Abstract
Biological N2 fixation is an important nitrogen source for surface ocean microbial communities. However, nearly all information on the diversity and gene expression of organisms responsible for oceanic N2 fixation in the environment has come from targeted approaches that assay only a small number of genes and organisms. Using genomes of diazotrophic cyanobacteria to extract reads from extensive meta-genomic and -transcriptomic libraries, we examined diazotroph diversity and gene expression from the Amazon River plume, an area characterized by salinity and nutrient gradients. Diazotroph genome and transcript sequences were most abundant in the transitional waters compared with lower salinity or oceanic water masses. We were able to distinguish two genetically divergent phylotypes within the Hemiaulus-associated Richelia sequences, which were the most abundant diazotroph sequences in the data set. Photosystem (PS)-II transcripts in Richelia populations were much less abundant than those in Trichodesmium, and transcripts from several Richelia PS-II genes were absent, indicating a prominent role for cyclic electron transport in Richelia. In addition, there were several abundant regulatory transcripts, including one that targets a gene involved in PS-I cyclic electron transport in Richelia. High sequence coverage of the Richelia transcripts, as well as those from Trichodesmium populations, allowed us to identify expressed regions of the genomes that had been overlooked by genome annotations. High-coverage genomic and transcription analysis enabled the characterization of distinct phylotypes within diazotrophic populations, revealed a distinction in a core process between dominant populations and provided evidence for a prominent role for noncoding RNAs in microbial communities.
Introduction
The productivity of a large fraction of the ocean's surface waters is limited by the availability of fixed inorganic nitrogen (N) (Zehr and Kudela, 2011). Some organisms, termed diazotrophs, have the ability to assimilate, or fix, N2 gas, thus avoiding N limitation. N2 fixation is an important source of ‘new' N to maintain primary production in oligotrophic oceans (Dugdale and Goering, 1967).
Diazotrophic cyanobacteria have been shown to comprise a large fraction of microbial communities in the Amazon River plume and surrounding waters (Foster et al., 2007; Goebel et al., 2010). As the high-nutrient riverine water mixes with oligotrophic oceanic waters, NO3− and NO2− are rapidly taken up by microbial communities dominated by coastal diatoms (Shipe et al., 2007; Subramaniam et al., 2008; Goes et al., 2014). Further along the mixing gradient, some nutrients (Si, P and Fe) persist in relatively high concentrations, but N is depleted, providing an advantage to the diazotrophs (Foster et al., 2007; Shipe et al., 2007; Subramaniam et al., 2008; Goes et al., 2014). The cyanobacterium Richelia, located within the cell wall of the diatom Hemiaulus, is the most abundant N2 fixer in transitional waters (30–35 PSU (practical salinity unit)), whereas the colony-forming, filamentous Trichodesmium is the dominant diazotroph in more oceanic waters (>35 PSU) (Carpenter et al., 1999; Subramaniam et al., 2008). The free-living unicellular cyanobacterium Crocosphaera, the picoeukaryotic alga-associated UCYN-A and Richelia associated with the diatom Rhizosolenia have also been detected in and around the Amazon River plume (Foster et al., 2007; Goebel et al., 2010).
The abundance of diazotrophic cyanobacteria strongly influences surface communities and nutrient cycling in this area. A bloom of Richelia-harboring Hemiaulus in transitional waters, accompanied by Trichodesmium, accounted for an estimated input of nearly 0.5 Tg N to the surface community over just a 10-day period (Carpenter et al., 1999). Another study found that the particulate export at transitional stations was dominated by Richelia–Hemiaulus associations that were estimated to be responsible for the sequestration of 20 Tg Carbon (C) to the deep ocean annually (Subramaniam et al., 2008). These studies show the significance of the Amazon River plume diazotroph community, as a whole, but provide little information about the organisms that comprise the populations within that community.
Prior studies of oceanic diazotroph diversity, abundance and activity have mostly been based on microscopic observations or molecular biology methods targeting a specific gene (for example, nifH, hetR). In contrast, metatranscriptomics avoids potential bias stemming from targeting predetermined organisms or processes while providing a full transcription snapshot of microorganisms comprising the entire microbial community. Studying metatranscriptomes of marine microbial communities, in general, have revealed the abundance of novel transcripts and small RNAs (Gilbert et al., 2008; Shi et al., 2009), the intricacies of diatom population response to iron limitation (Marchetti et al., 2012) and the synchronicity of diel transcription among bacterial and archaeal populations (Ottesen et al., 2013, 2014). In addition, sequences implicating a novel bacterial group and a euryarchaeal population in deep-sea N and C cycling were found to be abundant in a Gulf of California metatranscriptome (Baker et al., 2013).
Although more community-based research is enabled through the use of metatranscriptomes, only a few studies have utilized this tool to elucidate the physiological state of cells within diazotrophic populations. Important information such as the expression of key nutrient limitation response genes, as well as highly expressed genes of unknown function, were obtained from metatranscriptomic analyses of Crocosphaera (Hewson et al., 2009a) and Trichodesmium populations (Hewson et al., 2009b). In the current study, we coupled metatranscriptomic and metagenomic approaches to analyze the N2-fixing community that drives new production in the Amazon River plume.
Materials and methods
Sample collection
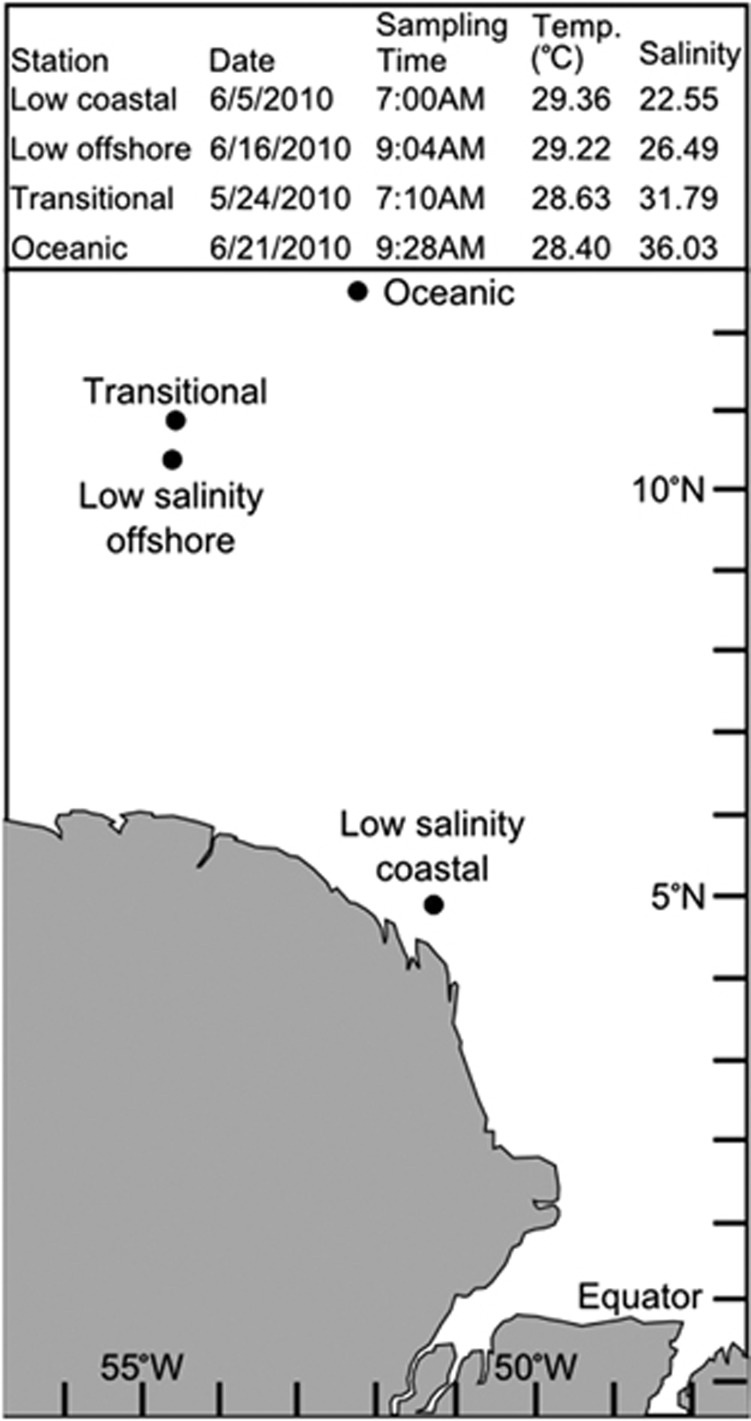
Samples were collected in May–June 2010 as part of the Amazon Influence on the Atlantic: Carbon Export from Nitrogen Fixation by Diatom Symbioses (ANACONDAS) project. Surface waters were sampled aboard the R/V Knorr from four stations (Figure 1). Samples (20 l) were taken in duplicate for each of the sample types described below (DNA, RNA and poly(A)-RNA) and prefiltered (156 μm) to remove grazers before filtration through a 2.0 μm pore-size, 142 mm diameter polycarbonate membrane filter (Sterlitech Corporation, Kent, WA, USA). For all samples but the poly(A)-RNA, the 2.0 μm filter was in-line with a 0.22 μm pore-size, 142 mm diameter Supor membrane filter (Pall, Port Washington, NY, USA). Immediately after filtration, and within 30 min of water collection, filters were stored in RNAlater (Applied Biosystems, Austin, TX, USA). They were incubated overnight at room temperature and stored at −80 °C.
