LinA-initiated degradation pathways (Kaur et al. 2013).
Strain LL02 is similar to LL03 in that degradation of α-, γ-, and δ-HCH all appears to be LinA-initiated, producing their respective PCCHs, and no degradation of β-HCH is observed. The largest difference between LL02 and LL03 comes in the further degradation of γ-PCCH, with LL03 (and UT26) accumulating 1,2,4-TCB as a dead end product, whereas LL02 accumulates 1,2,3-TCB, 1,2,4-TCB, and 1,3,5-TCB. This is the first report of the 1,2,3- and 1,3,5-TCB isomers being observed from γ-HCH degradation. It is unclear why different TCB isomers would be produced in this degradation, as 1,2,4-TCB is proposed to be spontaneously produced from tetrachlorocyclohexadiene, the putative final product of LinA activity (Nagata et al. 1993b). However, as 1,2,4-TCB is identified in degradation assays with purified LinA enzyme, the altered metabolite profile provides additional evidence for the presence of a novel LinA variant in strain LL02.
Genome sequences of Czech strains
For each strain, short and long insert libraries were prepared and sequenced using Illumina HiSequation 2000. After quality control, the short insert sequences were assembled using the ABySS assembler and the best assembly for each strain was selected for scaffolding and gap filling using the matepair reads. The final assemblies produced draft genomes comprising 21, 26, and 82 contigs for LL01, LL02, and LL03, respectively (Table 1). These contigs were assembled into 18, 19, and 11 scaffolds, respectively. The LL01 assembly contains four chromosomal scaffolds (totaling 4.1 Mb in size) and a plasmid scaffold of 186 kb. The LL02 assembly contains 10 chromosomal scaffolds and a plasmid scaffold of 242 kb. In the LL03 assembly, 98% of the genome is contained in four circular scaffolds, comprising two chromosomal scaffolds (3.4 Mb and 641 kb) and two plasmid scaffolds (510 kb and 34 kb). The total genome size and GC% content for each assembly is consistent with previously sequenced genomes from the relevant genus (Copley et al. 2012; Dong et al. 2014; Gan et al. 2012; Luo et al. 2012; Masai et al. 2012). Annotation of each genome was performed using the PGAAP (Angiuoli et al. 2008).
Table 1. Assembly summary: summary of the final genome assemblies produced for this study.
| Strain | Number of Contigs | Contig N50 | Number of Scaffolds | Final Assembly Size, Mbp | G/C Content, % | Chromosomal Scaffolds, (Total Size (Mbp) | Plasmid Scaffolds, Total Size (kbp) |
|---|---|---|---|---|---|---|---|
| LL01 | 21 | 480 | 18 | 4.6 | 63.6 | 4 (4.1) | 1 (186) |
| LL02 | 26 | 797 | 19 | 5.3 | 64.0 | 10 (4.9) | 1 (242) |
| LL03 | 82 | 127 | 11 | 4.7 | 63.5 | 2 (4) | 2 (544) |
lin genes and IS6100 elements in LL01, LL02, and LL03
We identified the lin genes present in LL01, LL02, and LL03 by homology to the protein sequences from UT26 using the top BLAST hits (E-value < 1e-20) (Table S1). Each potential hit was then used as a BLAST query against the NCBI nr database to confirm that its closest match was a lin gene. There is consistent representation of the conserved downstream elements of the pathway representing core sphingonomad functional systems, with each strain containing a linKLMN cluster encoding an ABC transporter and most components of the linGHIJ cluster encoding the β-ketoadipate pathway (Nagata et al. 2010). In strain LL01, however, the linGHIJ cluster is disrupted by an IS6100 element, causing the transcriptional regulator linI to be interrupted and linJ (encoding an acetyl-CoA acetyltransferase) to be lost.
Unlike the core functional genes, there is considerable variation in the presence of the remaining lin genes (linA-F) in the three strains. In LL01, we identified a complete upstream pathway, with linA, linB (two copies) and linC all present; however, linRED and linF are missing. In LL02 we only identified linA and an IS6100-truncated linF, whereas in LL03 we only identified linA, linF, and a disrupted linR. These variable genes (linA-F) comprise the HCH-specific component of the pathway, in contrast to the more general role performed by the more consistently represented core functional genes. The presence or absence of linA and linB in each of the three strains is sufficient in these cases to explain which HCH isomers each strain could degrade. Further sequencing of additional isolates would identify the extent of upstream lin gene variation and its association with HCH isomer degradation.
We also assessed each genome for the prevalence of IS6100 elements, given their known association with the lin genes (Dogra et al. 2004). We identified 14, 19, and 24 IS6100 elements in LL01, LL02, and LL03, respectively, either confirmed through a match to the IS6100 transposase (accession BAI98840) or inferred by a match to two 15 bp IS6100 inverted repeats (GGCTCTGTTGCAAA) across contig boundaries (Mahillon and Chandler 1998). These numbers fall within the range seen among sequenced HCH-degrading strains (13−26) and are much greater than seen in the three non−HCH-degrading strains from the same genera used in our comparative analysis; Novosphingobium aromaticivorans DSM12444 (0), Sphingomonas wittichii RW-1 (1), and Sphingobium sp. SYK-6 (0) (Table 2). IS6100 is not a lin-gene−specific transposable element, having also been associated with catabolic genes for nylon (Flavobacterium sp. KI72, Pseudomonas sp. NK87), carbazole (Sphingomonas sp. XLDN2-5), and nonyl-phenols (Sphingomonas sp. NP5), among others, however, only small numbers of IS6100 elements (2−5) have been identified in these cases (Gai et al. 2010; Kato et al. 1994; Takeo et al. 2012; Yasuhira et al. 2007). The IS6100 elements in LL01, LL02, and LL03 appear to be highly clustered around the lin genes, supporting the proposed involvement of IS6100 in lin gene recruitment. In LL03 for example, the 19 IS6100 elements found on the 3.4 Mb main chromosome scaffold lie at a median distance of 25.5 kb from the nearest lin gene, with five found near linGHIJ, four near linX and linA, two near linKLMN, and seven near linF (Figure 2).
Table 2. IS6100 elements in the studied genomes.
| Confirmed | Putative | Total | |
|---|---|---|---|
| LL01 | 12 | 2 | 14 |
| LL02 | 17 | 2 | 19 |
| LL03 | 24 | 0 | 24 |
| B90A | 0 | 20 | 20 |
| HDIP04 | 4 | 9 | 13 |
| RL-3 | 16 | 7 | 26 |
| P25 | 23 | 2 | 25 |
| IP26 | 16 | 10 | 26 |
| UT26 | 14 | NAa | 14 |
| MM-1 | 14 | NAa | 14 |
| RW-1 | 1 | NAa | 1 |
| SYK-6 | 0 | NAa | 0 |
| DSM12444 | 0 | NAa | 0 |
Number of confirmed and predicted IS6100 elements in the 13 genomes studied in this article. Confirmed elements were identified by a match to the IS6100 transposase and putative elements were predicted from the presence of two IS6100 terminal inverted repeat sequences across contig boundaries. In completed genomes, the inverted repeat sequence is only found associated with IS6100 elements. NA, not available.
Complete genome, no contig boundaries to assess putative IS6100 elements.
