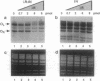
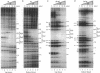
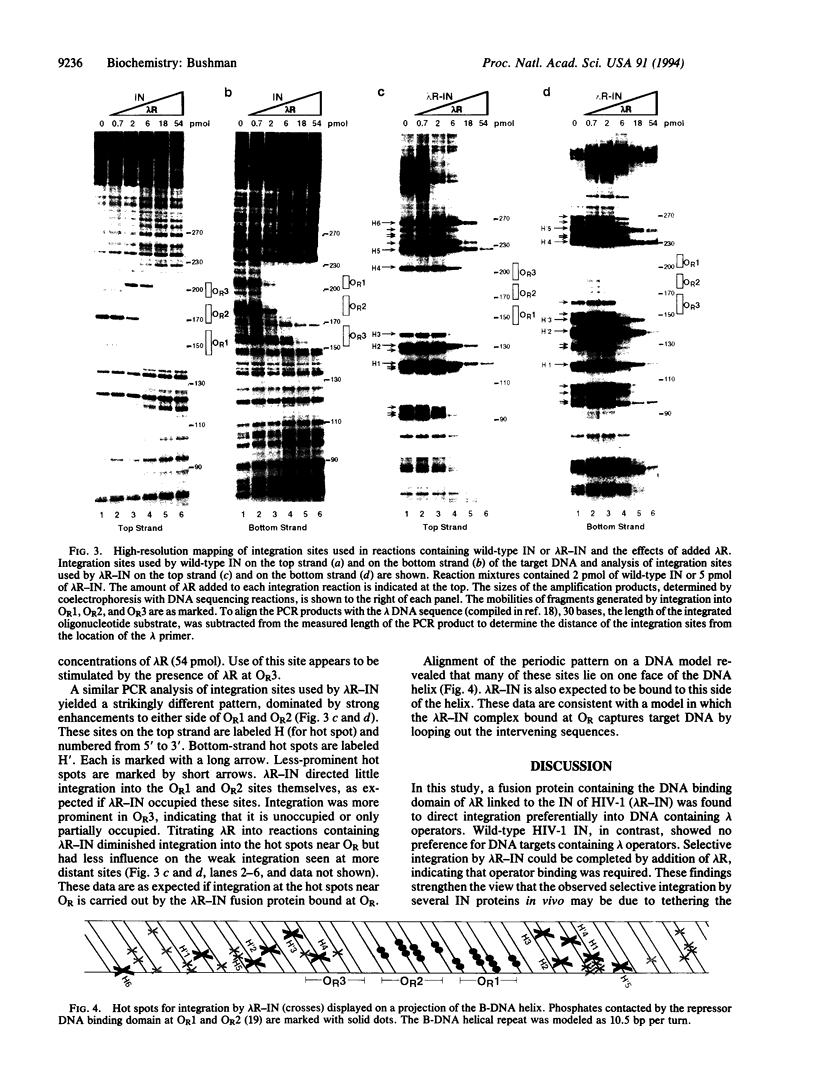
Abstract
Certain retrovirus and retrotransposons display strong biases in the selection of host DNA sites for integration. To probe the possibility that simple tethering of the retroelement integrase protein to a target DNA site is sufficient to direct integration, the activities of a hybrid composed of human immunodeficiency virus 1 integrase and lambda repressor were analyzed. In in vitro reactions containing several target DNAs, the lambda repressor-integrase hybrid was found to direct integration selectively to targets containing lambda operators. Addition of lambda repressor blocked selective integration, indicating that binding to the operators was required. The lambda repressor-integrase hybrid protein directed integration primarily to sites near the operators on the same face of the B-DNA helix, indicating that target DNA was probably captured by looping out the intervening sequences. Such hybrid integrase proteins may be useful for directing retroviral integration to specific sequences in vivo.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beamer L. J., Pabo C. O. Refined 1.8 A crystal structure of the lambda repressor-operator complex. J Mol Biol. 1992 Sep 5;227(1):177–196. doi: 10.1016/0022-2836(92)90690-l. [DOI] [PubMed] [Google Scholar]
- Bushman F. D., Craigie R. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F. D., Engelman A., Palmer I., Wingfield P., Craigie R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F. D., Fujiwara T., Craigie R. Retroviral DNA integration directed by HIV integration protein in vitro. Science. 1990 Sep 28;249(4976):1555–1558. doi: 10.1126/science.2171144. [DOI] [PubMed] [Google Scholar]
- Bushman F. D. The bacteriophage 434 right operator. Roles of O(R)1, O(R)2 and O(R)3. J Mol Biol. 1993 Mar 5;230(1):28–40. doi: 10.1006/jmbi.1993.1123. [DOI] [PubMed] [Google Scholar]
- Chalker D. L., Sandmeyer S. B. Ty3 integrates within the region of RNA polymerase III transcription initiation. Genes Dev. 1992 Jan;6(1):117–128. doi: 10.1101/gad.6.1.117. [DOI] [PubMed] [Google Scholar]
- Engelman A., Bushman F. D., Craigie R. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 1993 Aug;12(8):3269–3275. doi: 10.1002/j.1460-2075.1993.tb05996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S. P. Genetics of retroviral integration. Annu Rev Genet. 1992;26:527–544. doi: 10.1146/annurev.ge.26.120192.002523. [DOI] [PubMed] [Google Scholar]