Figure 1.
Amazon River plume stations.
Sample preparation for DNA sequencing
DNA extraction and purification was conducted as previously described (Zhou et al., 1996; Crump et al., 1999, 2003) with some modification. Briefly, once each filter thawed, it was removed from RNAlater. In order to clean any residual RNAlater, the filter was rinsed three times in autoclaved, filter-sterilized, 0.1% phosphate-buffered saline. In order to prevent the loss of any material that washed off of the filter, the liquid from the rinses was pooled with the RNAlater used for storage and pushed through a 0.2 μm Sterivex-GP filter capsule (Millipore, Darmstadt, Germany). The filter capsule was then triple-rinsed with phosphate-buffered saline using a sterile syringe. Once the filters and the filtered suspension material were thoroughly rinsed, they were either broken or sliced into smaller pieces (see below) and recombined in DNA extraction buffer (DEB: 0.1 M Tris-HCl (pH 8), 0.1 M Na-EDTA (pH 8), 0.1 M Na2H2PO4 (pH 8), 1.5 M NaCl and 5% CTAB). The 142 mm, 0.22 μm Supor filters were placed in Whirl-Pak bags (Nasco, Fort Atkinson, WI, USA), flash-frozen in liquid nitrogen and broken into small pieces using a rubber mallet. The 2.0 μm pore-size, 142 mm diameter polycarbonate membrane filters were sliced on a sterile cutting board with the filter folded in to prevent the cells from sliding off the surface of the filter. For Sterivex filters, the filter was removed from the casing by cracking the housing with pliers, sliced on a sterile cutting board and added to the DNA extraction buffer with the original membrane filter. An internal genomic DNA standard (Thermus thermophilus HB8 genomic DNA) was also added as a means to normalize sequencing coverage across samples (Satinsky et al., 2014). The standard genomic DNA was spiked into each individual sample in a known abundance (8.4 ng l−1 filtered) before the initiation of cell lysis. The samples were then extracted as previously described (Crump et al., 2003) with adjustments for the larger volumes associated with 142 mm filters.
Sample preparation for total community RNA
RNA extraction and DNA removal were carried out as previously described (Poretsky et al., 2009a, 2009b; Gifford et al., 2010). In brief, after the filters were broken, as described above for DNA sample filters, they were transferred to a lysis solution consisting of 8 ml of RLT Lysis Solution (Qiagen, Valencia, CA, USA) and 3 g of RNA PowerSoil beads (Mo-Bio, Carlsbad, CA, USA). Two synthesized mRNA standards, which were 916 and 970 nt in length, were synthesized from the commercial vectors pTXB1 vector (New England Biolabs, Ipswich, MA, USA) and pFN18A Halotag T7 Flexi Vector (Promega, Madison, WI, USA) respectively, and were added individually to the prepared lysis tubes in known copy numbers (pTXB1=2.104 × 1010 copies; pFN18A=1.172 × 1010 copies) before initiation of cell lysis (Satinsky et al., 2014). Tubes containing the filter pieces and lysis solution were vortexed for 10 min, and RNA was purified from cell lysate using the RNeasy Kit (Qiagen). To remove residual DNA, the Turbo DNA-free kit (Invitrogen, Carlsbad, CA, USA) was used and two aliquots of Turbo DNase were added at different times to the samples in order to improve DNA removal. Ribosomal RNA (rRNA) was removed using community-specific probes prepared with DNA from a simultaneously collected sample (Stewart et al., 2010). Biotinylated rRNA probes were synthesized for bacterial and archaeal 16S and 23S rRNA and eukaryotic 18S and 28S rRNA, and probe-bound rRNA was removed via hybridization to streptavidin-coated magnetic beads (New England Biolabs). Successful removal of rRNA from the samples was confirmed using either an Experion automated electrophoresis system (Bio-Rad Laboratories, Hercules, CA, USA) or a Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Samples were then linearly amplified using the MessageAmp II-Bacteria Kit (Applied Biosystems). Low sequencing yield has previously been attributed to this kit (Shi et al., 2010b), but multiple studies have reported high reproducibility (Francois et al., 2007; Frias-Lopez et al., 2008). Random primers were used with the Superscript III first-strand synthesis system (Invitrogen) to copy the amplified mRNA to complementary DNA (cDNA), followed by the NEBnext mRNA second-strand synthesis module (New England Biolabs). The QIAquick PCR purification kit (Qiagen) was used to purify the double-stranded cDNA, followed by ethanol precipitation. The nucleic acids were resuspended in 100 μl of TE buffer and stored at −80 °C.
Sample preparation for poly(A) tail-selected RNA
An additional metatranscriptome protocol that selectively sequenced RNA sequences with poly(A) tails was conducted on the 2.0 μm pore-size filter samples only. The samples were prepared as described above for the total community RNA samples with the following exceptions. The lysis solution for poly(A) tail-selected RNA contained 9 ml of RLT Lysis Solution, 250 μl of zirconium beads (OPS Diagnostics, Lebanon, NJ, USA) and an internal poly(A)-tailed mRNA standard (2.0 × 109 copies per tube) (Satinsky et al., 2014). The poly(A) standard was created from an HAP-1 Protolomerase viral gene. An amplicon (544 bp) with a poly(A) tail and a T7 promoter was synthesized through PCR from the template DNA. The amplicon was then used as template for an in vitro transcription reaction to produce the standard sequence with a poly(A) tail. The Oligotex mRNA kit (Qiagen) was used to isolate poly(A)-tailed mRNA from total RNA. The poly(A)-tailed mRNA was then linearly amplified with the MessageAmp II-aRNA Amplification Kit (Applied Biosystems). Double-stranded cDNA was prepared as described above for total community RNA with the exception that no ethanol precipitation was done.