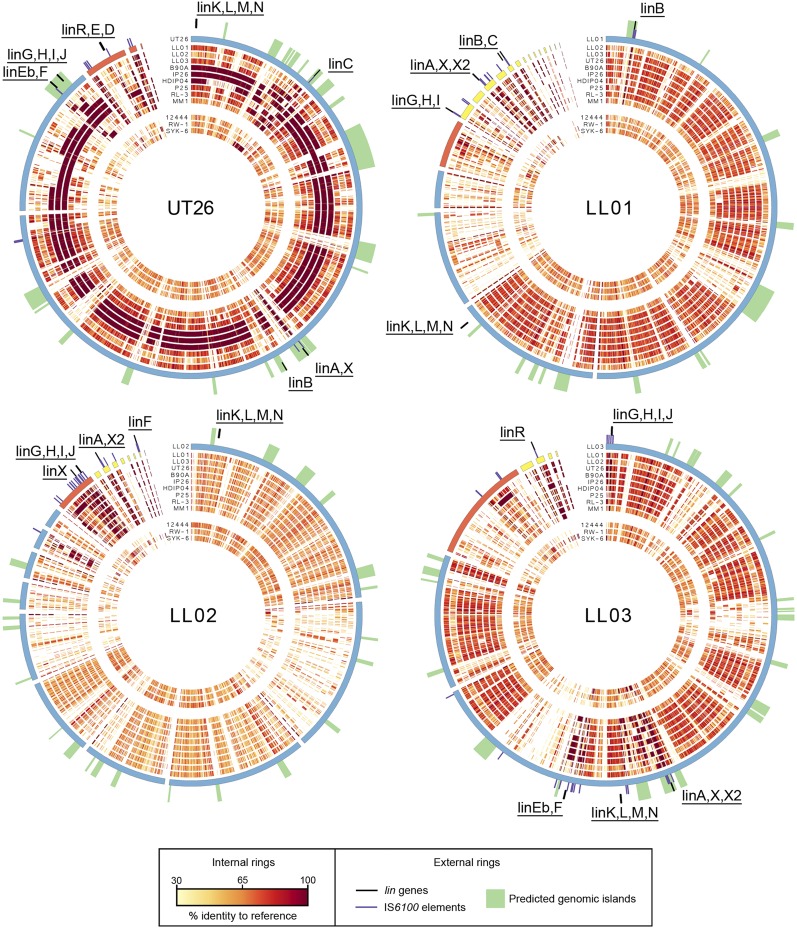
Figure 2.
Whole-genome alignments of sequenced strains. Whole-genome alignments of the other 12 genomes (see the Materials and Methods section for names and accession numbers) to each of the UT26, LL01, LL02, and LL03 genomes. The color gradient of the inner rings indicates homology to the reference genome sequence. On display starting from the innermost ring are three non−HCH-degrading strains, then 10 HCH-degrading strains, with the reference genome the outermost ring. Reference chromosomes or scaffolds predicted to be from chromosomes are colored blue, plasmids are colored red, and small scaffolds that cannot be unambiguously determined to be from either chromosomal or plasmid origin are colored yellow. The positions of IS6100 elements (purple) and lin genes (black) in the reference genome are indicated. Predicted genomic islands are highlighted in green. HCH, hexachlorocyclohexane.
Comparison of Czech strains to other HCH-degrading strains
Previous genomic comparisons between UT26 and B90A have suggested that these two strains may have been derived from a common ancestor containing most of the lin genes (Sangwan et al. 2014). However, a similar analysis between UT26 and three other HCH-degrading strains (Sphingobium sp. TKS, Sphingobium sp. MI1205 and Sphingomonas sp. MM-1) suggested the independent acquisition of HCH degradation in each of the four strains (Nagata et al. 2014). The expanding number of genomes available for HCH-degrading strains allows us to test the relative contribution of these two modes of evolution more broadly in lin pathway evolution.
Whole-genome alignments of nine HCH-degrading strains (S. czechense LL01, N. barchaimii LL02, S. baderi LL03, S. indicum B90A, S. chinhatense IP26, Sphingobium sp. HDIP04, S. quisquiliarum P25, S. ummariense RL-3, and Sphingomonas sp. MM-1), and the three non−HCH- degrading strains mentioned previously, to UT26 reveal both modes of evolution noted previously (Figure 2). Three of the other HCH degraders, B90A, IP26, and HDIP04, all have very high homology (ANI of 97.1–97.7%) to large regions of the UT26 genome, indicating a recent common ancestor from which they may have obtained most lin genes. However, the remaining six strains, including all three Czech strains, show smaller regions of homology, at lower levels of identity to UT26 (ANI 75.3–81.9%), indicating more divergence among the shared set of genes common to all sphingomonads. Compared with this low overall level of homology, the high identity (generally >97%) of lin genes in these strains to UT26 indicates that they have likely been acquired horizontally, rather than through shared ancestry with UT26. The sequence composition of the genomic regions containing lin genes also suggests the acquisition by horizontal transfer; the GIST (Hasan et al. 2012) identified predicted genomic islands which contain all lin genes in UT26 except linKLMN.
The aforementioned alignments were repeated with each of LL01, LL02, and LL03 as a reference genome to determine whether these strains acquired their lin genes independently of each other (Figure 2). Unlike what was observed in the comparisons of UT26, these three showed no close relationship to any of the other genomes studied (LL01 ANI: 73.6–79.9%, LL02 ANI: 73.0–75.1%, LL03 ANI: 75.7–81.8%). Where small areas of strong homology exist between the strains, they are associated with the lin genes, IS6100 elements or genomic islands predicted by GIST. This suggests that the distinct lin gene complements of each of these strains may reflect independent horizontal gene transfer events. This whole-genome analysis is consistent with phylogenies based on 16S sequences that place LL01, LL02, and LL03 relatively distant from other HCH-degrading strains (Kaur et al. 2013; Niharika et al. 2013a; Niharika et al. 2013b).
As indicated previously, there is considerable variation in lin gene complement among the 10 HCH-degrading genomes considered here (Figure 3). The four very closely related strains (UT26, B90A, IP26, and HDIP04) each have a complete lin pathway, suggesting that it was acquired from their common ancestor. Of the remaining strains, only MM-1 has a complete pathway, whereas LL01, LL02, LL03, RL-3, and P25 have missing components. The selective pressure to acquire a lin pathway in HCH-contaminated soil suggests that these strains with partial lin pathways are likely in the early stages of pathway assembly, having acquired only a few of the required genes, but we cannot discount the possibility that some represent instances of gene loss (Sangwan et al. 2012).
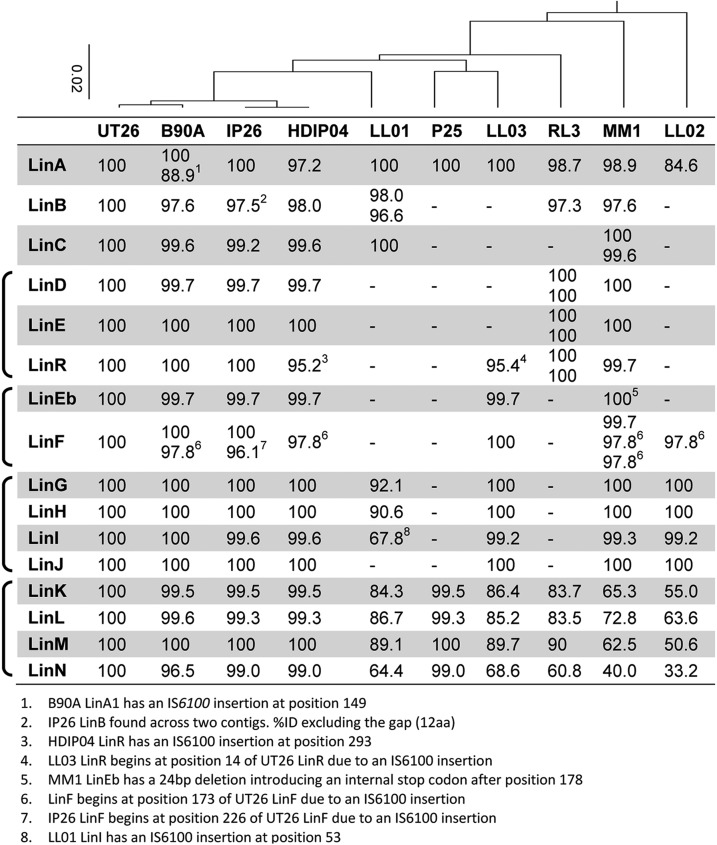
Figure 3.