- Hu J. C., O'Shea E. K., Kim P. S., Sauer R. T. Sequence requirements for coiled-coils: analysis with lambda repressor-GCN4 leucine zipper fusions. Science. 1990 Dec 7;250(4986):1400–1403. doi: 10.1126/science.2147779. [DOI] [PubMed] [Google Scholar]
- Ji H., Moore D. P., Blomberg M. A., Braiterman L. T., Voytas D. F., Natsoulis G., Boeke J. D. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell. 1993 Jun 4;73(5):1007–1018. doi: 10.1016/0092-8674(93)90278-x. [DOI] [PubMed] [Google Scholar]
- Kitamura Y., Lee Y. M., Coffin J. M. Nonrandom integration of retroviral DNA in vitro: effect of CpG methylation. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5532–5536. doi: 10.1073/pnas.89.12.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss D., Bushman F. D., Wolffe A. P. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc Natl Acad Sci U S A. 1994 Jun 21;91(13):5913–5917. doi: 10.1073/pnas.91.13.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryciak P. M., Varmus H. E. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell. 1992 May 29;69(5):769–780. doi: 10.1016/0092-8674(92)90289-o. [DOI] [PubMed] [Google Scholar]
- Ptashne M., Jeffrey A., Johnson A. D., Maurer R., Meyer B. J., Pabo C. O., Roberts T. M., Sauer R. T. How the lambda repressor and cro work. Cell. 1980 Jan;19(1):1–11. doi: 10.1016/0092-8674(80)90383-9. [DOI] [PubMed] [Google Scholar]
- Sherman P. A., Fyfe J. A. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc Natl Acad Sci U S A. 1990 Jul;87(13):5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih C. C., Stoye J. P., Coffin J. M. Highly preferred targets for retrovirus integration. Cell. 1988 May 20;53(4):531–537. doi: 10.1016/0092-8674(88)90569-7. [DOI] [PubMed] [Google Scholar]
- Vijaya S., Steffen D. L., Robinson H. L. Acceptor sites for retroviral integrations map near DNase I-hypersensitive sites in chromatin. J Virol. 1986 Nov;60(2):683–692. doi: 10.1128/jvi.60.2.683-692.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent D. C., Groeneger A. A., Plasterk R. H. Mutational analysis of the integrase protein of human immunodeficiency virus type 2. Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9598–9602. doi: 10.1073/pnas.89.20.9598. [DOI] [PMC free article] [PubMed] [Google Scholar]