Sequencing and post-sequencing screening
Nucleic acids from all samples were ultrasonically sheared to fragments (∼200–250 bp) and TruSeq libraries (Illumina Inc., San Diego, CA, USA) were constructed for paired-end sequencing (2 × 150 bp) using the Illumina Genome Analyzer IIx sequencing platform (Illumina). SHE-RA (Rodrigue et al., 2010) was used to join paired-end reads with a quality metric score of 0.5, and paired reads were then trimmed using SeqTrim (Falgueras et al., 2010). A BLAST analysis of metatranscriptome reads was conducted against a database containing representative rRNA sequences along with the internal standard sequences (blastn, bit score >50) (Gifford et al., 2010). The cDNA reads with BLAST hits were removed from the data set (Supplementary Table S1). To remove internal standard sequences from the metagenome reads, DNA reads with a BLAST hit against the T. thermophilus HB8 genome (blastn, bit score >50) were queried against the RefSeq protein database. Reads with a BLAST hit matching a T. thermophilus protein (blastx, bit score >40) were designated as internal standard and removed.
More than 39 million DNA sequence reads were obtained, with more than 27 million reads remaining after sequence trimming and removal of standards (Supplementary Table S1). A total of 162 million cDNA reads were sequenced from the four stations, and over 53 million reads remained after trimming and removal of standards, rRNA and transfer RNA reads (Supplementary Table S1). The DNA sequence reads, as well as the cDNA reads from the 0.2 μm size fraction, from the low-salinity offshore station were unavailable at the time of the writing of this report, and thus are not included in this study. DNA reads were an average of 190 bp long, whereas cDNA averaged 173 bp each. An earlier version of these data than those deposited at National Center for Biotechnology Information (NCBI; PRJNA237344) was used for this study.
Identification and analysis of diazotroph reads
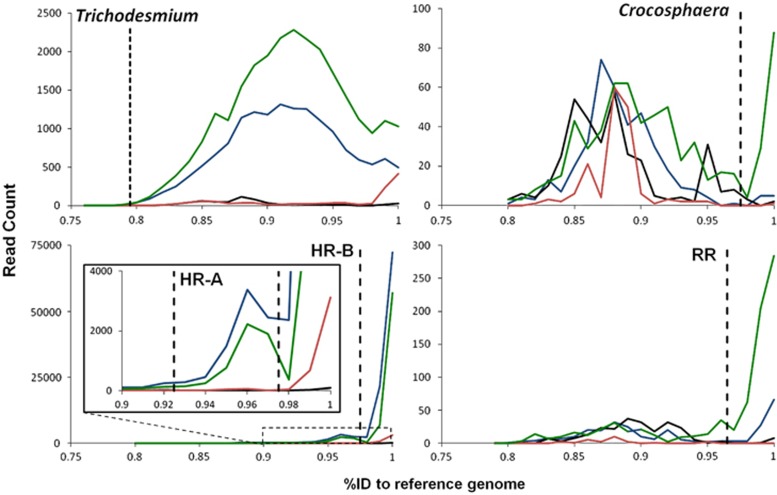
A BLAST analysis of the DNA and cDNA reads against the genomes of six oceanic N2-fixing cyanobacteria (Table 1) was conducted (blastn, bit score >50). The whole-genome sequences were used in order to analyze the organisms in the context of all cellular processes rather than target specific pathways (for example, N2 fixation). In addition, given that diversity varies depending on the open-reading frame (ORF) or intergenic spacer region (IGS), the inclusion of the whole genomes prevented a strong bias from any predetermined gene groups. Replicate reads, defined as those that matched another read from the same sample across the first 100 bp, were removed. A BLAST analysis of nonduplicate potential diazotrophic reads was then conducted against the nr/nt database (NCBI, blastn, e-value ≤10, hit length ≥50 bp). The percent identities of each read with a top BLAST hit to one of the diazotrophic cyanobacterial genomes was plotted in order to determine a cutoff percent identity value for each organism (Figure 2). DNA reads with hits above these cutoff values for each organism at each station were summed and normalized to the internal standard recovery percentage for that sample and the genome length of the organism, resulting in genome copies l−1 kbp−1. A BLAST analysis of the cDNA reads above the percent identity cutoff for a given organism was conducted against a database of ORFs and IGSs of that organism (blastn) in order to assign each read to a functional region. An ORF or IGS was considered to be detected in the data set if at least one read was assigned to it. For each detected ORF, the number of reads assigned was normalized for the gene length and the sample internal standard, as described above, to arrive at transcript copies l−1 kbp−1. When transcript abundances are discussed throughout this study, they are presented in these units because the normalization provides absolute estimates and, thus, tracks the relative number of reads that cover a given transcript just as sequence coverage depth, but can more appropriately be used to compare whole transcriptome expression of individual populations across several stations. For IGSs with <10 reads assigned, the entire IGS length was used for normalization. For those IGSs with at least 10 reads assigned, reads were mapped to the IGS in order to get a more accurate transcript length. The mapping was done using the GS Reference Mapper (Roche, Nutley, NJ, USA) with default settings. Mapping of cDNA reads to the gene sequence was done in the same manner for abundant diazotroph transcripts.
Table 1. Oceanic cyanobacterial diazotroph genomes.
| Diazotrophic Cyanobacterium | Genome Size (Mb) | Morphology | Lifestyle |
|---|---|---|---|
| Richelia intracellularis HH01 | 3.2 | Filamentous, heterocyst-forming | Hemiaulus-associated |
| Richelia intracellularis RC01 | 5.5 | Filamentous, heterocyst-forming | Rhizosolenia-associated |
| Trichodesmium erythraeum IMS101 | 7.8 | Filamentous | Free living |
| Crocosphaera watsonii WH 8501 | 6.2 | Unicellular | Free living |
| *Calothrix rhizosoleniae SC01 | 6.0 | Filamentous, heterocyst-forming | Chaetoceros-associated |
| *UCYN-A | 1.4 | Unicellular | Prymnesiophyte-associated |
The four diazotrophic cyanobacterial genomes used as references for the Amazon River plume populations, and two additional genomes (*) that were not found in these data.
Figure 2.
Natural population similarity to genomes. Histograms of the percent identity of reads with a top hit to each of four diazotrophic cyanobacteria genomes from the transitional (blue), oceanic (green), low-salinity offshore (red) and low-salinity coastal (black) stations. The dotted lines mark the cutoff used in this study for each population. HR, Hemiaulus-associated Richelia (split in ‘A' and ‘B' populations as discussed in the text); RR, Rhizosolenia-associated Richelia.
A BLAST analysis of the nonduplicate reads that were not assigned to one of the six genomes was conducted against the nr database (NCBI, blastx, e-value ≤10, hit length ≥17 amino acids). The reads with a top BLAST hit in the nr database to a nifH gene sequence were pulled to assess the non-cyanobacterial diazotrophic populations in the data set.