lin pathway composition of studied strains. Presence of lin genes in the 10 HCH-degrading strains studied, along with the percent identity of each encoded protein to the equivalent in UT26. Genes grouped in operons are indicated with braces to the left of the gene names. Multiple values indicate duplicate copies of the gene in a strain. The neighbor-joining phylogeny above the strain names was produced from a clustalw alignment of the 16S rDNA sequences (UT26: AF039168, B90A: AY519129, IP26: EF190507, HDIP04: EF424393, LL01: JN646865, P25: EU781657, LL03: JN695620, RL-3: EF207155, MM-1: CP004036;G432_r19183, LL02: JN695619) of each strain with E. coli (ECK3843) as an outgroup (not shown). HCH, hexachlorocyclohexane.
There are also instances of multiple copies of the linA-F genes, with two copies of linA in B90A, two of linB in LL01, two of linC in MM1, two of linRED in RL-3, two of linF in B90A and IP26, and three of linF in MM1. In the case of linF, however, all extra copies are disrupted by IS6100 elements, and it is unknown if they are functional. Multiple copies of the upstream genes may relax selective constraints, allowing functional specialization of the duplicated copies as seen in the enantioselective degradation of α-HCH by LinA1/LinA2 in B90A (Suar et al. 2005), or it may increase the expression of the gene, with linA, linB, and linC all being expressed constitutively (Suar et al. 2004). Little variation is seen in protein sequences for these genes however, with almost all copies observed having very high identities to the UT26 sequences (Figure 3). This paucity of variation is observed in all lin genes except linKLMN, further supporting the prediction that these genes, including core downstream components such as the β-ketoadipate pathway encoding linGHIJ, have been acquired by horizontal transfer. The greater variation among linKLMN sequences suggests that this cluster has not undergone horizontal transfer, but has been acquired from a common ancestor.
Genomic organization of lin genes in HCH-degrading strains
We looked at microsynteny at the lin gene loci to investigate the extent of the horizontally transferred regions and their ongoing rearrangement (Figure 4 and Figure 5). Alignments of genomic regions from the above ten genome sequences plus a linB containing plasmid isolated exogenously from HCH-contaminated soil (Miyazaki et al. 2006) revealed high levels of conservation around the lin genes, generally delimited by IS6100 elements. These regions range from many kilobases in length in the case of the downstream lin genes to just a few hundred base pairs for some of the upstream lin genes. Because longer horizontally transferred sequences are more likely to have a higher fitness cost, the size of these conserved regions may be related to the extent of horizontal transfer of each region. The high level of conservation of gene content and intergenic sequence around all lin genes except linKLMN (Supporting Information, Figure S2) suggests that each has been derived from a single original source.
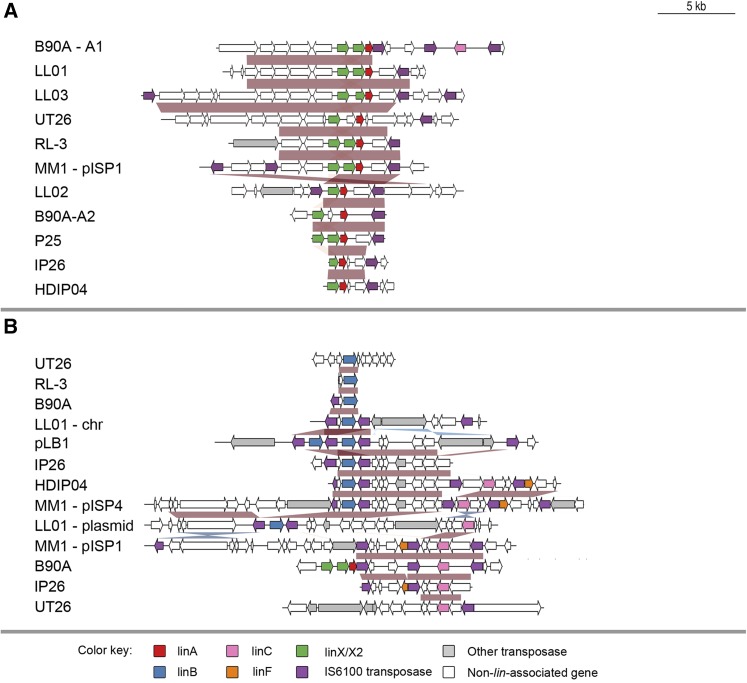
Figure 4.
Genomic organization of upstream lin genes. Alignments detailing strong conservation and IS6100 involvement immediately surrounding the upstream lin genes in HCH-degrading strains. Alignments of the regions surrounding (A) linA and (B) linB and linC were performed with MEGABLAST and ordered to give maximal pairwise alignment lengths. Conserved regions are indicated by red shading where the matches are in the same orientation and by blue shading where the matches are in reverse orientation. Also found associated with these genes are an IS6100 truncated linF and linX/linX2 (linC-like sequences not necessary for HCH degradation). HCH, hexachlorocyclohexane.
Figure 5.
Genomic organization of downstream lin genes. Alignments detailing conservation and rearrangements in the genomic regions of the downstream lin genes. Alignments of the regions surrounding (A) linDER and (B) linFEb and linGHIJ, were performed with MEGABLAST and ordered to give maximal pairwise alignment lengths. Conserved regions are indicated by red shading where the matches are in the same orientation and by blue shading where the matches are in reverse orientation. Note that association of IS6100 elements with linGHIJ is unique to strains LL01, LL02 and LL03.
One major difference between strains is the location of IS6100 insertions around certain lin gene regions, in particular linA and linRED. In the case of linA, we identified five different insertion sites downstream of linA. The UT26 sequence has an insertion 3638 bp downstream of linA, while there are also insertions at 1550 bp (LL01, LL02, LL03, RL-3, MM1, B90A-A2, and P25), 1254 bp (IP26), and 1197 bp (HDIP04) downstream of linA, as well as the B90ALinA1 insertion 444 bp into the linA gene (Figure 4A). Of the eight linRED clusters identified, five have an IS6100 element upstream of the cluster (UT26, IP26, HDIP04, and two clusters in RL-3). Each of these IS elements is located in a different position (Figure 4C). These examples show the continuing activity of IS6100 in HCH-degrading strains and their potential for ongoing involvement in the transfer and evolution of the lin pathway.
Not all lin genes show such variability, however. The location of IS6100 around linB is quite consistent in all strains, with insertion sites 307 bp upstream and 105 bp downstream of the gene (Figure 4B). The only exceptions to this are found in IP26, where the insertion site is 387 bp upstream, and in UT26, which lacks any flanking IS elements around linB. As will be discussed below, this lack of variability in IS6100 arrangement around linB may be because it is a recent acquisition that has been selectively favored to increase linB transmissibility.