KEGG (Kyoto Encyclopedia of Genes and Genomes) orthology K numbers were assigned to Richelia intracellularis HH01 ORFs by submitting the protein sequences to the KEGG Automatic Annotation Server (KAAS) (Moriya et al., 2007) using the best bidirectional hit method. The Trichodesmium K numbers were obtained through the DOE Joint Genome Institute Integrated Microbial Genomes (img) annotation table for Trichodesmium erythraeum IMS101. The transcript abundance for each KEGG pathway was then calculated by summing the normalized transcript abundances of all the ORFs assigned to the given pathway in that organism.
Results
The four stations sampled were classified by the sea surface salinity at each, and referred to as oceanic (36.03), transitional (31.79) and low salinity (26.49 offshore and 22.55 coastal; Figure 1). The sea surface temperatures ranged between 28.4 °C (oceanic) and 29.36 °C (coastal) and all samples were taken in the morning between 0700 and 0930 h within a 1-month span (Figure 1).
Environmental sequence similarity to references
Most of the reads that had a top BLAST hit to one of the diazotroph genomes aligned best with either the R. intracellularis HH01 genome (71.8%) or the T. erythraeum IMS101 genome (19.2%). The reads that had a top BLAST hit to the R. intracellularis HH01 genome (163 293 DNA, 16 211 cDNA) were split into two populations, with 91.5% of those reads at least 98% identical (nucleotides) to the genome sequence and referred to as the Hemiaulus–Richelia (HR)-B population (Figure 2). An additional 7.6% of the R. intracellularis HH01 reads fell within the range of a secondary peak between 93% and 97% identity, which we termed the HR-A population (Figure 2). The diazotroph sequence reads that had a top BLAST hit to the T. erythraeum IMS101 genome (33 038 DNA, 10 851 cDNA) exhibited a peak at 92% identity. All but 26 reads were above the determined cutoff of 80% identity to the genome sequence (Figure 2). Fewer reads had a top BLAST hit to the Crocosphaera watsonii WH8501 genome (998 DNA, 532 cDNA) or the Rhizosolenia-associated R. intracellularis RC01 genome (907 DNA, 440 cDNA), but both sets of reads had a peak at 100% identity to genome sequences (Figure 2). The Crocosphaera population consisted of reads that were at least 98% identical to the C. watsonii WH8501 genome. Reads at least 97% identical to the R. intracellularis RC01 genome were analyzed for the Rhizosolenia–Richelia (RR) population. A fraction of reads had a top BLAST hit to the unicellular haptophyte-associated UCYN-A cyanobacteria genome (664 DNA, 488 cDNA) and the heterocyst-forming external diatom symbiont Calothrix rhizosoleniae SC01 genome (591 DNA, 215 cDNA), but neither had more than 50 reads at least 95% identical to the genome sequence (data not shown). These reads were not analyzed further.
Diazotroph metagenomes
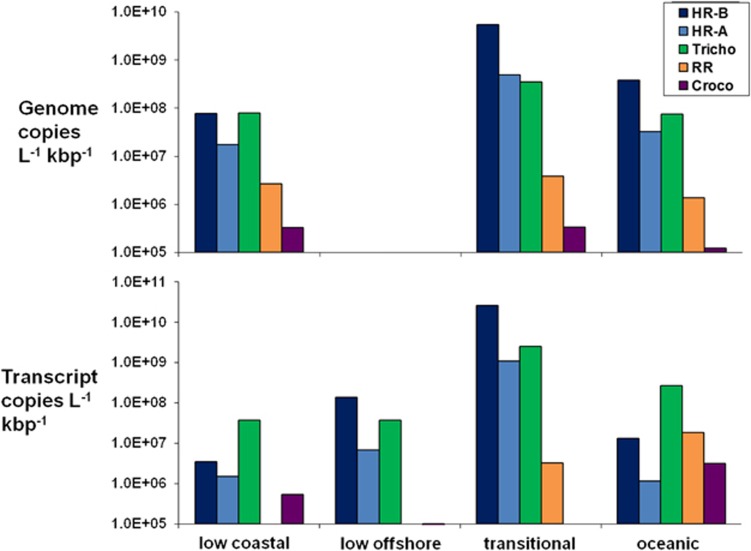
The oceanic metagenome consisted of 0.95% diazotroph reads (89 683 reads), and 1.17% of the transitional metagenome comprised diazotrophic reads (105 153 reads). The low-salinity coastal metagenome was 0.01% diazotrophic reads (514 reads). Total normalized diazotrophic cyanobacterium DNA from three stations was 7.1 × 109 genome copies l−1 kbp−1, with the majority at the transitional station (6.4 × 109 genome copies l−1 kbp−1) (Figure 3). Overall, the sequences from the HR-B population (98–100% identity to the genome) were the most abundant (6.0 × 109 genome copies l−1 kbp−1), and an order of magnitude greater than the sequences from the HR-A population (94–97% identity, 5.4 × 108 genome copies l−1 kbp−1) and the Trichodesmium population (5.1 × 108 genome copies l−1 kbp−1). RR population sequences were present at a lower abundance (7.9 × 106 genome copies l−1 kbp−1), and Crocosphaera population sequences were the least abundant in the diazotrophic cyanobacterium data set (7.9 × 105 genome copies l−1 kbp−1).
Figure 3.
Diazotrophic cyanobacterial DNA and cDNA abundance. Normalized DNA (above) and cDNA (below) data for the five diazotrophic cyanobacterial populations at each of the four stations, with the exception of the low offshore station (DNA not sampled).
Diazotroph transcriptomes
Diazotroph reads (14 557 reads) were 0.10% of the transitional metatranscriptome, whereas 0.05% of each of the low-salinity offshore and oceanic metatranscriptomes were diazotroph reads (5132 reads and 6230 reads, respectively). Less than 0.01% of the reads in the low-salinity coastal metatranscriptome were diazotrophic (281 reads). The total normalized diazotrophic cDNA from four stations was 3.01 × 1010 gene copies l−1 kbp−1, and nearly all of that was from the transitional station (2.96 × 1010 gene copies l−1 kbp−1). Similar to the normalized DNA abundance, normalized HR-B population cDNA from the four stations (2.6 × 1010 gene copies l−1 kbp−1) was one order of magnitude greater than that of the HR-A population (1.1 × 109 gene copies l−1 kbp−1) or Trichodesmium (2.9 × 109 gene copies l−1 kbp−1). RR population cDNA (2.2 × 107 gene copies l−1 kbp−1) and Crocosphaera cDNA (3.7 × 106 gene copies l−1 kbp−1) were present at lower abundances.