N. barchaimii LL02 contains an uncharacterized LinA variant
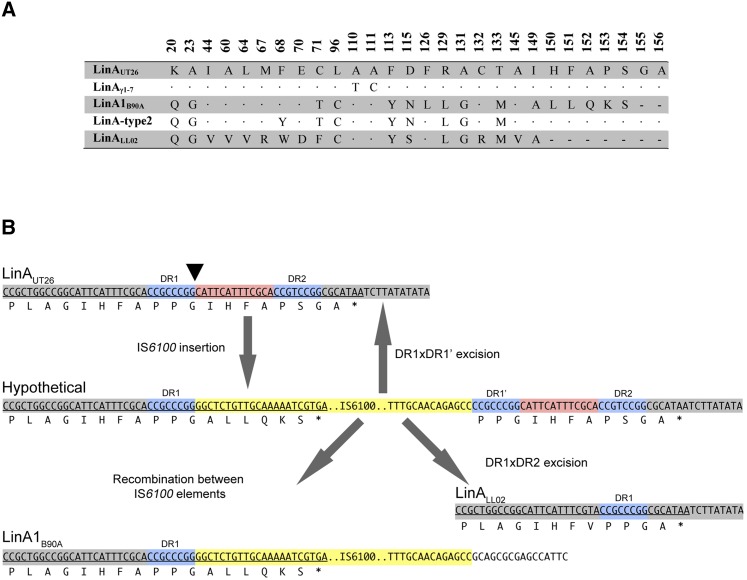
Closer analysis of the linA gene sequence in N. barchaimii revealed that it encodes an uncharacterized LinA enzyme (Figure 6A). Compared with the two B90A variants LinA1 (accession number AAN64239) and LinA2 (ACM48253), the LinALL02 contains 11 and 17 amino acid differences respectively, as well as a seven amino acid deletion at the C-terminal end. Many of the mutations in LinALL02 are clustered near the catalytic histidine (H73) and affect amino acids predicted to form the active site (Okai et al. 2010). In other characterized LinA enzymes, small changes to the amino acid sequence are capable of altering the α-HCH enantiomer preference or the α-/γ-HCH isomer preference. The mutations in LinALL02, therefore, provide candidates for the shift in isomer preference that sees strain LL02 preferentially degrading δ-HCH. Further work characterizing LinALL02 would confirm this hypothesis and identify which mutations are necessary for the altered phenotype. A sequence identical to LinALL02 has also been recently identified in a metagenomic screen of Indian soil samples, indicating that it is not a geographically restricted variant (Lal et al. 2013).
Figure 6.
Comparison of LinA sequence variants. (A) Amino acid differences between major LinA variants and the uncharacterized LinA of LL02. (B) Proposed mechanism for the IS6100-mediated 21-bp deletion in LL02. The initial insertion site of the IS6100 cointegrates (yellow) in a LinAUT26-like sequence is marked with a black triangle. The insertion induces the duplication of 8bp at the insertion site (DR1 and DR1´ in blue). IS6100 elements are capable of reversion through homologous recombination between the induced direct repeats (DR1×DR1´). A near-identical sequence (DR2) is slightly further downstream of the insertion, however, and recombination between DR1 and DR2 gives rise to the sequence observed in LL02, with the deletion of one DR and the intervening sequence (colored red). Also shown is the sequence of LinA1 in B90A, containing the IS6100 insertion and a different downstream sequence. This has likely arisen through homologous recombination between IS6100 elements, deleting the intervening region containing the original C-terminus of LinA.
Discussion
Recruitment of lin genes in sequenced genomes
Until recently, almost all characterized HCH-degrading strains contained a full complement of lin genes [Sphingomonas sp. γ12-7, the only exception, being deficient in linRED (Böltner et al. 2005)]. However, emerging evidence suggests that there is considerable variability in the recruitment of the pathway, with eight of the ten genomes studied here either missing or containing multiple copies of the linA-F genes. Of these variable genes, only linA is present in all strains, suggesting that it is necessary for a strain to be isolated in an enrichment screen for HCH degradation. linA also appears to be sufficient for isolation in an enrichment screen, as it is the only upstream lin gene found in S. quisquiliarum P25. The remaining genes (linB-F) are each absent in several strains and, given the horizontal transfer involved in pathway acquisition, likely represent early stages of pathway evolution, having acquired some lin genes but not all. These strains may therefore provide a valuable resource for studying the gradual accumulation of the lin genes and what function individual elements may serve in the absence of a full pathway.
We cannot be certain that these strains are yet to acquire some lin genes, however, because they are known to be highly mobile as the result of IS6100-mediated genome rearrangements (Nagata et al. 2011). Laboratory studies have shown that lin genes are lost rapidly when the strain is no longer exposed to HCH, with Pseudomonas aeruginosa ITRC-5 losing linA-E when grown in rich media (Singh et al. 2007). Some of the cases of missing lin genes may therefore be due to gene loss events, rather than the strain not yet acquiring the gene. This is the most likely explanation for some genes in LL03, as an IS6100-interrupted linR suggests that an IS6100-mediated rearrangement is the cause of linED missing from the genome. The situation in the Czech strains is further complicated because preliminary data suggested that they were able to grow on HCH as a sole carbon source (Kaur et al. 2013; Niharika et al. 2013a,b); however, we have been unable to replicate this phenotype. This finding suggests that these strains may have undergone secondary loss of some lin genes, during either preservation at the stock center or in the process of resuscitation, making it impossible to determine the original lin complement of these lines. The analysis of isolated cultures may therefore provide an incomplete picture of lin pathway evolution and need to be interpreted in light of other approaches, such as metagenome sequencing, that look at pathway organization in soil populations.
Where instances of missing lin genes can be confirmed in natural populations, there are two main proposals for how these strains are able to degrade HCH. It is possible that the functions performed by missing lin genes may be carried out by other unknown enzymes in the organism. As an alternative, the complementing functions may be carried out by other members of the microbial community. Both of these hypotheses have been supported in different strains. Xanthomonas sp. ICH12 provides an example of the former, with the identification of a novel metabolite after PCCH production (Manickam et al. 2007). The latter hypothesis is supported by studies into microbial consortia isolated from HCH-contaminated Indian soil samples, in which the community was able to degrade HCH far more effectively than any of the individually isolated elements (Elcey and Kunhi 2010; Manonmani et al. 2000).
Also informative in respect of the lin pathway’s origins are the genomic regions immediately surrounding the lin genes. In almost all cases, the genomic context of each lin gene (including flanking genes and intergenic regions) is highly conserved (e.g., 100% match for the 1 kb downstream of linA). The conserved region generally extends out to a mobile genetic element, usually an IS6100 element, and then abruptly ceases. Together with the whole-genome analysis suggesting multiple, independent acquisitions of the lin genes, these data suggest that each lin gene has a single evolutionary origin and then is rapidly distributed across species through horizontal transfer events.
linB is an interesting case because there appear to be two potential mechanisms of transfer. In UT26, linB is situated chromosomally in a GIST-predicted genomic island (Figure 2). Closer inspection of the predicted region identified it as a putative 73-kb integrative and conjugative element (ICE). ICEs (also known as conjugative transposons) are widely dispersed, horizontally transferred elements that encode their own conjugative transfer and integration/excision from a host genome (Guglielmini et al. 2011; Wozniak and Waldor 2010). This 73-kb region on the UT26 first chromosome contains the integrase, relaxase, and recombination directionality factor required for excision and integration of the region (Ramsay et al. 2006), as well as a type IV secretory system that conjugatively transfers the excised element between cells. These elements are known to carry accessory genes responsible for a range of functions including pathogenicity (Chuzeville et al. 2012; Seth-Smith et al. 2012), symbiosis (Sullivan et al. 1995; Sullivan et al. 2002), antibiotic resistance (Michael et al. 2012; Rodríguez-Blanco et al. 2012), and xenobiotic catabolism (Gaillard et al. 2006; Shettigar et al. 2012). In all other sequenced genomes containing a linB, however, the gene is flanked by two IS6100 elements, with intergenic sequence 307 bp upstream and 105 bp downstream identical to the UT26 sequence (Figure 4B). As IS6100 elements only very rarely excise themselves without leaving a copy of the element, it is highly likely that the IS6100-linB-IS6100 element is derived from the linB containing ICE seen in UT26. Thus, Although linB may initially have been acquired by conjugation and integration of an ICE, it appears that the acquisition of flanking IS6100 elements has subsequently been key to the widespread prevalence of linB in isolated strains. With high HCH concentrations appearing to impart a selective pressure for acquisition of the lin genes (Sangwan et al. 2012), an increase in IS6100 activity could provide a possible mechanism.