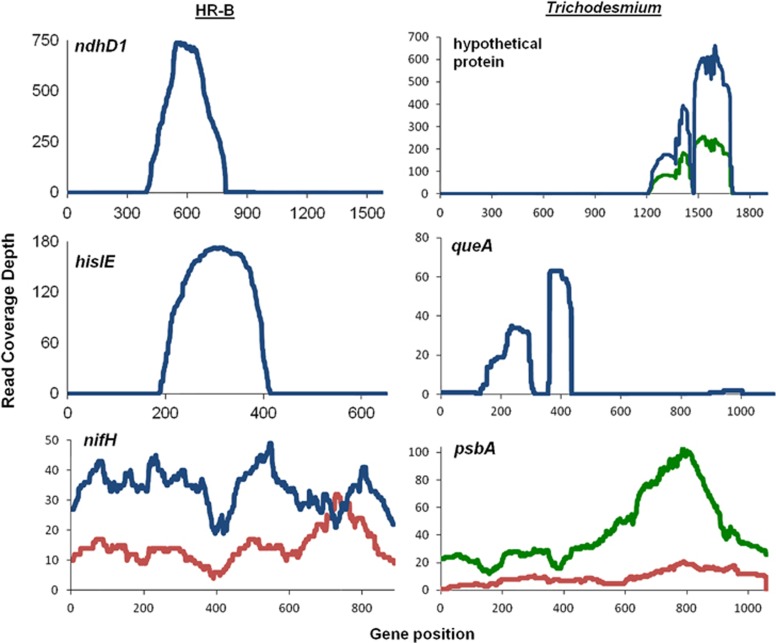
The R. intracellularis HH01 genome contains 2278 genes and 1590 of them (69.8%) were detected in the HR-B population transcriptomes (15 311 reads) (Supplementary Figure S1). In contrast, 2233 of the R. intracellularis HH01 genes (98.0%) were detected in the metagenomes (148 968 reads). Most of the genes not found in the transcriptomes were hypothetical proteins (401 out of 688). There were also 689 IGSs with at least one cDNA read, including several that were among the most abundant transcripts. The two most abundant ORFs at the transitional station were ndhD1 (RintHH_21740), which encodes the D1 subunit of NADH dehydrogenase I, and hisIE (RintHH_14390), which encodes a fused phosphoribosyl-AMP cyclohydrolase/phosphoribosyl-ATP pyrophosphatase gene. A total of 1081 reads from the transitional station were assigned to ndhD1 and they mapped mostly to a 397-bp region in the middle of the 1572 bp gene sequence (Figure 4). Similarly, all 171 hisIE reads from the transitional station covered only 232 bp of the 651 bp gene (Figure 4). HR-B population nifH cDNA reads from the transitional and low-salinity offshore stations displayed even distribution along the gene, relative to ndhD1 and hisIE (Figure 4).
Figure 4.
Transcript coverage of abundant genes. cDNA reads from the transitional (blue), oceanic (green) and low-salinity offshore (red) stations mapped to abundant genes from the HR-B (left column) and Trichodesmium (right column) metatranscriptomes. ndhD1 (RintHH_21740, NADH dehydrogenase I subunit D1), hisIE (RintHH_14390, fused phosphoribosyl-AMP cyclohydrolase/phosphoribosyl-ATP pyrophosphatase), nifH (RintHH_3070, nitrogenase iron protein), hypothetical protein (Tery_2611, FHA domain containing protein), queA (Tery_0731, S-adenosylmethionine–transfer RNA-ribosyltransferase isomerase) and psbA (Tery_4763, photosystem II protein D1).
Only 265 R. intracellularis HH01 genes and 85 IGSs were found in the HR-A population transcriptomes (659 reads). Just 1177 of the 2278 R. intracellularis HH01 genes (51.7%) were detected in the metagenomes (1177 reads). Of the HR-A transcripts detected, 85 genes and 39 IGSs did not appear among the HR-B population transcript sequences. The most abundant transcript was at the transitional station and coded a cyanobacteria-specific hypothetical protein (RintHH_13740).
The RR population transcriptomes consisted of 253 reads, which were assigned to 129 ORFs and 46 IGSs. Just six RR cDNA reads were found in the transitional station sequences, whereas the rest were from the oceanic metatranscriptome. The most abundant transcripts found in the RR population at the oceanic station were a hypothetical protein (RintRC_2139) and the photosystem (PS)-II psbA gene (RintRC_7737).
The Trichodesmium transcriptomes (9892 reads) contained transcripts for 1634 genes out of 5076 in the T. erythraeum IMS101 genome (32.2%) and 247 IGSs (Supplementary Figure S1). The Trichodesmium metagenomes (33 017 reads) contained 3772 of the genes in the genome (74.3%). The most abundant Trichodesmium transcript at each of the transitional and oceanic stations was a hypothetical protein (Tery_2611). Reads from each of those stations only mapped to a small region of the gene (Figure 4). Reads assigned to a gene that encodes an S-adenosylmethionine–transfer RNA-ribosyltransferase isomerase (queA, Tery_0731) were found mostly at the transitional station, and also mapped to just a small portion of the gene (Figure 4). Genes involved in gas vesicles (Tery_2324, Tery_2325), PS-II (Tery_4763) and other hypothetical proteins (Tery_0654, Tery_0835) were among the most abundant Trichodesmium transcripts at each station. Oceanic and low-salinity offshore station reads from a PS-II gene (Tery_4763) transcript were evenly distributed along the gene (Figure 4).
The transcriptomes of the unicellular Crocosphaera comprised 85 reads, 80 of which are from the oceanic station. Hypothetical proteins (CwatDRAFT_4329, CwatDRAFT_2191) and genes involved in photosynthesis (CwatDRAFT_0162, CwatDRAFT_1423) were the most abundant Crocosphaera transcripts at the oceanic station.
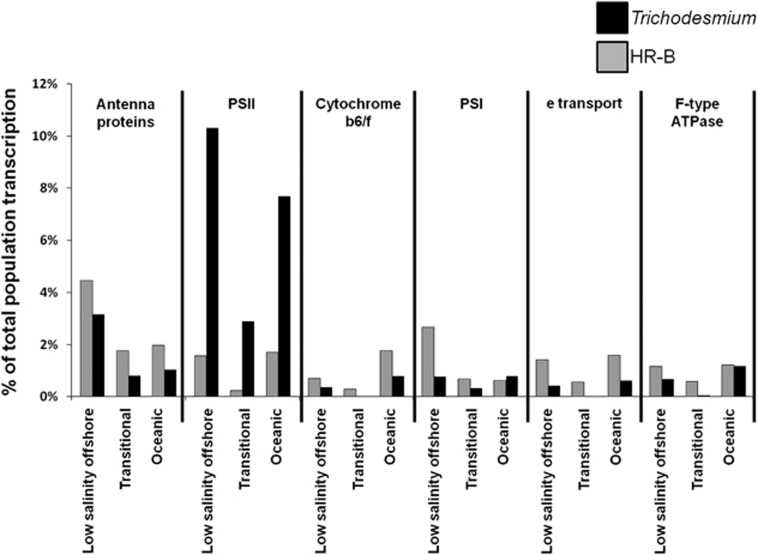
Given the low coverage of the transcripts from the HR-A, RR and Crocosphaera populations, the transcription profiles of only the HR-B and Trichodesmium populations were compared more closely. On account of the lack of diazotrophic abundance in the low-salinity coastal data sets, the populations were compared only among the other three stations. KEGG pathways were identified for the ORFs of 10 560 HR-B reads and 5001 Trichodesmium reads within the three metatranscriptomes. Photosynthesis was the most abundant KEGG pathway in the HR-B and Trichodesmium metatranscriptomes at each station. Within the photosynthesis pathway, antenna proteins were 1.8–4.5% of HR-B transcription, and PS-I proteins were 2.7% at the low-salinity offshore station (Figure 5). PS-II genes were the most abundant photosynthesis group in Trichodesmium transcription at each station (2.9–10.3%), and antenna proteins were also abundant (0.8–3.2%) (Figure 5). All other gene groups for each population were no more than 2.0% of population transcription at any station (Figure 5).
Figure 5.
Photosynthesis component transcription. The normalized abundance of transcripts within six KEGG-defined photosynthesis components relative to the total normalized transcript abundance for a population at a given station.
nifH sequences
Three cDNA reads at the low-salinity offshore station had top BLAST hits to gammaproteobacteria nifH genes, compared with 99 nifH transcript reads at that station that were assigned to a diazotrophic cyanobacteria genome. An additional three cDNA reads were found at the oceanic station with top hits to gammaproteobacteria nifH genes, whereas cyanobacteria nifH transcripts accounted for 43 reads at that station. None of the 214 nifH transcript reads at the transitional station, and no DNA reads, were attributed to heterotrophic nifH genes.
Discussion
At the time of sampling, the Amazon River plume had its maximum discharge rate for 2010 (Yeung et al., 2012). The plume flowed northwest and was defined by reduced sea surface salinity and elevated chlorophyll-a relative to surrounding water (Yeung et al., 2012; Goes et al., 2014). The riverine discharge had low concentrations of NO3− and NO2−, but SiO32− and PO43− within the plume were higher than surrounding waters (Goes et al., 2014). In addition, there was a coupling between the diatom-associated diazotrophs, drawdown of C and Si, and export efficiency (Yeung et al., 2012).
Cyanobacteria comprised the majority of the diazotrophic community in the sequence data set, and the distributions of the individual diazotroph populations in our study largely agree with previous observations from this region. However, it is possible that the 156 μm prefiltration may have removed some long-chain diatoms harboring diazotrophs and large Trichodesmium colonies from the sequenced samples, altering the representation of these populations in our data. The riverine fixed N concentration is high enough in low-salinity waters to negate the advantage of N2 fixation (Subramaniam et al., 2008), and thus fewer diazotrophs are found in these waters. Furthest from the Amazon River influence, Trichodesmium is the dominant diazotroph in the more oceanic environment, as has been observed previously (Foster et al., 2007; Turk-Kubo et al., 2012). In transitional waters between the river input and open ocean, enough fixed N has been assimilated by the community, but riverine P, Fe and Si are still in sufficiently high concentrations to create ideal conditions for diazotrophs, especially those in association with diatoms (Yeung et al., 2012; Goes et al., 2014).
The two most prominent diatom symbionts in our data were each associated with diatoms of the genus Hemiaulus. These two distinct symbiont populations were separated by a slight difference in sequence similarity, and likely represent symbionts of different Hemiaulus species. The use of the Hemiaulus hauckii symbiont as the reference genome, and the high similarity between it and the Hemiaulus membranaceus symbiont genome (Hilton et al., 2013), place the symbionts of these two diatoms within the high percent identity range of the Amazon River plume HR-B population. The less similar HR-A population was likely made up of the symbionts of Hemiaulus indicus and/or Hemiaulus sinensis, each of which have also been observed harboring heterocyst-forming symbionts (Sundström, 1984; Villareal, 1991). Previous phylogenetic analysis has reported two distinct clades within the Hemiaulus symbionts, het2A and het2B, that exhibit a similar genetic distance as HR-A and HR-B (Janson et al., 1999b; Foster and Zehr, 2006). All of the HR-B reads that aligned with the hetR region used in these previous studies (49 DNA, 6 cDNA reads) exhibited more similarity to het2B sequences than het2A sequences. However, no HR-A population DNA or cDNA reads mapped to the hetR region amplified in these studies, and hence we were not able to confirm that this population is within the het2A clade.
The high coverage of the HR-B and Trichodesmium metagenomes across their respective genomes shows that these populations were well represented in the sampled data. The relatively lower similarity between the Trichodesmium populations and the representative genome is similar to previous studies that investigated the diversity of Trichodesmium hetR gene fragments (Janson et al., 1999a; Lundgren et al., 2005; Hynes et al., 2012). In addition, if the gene content of the Trichodesmium populations varies from the T. erythraeum IMS101 reference genome just as the percent identity does, some of the Trichodesmium genes may be absent from the metagenome because they are not present in the genomes of the natural populations. Thus, the Trichodesmium population coverage may actually be higher than the metagenomic coverage indicates. The diversity of the Trichodesmium populations relative to other reference sequences is explored in Supplementary Materials. The metatranscriptomics analysis was focused on the two populations that were well represented in the data sets. It should be noted that although the presence of Crocosphaera was anticipated, the unicellular cyanobacterium fixes N2 at night (Mohr et al., 2010; Shi et al. 2010a), and thus N2 fixation gene transcripts from this population were not expected to be found in the morning samples.
The HR-B and Trichodesmium populations exhibited very different abundances of PS-II gene transcripts relative to the total normalized transcription abundance for the given population in three different environments, making it more likely that this is a trend with biological implications rather than a chance sampling occurrence. Two Trichodesmium psbA copies, coding the PS-II D1 subunit, were among the 11 most abundant transcripts in the Trichodesmium low-salinity offshore and oceanic transcriptomes. In addition, one of the psbA copies was the fourteenth most abundant gene in the Trichodesmium transitional transcriptome. High expression of PS-II genes, relative to other photosynthesis genes, has been commonly observed (Levitan et al., 2010; Mohr et al., 2010) because of a high rate of PS-II protein turnover as a result of photodamage (Aro et al., 1993). Only one psbA gene copy is present in the R. intracellularis HH01 genome assembly, but it is alone on a contig. This is indicative that it could not be assembled among other sequences because it has multiple gene copies in the genome. The transcripts of psbA were among the 15 most abundant transcripts in the HR-B low-salinity offshore and oceanic transcriptomes and detected in the transitional transcriptome, although at low abundance. However, PS-II genes psbH and psbK were not detected in any HR-B transcriptome, despite psbH transcripts among the 18 most abundant Trichodesmium transcripts in each of the low-salinity offshore and transitional transcriptomes. In addition, psbH and psbK were each detected in the Trichodesmium oceanic transcriptome. In the diazotrophic cyanobacterium Synechocystis, neither psbH nor psbK were essential to photoautotrophic growth, but the loss of either resulted in reduced growth rates (Ikeuchi et al., 1991; Mayes et al., 1993). The PS-II transcript differences may reflect the morphological difference between Richelia and Trichodesmium, or indicate the Hemiaulus symbiont has reduced growth rates, as seen with heterocyst-forming cyanobacteria in other associations (Peters and Meeks, 1989; Adams et al., 2006). It is also possible that Richelia is better protected from photodamage within the diatom, resulting in a lower PS-II protein turnover rate, and thus reduced PS-II gene expression relative to free-living oceanic cyanobacteria. However, psbH and psbK were each detected in one HR-A transcriptome, indicating that photosynthetic activity may differ between the two closely related Hemiaulus symbiont populations.
The transcripts within HR-B photosynthesis gene groups other than PS-II, however, were comparable, and often greater than that of Trichodesmium, relative to the total normalized transcription abundance for the given population. Thus, the HR-B populations may have been investing more energy toward cyclic electron transport around PS-I, rather than linear electron transport that requires PS-II activity. Cyclic electron transport can generate adenosine triphosphate (ATP) by recycling electrons through the reduction of NADPH by NADH dehydrogenase (Mi et al., 1995). Even though elevated transcription does not necessarily equate to increased activity, it is reasonable to assume that diatom symbionts may require additional ATP from cyclic electron transport. N2 fixation is an energetically expensive process (Ljones, 1979), and the symbionts increase N2 fixation not only to meet their own N needs, but also those of their host diatom (Foster et al., 2011).