IS6100 involvement in lin gene acquisition
The involvement of IS6100 in lin gene acquisition is still clearly an active process, with a variety of IS6100 element insertion sites identified. That these elements tended to define the limits of conserved regions around the lin genes is further evidence for the importance of IS6100 in their horizontal transfer, as noted above. In addition to the widespread association of IS6100 with the upstream lin genes, the new genomes sequenced for this study reveal the association of IS6100 elements with the genes of the β-ketoadipate pathway (linGHIJ). This is the first time that IS6100 elements have been found to be associated with these downstream components of the HCH degradation pathway. The fact that this arrangement is found in all three Czech strains suggests that, like the aforementioned linB, the acquisition of flanking IS6100 elements may be the key to widespread horizontal transfer.
The clustered arrangement of IS6100 with so many of the lin genes adds further evidence for a selectively favored increase in IS6100 activity and lin gene transmissibility. In HCH-contaminated soil, an increase in IS6100 activity would introduce a number of new elements in the genome. Any new IS6100 elements inserted near lin genes will be favored in due course as they increase the chance of lin genes being passed on to other community members. A pressure to acquire the upstream lin genes in HCH contaminated soil is therefore likely to explain the newly identified association with the downstream genes.
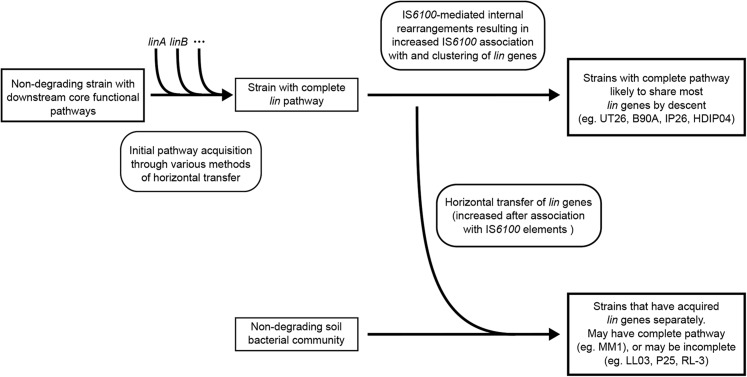
The increasing involvement of IS6100 with the lin genes may indicate the beginning of a second phase in the evolution of the pathway (Figure 7). The first phase of evolution, the initial formation of the pathway, is proposed to have occurred with the recruitment of the upstream genes (through various means) into an ancestral strain containing the downstream genes that make up core sphingomonad functions (Nagata et al. 2014). The current phase appears to involve a consolidation of the pathway, in which we see an increase in IS6100 activity that expands IS6100 association to genes that were initially transferred through other means (e.g., linB) and to downstream genes that were not previously known to be transferrable (linGHIJ). Although this appears to introduce more instability in the pathway than previously seen, it also increases the probability of the pathway being acquired after exposure to HCH. Indeed, the entire catalytic component of the lin pathway is now capable of IS6100-mediated transfer, opening up the possibility of transfer to species that lack the downstream components common to all sphingomonads.
Figure 7.
Hypothesized model of the lin pathway evolution. Proposed model of the processes involved in lin pathway evolution. Note that these steps are likely to be overlapping to some degree.
IS6100-mediated C-terminal truncation of LinALL02
In addition to influencing the pathway organization, transposition and recombination of IS6100 elements can affect the sequence of Lin enzymes. In the case of LinA, an IS6100 insertion is seen at nucleotide position 444 of LinA1 found in B90A, which alters the C-terminal sequence of the protein. In LinALL02, the seven amino acid C-terminal truncation is likely caused by imperfect excision of this IS6100 element (Figure 6B). Insertion of an IS6100 cointegrate is accompanied by an 8-bp direct repeat duplication of the insertion site (Herron et al. 1999). Homologous recombination can occur at this direct repeat, allowing reversion of the IS element insertion, although this occurs at very low rates (Weaden and Dyson 1998). Because this insertion in LinA occurs in a repeated GIHFAP motif, there is a nearly identical copy of the landing site direct repeat just downstream of the insertion (Figure 6B). An imperfect excision of the IS6100 element at this secondary direct repeat would give rise to the sequence observed in the genome of LL02.
The C-terminal extension is involved in stabilizing the LinA trimer and in closing off the active site once the substrate is bound (Macwan et al. 2013). The length of this extension is important for enzyme function, with two GIHFAP motifs producing enzymes with greater activity than those with one or three (Macwan et al. 2013). In the case of LinALL02, the predicted loss of activity due to the single repeat may be compensated for by the C132R substitution. This substitution was identified by an in vitro mutagenesis assay as being responsible for a sixfold increase in LinA expression and solubility in Escherichia coli cells (Mencía et al. 2006). LinALL02 is the first natural variant to be identified with this mutation.
Continuing pathway evolution
Many different models have been proposed for the evolution of new metabolic pathways (Fondi et al. 2009). The lin pathway provides an excellent model to study the early evolution of new pathways, given how recently it has arisen. Our analysis of 10 sequenced genomes suggests that the lin pathway is following the piecewise assembly model in which components of the pathway are pieced together from preexisting genes or operons (Fani et al. 2005). This approach is very similar to that proposed for the pentachlorophenol (PCP) degradation pathway of S. chlorophenolicum L1, which shares some downstream components of the lin pathway. S. chlorophenolicum L1 was found to share the downstream pathway (homologs of the linF and linEb genes) with a common ancestor and to then have acquired three upstream genes responsible for the specific degradation of PCP through two separate horizontal transfer events (Copley et al. 2012). One of the transfer events brought in two catabolic genes in a regulated operon (similar to the case of linRED) and one brought in a single catabolic gene (similar to the proposed acquisition of linA, linB, and linC). In the case of the PCP pathway, however, the lack of a known mechanism for the horizontal transfer precludes a discussion of the future evolutionary trajectory of the system.
In the case of the lin system, however, we can identify some of the mechanisms involved and their continuing effects. Various methods of horizontal transfer have been responsible for the initial recruitment of the upstream lin genes to strains already possessing a downstream component (Nagata et al. 2014) and this work implicates an ICE in the acquisition of linB. An initial association of IS6100 with some of the lin genes has spread such that the entire pathway (at least for the strains isolated from the Czech Republic) is now associated with IS6100 elements. The lin pathway therefore provides valuable insight into the intermediate steps of IS6100 activity, which has not been possible in previous investigations of IS6100-associated catabolic pathways. For example, in both the carbazole degradation pathway of Sphingomonas sp. XLDN2-5 and the 2-chloronitrobenzene degradation pathway of Pseudomonas stutzeri ZWLR2-1, the entire degradation pathway is organized into several large clusters flanked very closely by IS6100 elements (Gai et al. 2010; Liu et al. 2011). Although the origins of these clusters have been identified, no instances of intermediate pathway assembly have been observed. Here, we show the dynamic nature of IS6100-mediated pathway assembly, with many different insertion sites around the lin genes. This suggests that the lin pathway is still in the process of evolving and that IS6100 is playing an active role.
IS6100 activity also may be resulting in a clustering of the lin genes. Homologous recombination between directly repeated IS6100 elements can cause the deletion of the intervening genomic sequence (Mahillon and Chandler 1998). Although this is known to be responsible for deletions of lin genes (Nagata et al. 2011) it could also be responsible for bringing together upstream components of the pathway. In B90A we find a linA1-linF-linC cluster, and the linF-linC component also is found in IP26 and MM1-pISP1. In LL01 linB and linC are colocated, whereas HDIP04 and MM1-pISP4 each contain a linB-linC-linF cluster. This clustering is predicted by the Selfish Operon Theory, which posits that operon formation can be driven by selective pressure to reduce the number of horizontal transfer events required to acquire the pathway (Lawrence and Roth 1996). Although the contribution of this process to the formation of operons is disputed (Pál and Hurst 2004), the proposal that clustering of genes in the same pathway can result from horizontal transfer is supported by our observations of the lin pathway. Under this model, we expect to see additional clustering of the lin genes as strains continue to be exposed to HCH. Further sequencing of recently isolated strains will help determine if this predicted evolutionary trajectory is accurate.
Supplementary Material
Acknowledgments
This research was funded in part by CSIRO's Land and Water Flagship. SP was supported by Australian Postgraduate Award (APA).
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.114.015933/-/DC1
Communicating editor: S. I. Wright
Literature Cited
- Angiuoli S. V., Gussman A., Klimke W., Cochrane G., Field D., et al. , 2008. Toward an online repository of Standard Operating Procedures (SOPs) for (meta)genomic annotation. OMICS 12: 137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böltner D., Moreno-Morillas S., Ramos J. L., 2005. 16S rDNA phylogeny and distribution of lin genes in novel hexachlorocyclohexane-degrading Sphingomonas strains. Environ. Microbiol. 7: 1329–1338. [DOI] [PubMed] [Google Scholar]
- Breivik K., Pacyna J. M., Münch J., 1999. Use of alpha-, beta- and gamma-hexachlorocyclohexane in Europe, 1970–1996. Sci. Total Environ. 239: 151–163. [DOI] [PubMed] [Google Scholar]
- Chuzeville S., Puymège A., Madec J. Y., Haenni M., Payot S., 2012. Characterization of a new CAMP factor carried by an integrative and conjugative element in Streptococcus agalactiae and spreading in Streptococci. PLoS ONE 7: e48918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley S. D., Rokicki J., Turner P., Daligault H., Nolan M., et al. , 2012. The whole genome sequence of Sphingobium chlorophenolicum L-1: insights into the evolution of the pentachlorophenol degradation pathway. Genome Biol. Evol. 4: 184–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher A. L., Phillippy A., Carlton J., Salzberg S. L., 2002. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 30: 2478–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogra C., Raina V., Pal R., Suar M., Lal S., et al. , 2004. Organization of lin genes and IS6100 among different strains of hexachlorocyclohexane-degrading Sphingomonas paucimobilis: evidence for horizontal gene transfer. J. Bacteriol. 186: 2225–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C., Bai X., Lai Q., Xie Y., Chen X., et al. , 2014. Draft genome sequence of Sphingobium sp. strain C100, a polycyclic aromatic hydrocarbon-degrading bacterium from the deep-sea sediment of the Arctic Ocean. Genome Announc. 2: e01210–e01213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elcey C. D., Kunhi A. A. M., 2010. Substantially enhanced degradation of hexachlorocyclohexane isomers by a microbial consortium on acclimation. J. Agric. Food Chem. 58: 1046–1054. [DOI] [PubMed] [Google Scholar]
- Fani R., Brilli M., Liò P., 2005. The origin and evolution of operons: the piecewise building of the proteobacterial histidine operon. J. Mol. Evol. 60: 378–390. [DOI] [PubMed] [Google Scholar]
- Fondi M., Emiliani G., Fani R., 2009. Origin and evolution of operons and metabolic pathways. Res. Microbiol. 160: 502–512. [DOI] [PubMed] [Google Scholar]
- Gai Z., Wang X., Liu X., Tai C., Tang H., et al. , 2010. The genes coding for the conversion of carbazole to catechol are flanked by IS6100 elements in Sphingomonas sp. strain XLDN2–5. PLoS ONE 5: e10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard M., Vallaeys T., Vorhölter F. J., Minoia M., Werlen C., et al. , 2006. The clc element of Pseudomonas sp. strain B13, a genomic island with various catabolic properties. J. Bacteriol. 188: 1999–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan H. M., Chew T. H., Hudson A. O., Savka M. A., 2012. Genome sequence of Novosphingobium sp. strain Rr 2–17, a nopaline crown gall-associated bacterium isolated from Vitis vinifera L. grapevine. J. Bacteriol. 194: 5137–5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geueke B., Garg N., Ghosh S., Fleischmann T., Holliger C., et al. , 2013. Metabolomics of hexachlorocyclohexane (HCH) transformation: ratio of LinA to LinB determines metabolic fate of HCH isomers. Environ. Microbiol. 15: 1040–1049. [DOI] [PubMed] [Google Scholar]
- Guglielmini J., Quintais L., Garcillán-Barcia M. P., de la Cruz F., Rocha E. P. C., 2011. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 7: e1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy L., Kultima J. R., Andersson S. G. E., 2010. genoPlotR: comparative gene and genome visualization in R. Bioinformatics 26: 2334–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan M. S., Liu Q., Wang H., Fazekas J., Chen B., et al. , 2012. GIST: Genomic island suite of tools for predicting genomic islands in genomic sequences. Bioinformation 8: 203–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron P. R., Evans M. C., Dyson P. J., 1999. Low target site specificity of an IS6100 -based mini-transposon, Tn1792, developed for transposon mutagenesis of antibiotic producing Streptomyces. FEMS Microbiol. Lett. 171: 215–221. [DOI] [PubMed] [Google Scholar]
- Johri A. K., Dua M., Tuteja D., Saxena R., Saxena D. M., et al. , 1998. Degradation of alpha, beta, gamma and delta-hexachlorocyclohexanes by Sphingomonas paucimobilis. Biotechnol. Lett. 20: 885–887. [Google Scholar]
- Kato K., Ohtsuki K., Mitsuda H., Yomo T., Negoro S., et al. , 1994. Insertion sequence IS6100 on plasmid pOAD2, which degrades nylon oligomers. J. Bacteriol. 176: 1197–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur J., Moskalikova H., Niharika N., Sedlackova M., Hampl A., et al. , 2013. Sphingobium baderi sp. nov., isolated from a hexachlorocyclohexane dump site. Int. J. Syst. Evol. Microbiol. 63: 673–678. [DOI] [PubMed] [Google Scholar]
- Kohli P., Dua A., Sangwan N., Oldach P., Khurana J. P., et al. , 2013. Draft genome sequence of Sphingobium ummariense strain RL-3, a hexachlorocyclohexane-degrading bacterium. Genome Announc. 1: e00956–e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., et al. , 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Chaudhary P., Dwivedi M., Kumar R., Paul D., et al. , 2005. Enhanced biodegradation of beta- and delta-hexachlorocyclohexane in the presence of alpha- and gamma-isomers in contaminated soils. Environ. Sci. Technol. 39: 4005–4011. [DOI] [PubMed] [Google Scholar]