Intriguingly, the second most abundant transcript in HR-B transitional transcriptome may regulate cyclic electron transport. We hypothesize that this transcript is an antisense RNA (asRNA), as it had only partial coverage of the NADH dehydrogenase D1 subunit gene. The asRNAs are transcribed in the opposite direction to an mRNA target, can up- or downregulate that gene and require rho-independent termination mechanisms (Georg and Hess, 2011). A T-tail following a stem-loop secondary structure that could provide for such a termination mechanism was located by mfold (Zuker, 2003) near the predicted end of the HR-B ndhD1 asRNA. It is unclear whether this abundant transcript upregulates or downregulates the expression of ndhD1. In addition, NADH dehydrogenases have other functions in cyanobacteria (Ogawa and Mi, 2007), and thus it is unclear what affect the asRNA has on the symbiont or the association, as a whole. However, asRNAs have been identified for genes encoding other NADH dehydrogenase subunits in Synechocystis (Georg et al., 2009) and chloroplasts (Georg et al., 2010), indicating this level of regulation is not restricted to diatom symbionts.
Similar to HR-B ndhD1, other abundant transcripts in the Trichodesmium and HR-B transcriptomes showed only partial coverage on coding sequences. These reads may also belong to noncoding RNA transcripts, such as asRNAs. No stem-loop structure could be found near the end of the other transcripts in question, but other rho-independent termination mechanisms are possible (Georg and Hess, 2011). Significant expression has been observed for more than 400 asRNAs in Synechocystis (Mitschke et al., 2011), and thus it would not be surprising to detect additional regulatory transcripts in the cyanobacterial populations in our study.
The HR-B population transcriptomes were also characterized by an abundance of transcripts involved in N2 fixation. Both the Hemiaulus symbiont and Rhizosolenia symbiont genomes lack NH4+ transporters and the genes that encode the enzymes required to assimilate NO3−, NO2− and urease, limiting the N sources available to the symbionts (Hilton et al., 2013; Hilton, 2014). Two of the most abundant HR-B transcripts were nifH and nifD, encoding the iron protein and α-chain, respectively, of the MoFe protein of nitrogenase, the enzyme that catalyzes N2 fixation. Similarly, nifH was the ninth most abundant transcript in the RR transcriptome, highlighting the metabolic importance of N2 fixation in each diatom–diazotroph association.
Trichodesmium nitrogenase gene transcripts were detected in the transcriptome, but not in high abundance. However, there was little indication of Trichodesmium utilizing other nitrogen sources as NO3− and NO2− reductase genes were not detected in the transcript libraries. Furthermore, only one cDNA read was assigned to an NH4+ transporter transcript and one other cDNA read to a urease accessory protein, each at the oceanic station. Transcripts involved in important processes such as gas vesicle formation were more highly expressed in the Trichodesmium transcriptomes. Two of the most abundant transcripts in the low-salinity offshore, transitional and oceanic Trichodesmium transcriptomes were from gas vesicle protein genes adjacent to each other in the genome. Gas vesicles provide buoyancy to return to surface waters after Trichodesmium sinks to depth, possibly to acquire phosphorus (Villareal and Carpenter, 2003). Gas vesicles are important for remaining in the photic zone.
Unexpectedly, several of the highly abundant transcripts in the diazotroph metatranscriptomes corresponded to regions of the genome that have not been annotated as coding regions. Some of the IGS regions were between genes known to constitute an operon, and thus included in the transcript (for example, nifHDK). However, three of the top five most abundant transcripts in the HR-B transcriptome did not correspond to known operons. A BLAST analysis of these three IGS regions resulted in high similarity to a transfer mRNA (NZ_CAIY01000044_209707_211231), an RNA subunit of RNase P (NZ_CAIY01000027_241244_243250) and a leucine transfer RNA intron sequence (NZ_CAIY01000027_330123_331418). These functional regions have been poorly annotated in previously sequenced genomes, and thus were initially unidentified in the R. intracellularis HH01 genome. Similarly, an abundant Trichodesmium IGS region (NC_008312__1642616_1643889) showed similarity to transposases, which can be difficult to annotate, further demonstrating the value of transcription sequences in genome annotations.
The sequencing of metagenomes and metatranscriptomes in this study has made it possible to analyze diazotrophic populations that cannot be achieved through targeted assays such as PCR. With the ability to compare genetic markers from across the genome, we found that the majority of diazotroph populations in this environment were similar to the genomes currently available. However, the Trichodesmium population was an exception to this, and was not representative of T. erythraeum IMS101, the only currently sequenced Trichodesmium genome. This suggests that genomic sequencing of a variety of Trichodesmium species is needed to more accurately depict natural populations, their metabolic capabilities and their roles in surface communities. We also identified a need for studies on noncoding transcripts and their function in regulating a variety of metabolic processes of N2-fixing cyanobacteria, and of microbial communities, in general. In addition, our analysis revealed a stark contrast within the distribution of transcripts amongst vital cellular processes, such as photosynthesis and N2 fixation, between the free-living Trichodesmium and the diatom-associated Richelia. In this study, we utilized extensive community DNA and RNA sequencing to study individual diazotroph populations, and the metabolic pathways within those populations, to elucidate the community composition and cellular state of the diazotrophs in the Amazon River plume.