- Kumar Singh A., Sangwan N., Sharma A., Gupta V., Khurana J. P., et al. , 2013. Draft genome sequence of Sphingobium quisquiliarum strain P25T, a novel hexachlorocyclohexane (HCH)-degrading bacterium isolated from an HCH dumpsite. Genome Announc. 1: e00717–e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal D., Jindal S., Kumari H., Jit S., Nigam A., et al. , 2013. Bacterial diversity and real-time PCR based assessment of linA and linB gene distribution at hexachlorocyclohexane contaminated sites. J. Basic Microbiol. 53: 1–11. [DOI] [PubMed] [Google Scholar]
- Lal R., Dogra C., Malhotra S., Sharma P., Pal R., 2006. Diversity, distribution and divergence of lin genes in hexachlorocyclohexane-degrading sphingomonads. Trends Biotechnol. 24: 121–130. [DOI] [PubMed] [Google Scholar]
- Lal R., Pandey G., Sharma P., Kumari K., Malhotra S., et al. , 2010. Biochemistry of microbial degradation of hexachlorocyclohexane and prospects for bioremediation. Microbiol. Mol. Biol. Rev. 74: 58–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J. G., Roth J. R., 1996. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics 143: 1843–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Wang S.-J., Zhang J.-J., Dai H., Tang H., et al. , 2011. Patchwork assembly of nag-like nitroarene dioxygenase genes and the 3-chlorocatechol degradation cluster for evolution of the 2-chloronitrobenzene catabolism pathway in Pseudomonas stutzeri ZWLR2–1. Appl. Environ. Microbiol. 77: 4547–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y. R., Kang S. G., Kim S.-J., Kim M.-R., Li N., et al. , 2012. Genome sequence of benzo(a)pyrene-degrading bacterium Novosphingobium pentaromativorans US6–1. J. Bacteriol. 194: 907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macwan A. S., Srivastava N., Javed S., Kumar A., 2013. Role of a repeated hexapeptide motif GIHFAP near C-terminus in assembly, stability, and activity of “HCH dehydrochlorinase LinA”. Appl. Biochem. Biotechnol. 169: 1397–1404. [DOI] [PubMed] [Google Scholar]
- Mahillon J., Chandler M., 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62: 725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manickam N., Misra R., Mayilraj S., 2007. A novel pathway for the biodegradation of gamma-hexachlorocyclohexane by a Xanthomonas sp. strain ICH12. J. Appl. Microbiol. 102: 1468–1478. [DOI] [PubMed] [Google Scholar]
- Manonmani H. K., Chandrashekaraiah D. H., Sreedhar Reddy N., Elcey C. D., Kunhi A. A., 2000. Isolation and acclimation of a microbial consortium for improved aerobic degradation of alpha-hexachlorocyclohexane. J. Agric. Food Chem. 48: 4341–4351. [DOI] [PubMed] [Google Scholar]
- Masai E., Kamimura N., Kasai D., Oguchi A., Ankai A., et al. , 2012. Complete genome sequence of Sphingobium sp. strain SYK-6, a degrader of lignin-derived biaryls and monoaryls. J. Bacteriol. 194: 534–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencía M., Martínez-Ferri A. I., Alcalde M., De Lorenzo V., 2006. Identification of a -hexachlorocyclohexane dehydrochlorinase (LinA) variant with improved expression and solubility properties. Biocat. Biotrans 24: 223–230. [Google Scholar]
- Michael G. B., Kadlec K., Sweeney M. T., Brzuszkiewicz E., Liesegang H., et al. , 2012. ICEPmu1, an integrative conjugative element (ICE) of Pasteurella multocida: analysis of the regions that comprise 12 antimicrobial resistance genes. J. Antimicrob. Chemother. 67: 84–90. [DOI] [PubMed] [Google Scholar]
- Miyauchi K., Suh S. K., Nagata Y., Takagi M., 1998. Cloning and sequencing of a 2,5-dichlorohydroquinone reductive dehalogenase gene whose product is involved in degradation of gamma-hexachlorocyclohexane by Sphingomonas paucimobilis. J. Bacteriol. 180: 1354–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi K., Adachi Y., Nagata Y., Takagi M., 1999. Cloning and sequencing of a novel meta-cleavage dioxygenase gene whose product is involved in degradation of gamma-hexachlorocyclohexane in Sphingomonas paucimobilis. J. Bacteriol. 181: 6712–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki R., Sato Y., Ito M., Ohtsubo Y., Nagata Y., et al. , 2006. Complete nucleotide sequence of an exogenously isolated plasmid, pLB1, involved in gamma-hexachlorocyclohexane degradation. Appl. Environ. Microbiol. 72: 6923–6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee U., Kumar R., Mahato N. K., Khurana J. P., Lal R., 2013. Draft genome sequence of Sphingobium sp. strain HDIPO4, an avid degrader of hexachlorocyclohexane. Genome Announc. 1: e00749–e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadalin F., Vezzi F., Policriti A., 2012. GapFiller: a de novo assembly approach to fill the gap within paired reads. BMC Bioinformatics 13: S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata Y., Hatta T., Imai R., Kimbara K., Fukuda M., et al. , 1993a Purification and characterization of gamma-hexachlorocyclohexane (gamma-HCH) dehydrochlorinase (LinA) from Pseudomonas paucimobilis. Biosci. Biotechnol. Biochem. 57: 1582–1583. [DOI] [PubMed] [Google Scholar]
- Nagata Y., Nariya T., Ohtomo R., Fukuda M., Yano K., et al. , 1993b Cloning and sequencing of a dehalogenase gene encoding an enzyme with hydrolase activity involved in the degradation of gamma-hexachlorocyclohexane in Pseudomonas paucimobilis. J. Bacteriol. 175: 6403–6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata Y., Ohtomo R., Miyauchi K., Fukuda M., Yano K., et al. , 1994. Cloning and sequencing of a 2,5-dichloro-2,5-cyclohexadiene-1,4-diol dehydrogenase gene involved in the degradation of gamma-hexachlorocyclohexane in Pseudomonas paucimobilis. J. Bacteriol. 176: 3117–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata Y., Endo R., Ito M., Ohtsubo Y., Tsuda M., 2007. Aerobic degradation of lindane (gamma-hexachlorocyclohexane) in bacteria and its biochemical and molecular basis. Appl. Microbiol. Biotechnol. 76: 741–752. [DOI] [PubMed] [Google Scholar]
- Nagata Y., Ohtsubo Y., Endo R., Ichikawa N., Ankai A., et al. , 2010. Complete genome sequence of the representative γ-hexachlorocyclohexane-degrading bacterium Sphingobium japonicum UT26. J. Bacteriol. 192: 5852–5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata Y., Natsui S., Endo R., Ohtsubo Y., Ichikawa N., et al. , 2011. Genomic organization and genomic structural rearrangements of Sphingobium japonicum UT26, an archetypal γ-hexachlorocyclohexane-degrading bacterium. Enzyme Microb. Technol. 49: 499–508. [DOI] [PubMed] [Google Scholar]
- Nagata Y., Tabata M., Ohhata S., Tsuda M., 2014. Appearance and evolution of γ-hexachlorocyclohexane-degrading bacteria, pp. 19–41 in Biodegradative Bacteria: How Bacteria Degrade, Survive, Adapt, and Evolve, edited by Nojiri H., Tsuda M., Fukuda M., Kamagata Y. Springer, Japan. [Google Scholar]
- Niharika N., Moskalikova H., Kaur J., Khan F., Sedlackova M., et al. , 2013a Sphingobium czechense sp. nov., isolated from a hexachlorocyclohexane dump site. Int. J. Syst. Evol. Microbiol. 63: 723–728. [DOI] [PubMed] [Google Scholar]
- Niharika N., Moskalikova H., Kaur J., Sedlackova M., Hampl A., et al. , 2013b Novosphingobium barchaimii sp. nov., isolated from hexachlorocyclohexane-contaminated soil. Int. J. Syst. Evol. Microbiol. 63: 667–672. [DOI] [PubMed] [Google Scholar]
- Okai M., Kubota K., Fukuda M., Nagata Y., Nagata K., et al. , 2010. Crystal structure of γ-hexachlorocyclohexane dehydrochlorinase LinA from Sphingobium japonicum UT26. J. Mol. Biol. 403: 260–269. [DOI] [PubMed] [Google Scholar]
- Pál C., Hurst L. D., 2004. Evidence against the selfish operon theory. Trends Genet. 20: 232–234. [DOI] [PubMed] [Google Scholar]
- Ramsay J. P., Sullivan J. T., Stuart G. S., Lamont I. L., Ronson C. W., 2006. Excision and transfer of the Mesorhizobium loti R7A symbiosis island requires an integrase IntS, a novel recombination directionality factor RdfS, and a putative relaxase RlxS. Mol. Microbiol. 62: 723–734. [DOI] [PubMed] [Google Scholar]
- Richter M., Rosselló-Móra R., 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 106: 19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Blanco A., Lemos M. L., Osorio C. R., 2012. Integrating conjugative elements as vectors of antibiotic, mercury, and quaternary ammonium compound resistance in marine aquaculture environments. Antimicrob. Agents Chemother. 56: 2619–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangwan N., Lata P., Dwivedi V., Singh A., Niharika N., et al. , 2012. Comparative metagenomic analysis of soil microbial communities across three hexachlorocyclohexane contamination levels. PLoS One 7: e46219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangwan N., Verma H., Kumar R., Negi V., Lax S., et al. , 2014. Reconstructing an ancestral genotype of two hexachlorocyclohexane-degrading Sphingobium species using metagenomic sequence data. ISME J. 8: 398–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R., Edwards R., 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27: 863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo K., Wada H., 1989. Isolation and identification of an aerobic γ-HCH-decomposing bacterium from soil. Soil Sci. Plant Nutr. 35: 79–87. [Google Scholar]
- Seth-Smith H. M. B., Fookes M. C., Okoro C. K., Baker S., Harris S. R., et al. , 2012. Structure, diversity, and mobility of the Salmonella pathogenicity island 7 family of integrative and conjugative elements within Enterobacteriaceae. J. Bacteriol. 194: 1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P., Pandey R., Kumari K., Pandey G., Jackson C. J., et al. , 2011. Kinetic and sequence-structure-function analysis of known LinA variants with different hexachlorocyclohexane isomers. PLoS One 6: e25128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shettigar M., Pearce S., Pandey R., Khan F., Dorrian S. J., et al. , 2012. Cloning of a novel 6-chloronicotinic acid chlorohydrolase from the newly isolated 6-chloronicotinic acid mineralizing Bradyrhizobiaceae strain SG-6C. PLoS ONE 7: e51162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson J. T., Wong K., Jackman S. D., Schein J. E., Jones S. J. M., et al. , 2009. ABySS: a parallel assembler for short read sequence data. Genome Res. 19: 1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A. K., Chaudhary P., Macwan A. S., Diwedi U. N., Kumar A., 2007. Selective loss of lin genes from hexachlorocyclohexane-degrading Pseudomonas aeruginosa ITRC-5 under different growth conditions. Appl. Microbiol. Biotechnol. 76: 895–901. [DOI] [PubMed] [Google Scholar]
- Suar M., van der Meer J. R., Lawlor K., Holliger C., Lal R., 2004. Dynamics of multiple lin gene expression in Sphingomonas paucimobilis B90A in response to different hexachlorocyclohexane isomers. Appl. Environ. Microbiol. 70: 6650–6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suar M., Hauser A., Poiger T., Buser H. R., Müller M. D., et al. , 2005. Enantioselective transformation of alpha-hexachlorocyclohexane by the dehydrochlorinases LinA1 and LinA2 from the soil bacterium Sphingomonas paucimobilis B90A. Appl. Environ. Microbiol. 71: 8514–8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan J. T., Patrick H. N., Lowther W. L., Scott D. B., Ronson C. W., 1995. Nodulating strains of Rhizobium loti arise through chromosomal symbiotic gene transfer in the environment. Proc. Natl. Acad. Sci. USA 92: 8985–8989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan J. T., Trzebiatowski J. R., Cruickshank R. W., Gouzy J., Brown S. D., et al. , 2002. Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J. Bacteriol. 184: 3086–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata M., Endo R., Ito M., Ohtsubo Y., Kumar A., et al. , 2011. The lin genes for γ-hexachlorocyclohexane degradation in Sphingomonas sp. MM-1 proved to be dispersed across multiple plasmids. Biosci. Biotechnol. Biochem. 75: 466–472. [DOI] [PubMed] [Google Scholar]
- Tabata M., Ohtsubo Y., Ohhata S., Tsuda M., Nagata Y., 2013. Complete genome sequence of the γ-hexachlorocyclohexane-degrading bacterium Sphingomonas sp. strain MM-1. Genome Announc. 1: e00247–e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeo M., Maeda Y., Maeda J., Nishiyama N., Kitamura C., et al. , 2012. Two identical nonylphenol monooxygenase genes linked to IS6100 and some putative insertion sequence elements in Sphingomonas sp. NP5. Microbiology 158: 1796–1807. [DOI] [PubMed] [Google Scholar]
- Thomas J. C., Berger F., Jacquier M., Bernillon D., Baud-Grasset F., et al. , 1996. Isolation and characterization of a novel gamma-hexachlorocyclohexane-degrading bacterium. J. Bacteriol. 178: 6049–6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trantirek L., Hynková K., Nagata Y., Murzin A., Ansorgová A., et al. , 2001. Reaction mechanism and stereochemistry of gamma-hexachlorocyclohexane dehydrochlorinase LinA. J. Biol. Chem. 276: 7734–7740. [DOI] [PubMed] [Google Scholar]
- Vijgen J., Abhilash P. C., Li Y. F., Lal R., Forter M., et al. , 2011. Hexachlorocyclohexane (HCH) as new Stockholm Convention POPs—a global perspective on the management of Lindane and its waste isomers. Environ. Sci. Pollut. Res. Int. 18: 152–162. [DOI] [PubMed] [Google Scholar]
- Weaden J., Dyson P., 1998. Transposon mutagenesis with IS6100 in the avermectin producer Streptomyces avermitilis. Microbiol. 144: 1963–1970. [DOI] [PubMed] [Google Scholar]
- Wöhrnschimmel H., Tay P., von Waldow H., Hung H., Li Y. F., et al. , 2012. Comparative assessment of the global fate of α- and β-hexachlorocyclohexane before and after phase-out. Environ. Sci. Technol. 46: 2047–2054. [DOI] [PubMed] [Google Scholar]
- Wozniak R. A. F., Waldor M. K., 2010. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8: 552–563. [DOI] [PubMed] [Google Scholar]
- Wu J., Hong Q., Sun Y., Hong Y., Yan Q., et al. , 2007. Analysis of the role of LinA and LinB in biodegradation of delta-hexachlorocyclohexane. Environ. Microbiol. 9: 2331–2340. [DOI] [PubMed] [Google Scholar]
- Yasuhira K., Uedo Y., Takeo M., Kato D.-I., Negoro S., 2007. Genetic organization of nylon-oligomer-degrading enzymes from alkalophilic bacterium, Agromyces sp. KY5R. J. Biosci. Bioeng. 104: 521–524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.