Acknowledgments
This work was sponsored in part by the Gordon and Betty Moore Foundation (GBMF) Marine Investigator award (to JPZ), GMBF ROCA award (P Yager, PI), the UCSC Microbial Environmental, Genomics, Application, Modeling, Experimental, Remote sensing (MEGAMER) facility (supported by the GBMF) and the NSF Center for Microbial Oceanography: Research and Education (EF-0424599; to JPZ). We are very grateful to MA Moran, B Crump and J Paul for their assistance throughout this project and the writing of this report. We also thank D Bombar for comments on the manuscript, and I Shilova and J Robidart for technical help and discussions.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Adams DG, Bergman B, Nierzwicki-Bauer SA, Rai AN, Schüssler A.2006Cyanobacterial–plant symbiosesIn Dworkin M, Falkow S, Rosenberg E, Schleifer K, Stackebrandt E (eds),The Prokaryotes. A Handbook on the Biology of Bacteria vol. 1Springer Science: New York, NY; 331–363. [Google Scholar]
- Aro E-M, Virgin I, Andersson B. Photoinhibition of photosystem II. Inactivation, protein damage and turnover. Biochim Biophys Acta BBA Bioenerg. 1993;1143:113–134. doi: 10.1016/0005-2728(93)90134-2. [DOI] [PubMed] [Google Scholar]
- Baker BJ, Sheik CS, Taylor CA, Jain S, Bhasi A, Cavalcoli JD, et al. Community transcriptomic assembly reveals microbes that contribute to deep-sea carbon and nitrogen cycling. ISME J. 2013;7:1962–1973. doi: 10.1038/ismej.2013.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter EJ, Montoya JP, Burns J, Mulholland M, Subramaniam A, Capone DG. Extensive bloom of a N2 fixing symbiotic association in the tropical Atlantic Ocean. Mar Ecol Prog Ser. 1999;185:273–283. [Google Scholar]
- Crump BC, Armbrust EV, Baross JA. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl Environ Microbiol. 1999;65:3192–3204. doi: 10.1128/aem.65.7.3192-3204.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump BC, Kling GW, Bahr M, Hobbie JE. Bacterioplankton community shifts in an arctic lake correlate with seasonal changes in organic matter source. Appl Environ Microbiol. 2003;69:2253–2268. doi: 10.1128/AEM.69.4.2253-2268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugdale RC, Goering JJ. Uptake of new and regenerated forms of nitrogen in primary productivity. Limnol Oceanogr. 1967;12:196–206. [Google Scholar]
- Falgueras J, Lara AJ, Fernández-Pozo N, Cantón FR, Pérez-Trabado G, Claros MG. SeqTrim: a high-throughput pipeline for pre-processing any type of sequence read. BMC Bioinformatics. 2010;11:38. doi: 10.1186/1471-2105-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster RA, Kuypers MMM, Vagner T, Paerl RW, Musat N, Zehr JP. Nitrogen fixation and transfer in open ocean diatom–cyanobacterial symbioses. ISME J. 2011;5:1484–1493. doi: 10.1038/ismej.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster RA, Subramaniam A, Mahaffey C, Carpenter EJ, Capone DG, Zehr JP. Influence of the Amazon River plume on distributions of free-living and symbiotic cyanobacteria in the western tropical north Atlantic Ocean. Limnol Oceanogr. 2007;52:517–532. [Google Scholar]
- Foster RA, Zehr JP. Characterization of diatom-cyanobacteria symbioses on the basis of nifH, hetR and 16S rRNA sequences. Environ Microbiol. 2006;8:1913–1925. doi: 10.1111/j.1462-2920.2006.01068.x. [DOI] [PubMed] [Google Scholar]
- Francois P, Garzoni C, Bento M, Schrenzel J. Comparison of amplification methods for transcriptomic analyses of low abundance prokaryotic RNA sources. J Microbiol Methods. 2007;68:385–391. doi: 10.1016/j.mimet.2006.09.022. [DOI] [PubMed] [Google Scholar]
- Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW, et al. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA. 2008;105:3805. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georg J, Hess WR. cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev MMBR. 2011;75:286–300. doi: 10.1128/MMBR.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georg J, Honsel A, Voss B, Rennenberg H, Hess WR. A long antisense RNA in plant chloroplasts. New Phytol. 2010;186:615–622. doi: 10.1111/j.1469-8137.2010.03203.x. [DOI] [PubMed] [Google Scholar]
- Georg J, Voß B, Scholz I, Mitschke J, Wilde A, Hess WR. Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol Syst Biol. 2009;5:305. doi: 10.1038/msb.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford SM, Sharma S, Rinta-Kanto JM, Moran MA. Quantitative analysis of a deeply sequenced marine microbial metatranscriptome. ISME J. 2010;5:461–472. doi: 10.1038/ismej.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Field D, Huang Y, Edwards R, Li W, et al. Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities. PLoS One. 2008;3:e3042. doi: 10.1371/journal.pone.0003042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel NL, Turk KA, Achilles KM, Paerl R, Hewson I, Morrison AE, et al. Abundance and distribution of major groups of diazotrophic cyanobacteria and their potential contribution to N2 fixation in the tropical Atlantic Ocean. Environ Microbiol. 2010;12:3272–3289. doi: 10.1111/j.1462-2920.2010.02303.x. [DOI] [PubMed] [Google Scholar]
- Goes JI, Gomes H, do R, Chekalyuk AM, Carpenter EJ, Montoya JP, et al. Influence of the Amazon River discharge on the biogeography of phytoplankton communities in the western tropical north Atlantic. Prog Oceanogr. 2014;120:29–40. [Google Scholar]
- Hewson I, Poretsky RS, Beinart RA, White AE, Shi T, Bench SR, et al. In situ transcriptomic analysis of the globally important keystone N2-fixing taxon Crocosphaera watsonii. ISME J. 2009;3:618–631. doi: 10.1038/ismej.2009.8. [DOI] [PubMed] [Google Scholar]
- Hewson I, Poretsky RS, Dyhrman ST, Zielinski B, White AE, Tripp HJ, et al. Microbial community gene expression within colonies of the diazotroph, Trichodesmium, from the Southwest Pacific Ocean. ISME J. 2009;3:1286–1300. doi: 10.1038/ismej.2009.75. [DOI] [PubMed] [Google Scholar]
- Hilton JA. Ecology and evolution of diatom-associated cyanobacteria through genetic analyses. PhD, University of California: Santa Cruz, CA; 2014. [Google Scholar]
- Hilton JA, Foster RA, Tripp HJ, Carter BJ, Zehr JP, Villareal TA. Genomic deletions disrupt nitrogen metabolism pathways of a cyanobacterial diatom symbiont. Nat Commun. 2013;4:1767. doi: 10.1038/ncomms2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes AM, Webb EA, Doney SC, Waterbury JB. Comparison of cultured Trichodesmium (Cyanophyceae) with species characterized from the field. J Phycol. 2012;48:196–210. doi: 10.1111/j.1529-8817.2011.01096.x. [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Eggers B, Shen G, Webber A, Yu J, Hirano A, et al. Cloning of the psbK gene from Synechocystis sp. PCC 6803 and characterization of photosystem II in mutants lacking PSII-K. J Biol Chem. 1991;266:11111–11115. [PubMed] [Google Scholar]
- Janson S, Bergman B, Carpenter EJ, Giovannoni SJ, Vergin K. Genetic analysis of natural populations of the marine diazotrophic cyanobacterium Trichodesmium. FEMS Microbiol Ecol. 1999;30:57–65. [Google Scholar]
- Janson S, Wouters J, Bergman B, Carpenter EJ. Host specificity in the Richelia-diatom symbiosis revealed by hetR gene sequence analysis. Environ Microbiol. 1999;1:431–438. doi: 10.1046/j.1462-2920.1999.00053.x. [DOI] [PubMed] [Google Scholar]
- Levitan O, Sudhaus S, LaRoche J, Berman-Frank I. The influence of pCO2 and temperature on gene expression of carbon and nitrogen pathways in Trichodesmium IMS101. PLoS One. 2010;5:e15104. doi: 10.1371/journal.pone.0015104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljones T. Nitrogen fixation and bioenergetics: the role of ATP in nitrogenase catalysis. FEBS Lett. 1979;98:1–8. doi: 10.1016/0014-5793(79)80138-6. [DOI] [PubMed] [Google Scholar]
- Lundgren P, Janson S, Jonasson S, Singer A, Bergman B. Unveiling of novel radiations within Trichodesmium cluster by hetR gene sequence analysis. Appl Environ Microbiol. 2005;71:190–196. doi: 10.1128/AEM.71.1.190-196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti A, Schruth DM, Durkin CA, Parker MS, Kodner RB, Berthiaume CT, et al. Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability. Proc Natl Acad Sci USA. 2012;109:E317–E325. doi: 10.1073/pnas.1118408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayes SR, Dubbs JM, Vass I, Hideg E, Nagy L, Barber J. Further characterization of the psbH locus of Synechocystis sp. PCC 6803: inactivation of psbH impairs QA to QB electron transport in photosystem 2. Biochemistry (Mosc) 1993;32:1454–1465. doi: 10.1021/bi00057a008. [DOI] [PubMed] [Google Scholar]
- Mi H, Endo T, Ogawa T, Asada K. Thylakoid membrane-bound, NADPH-specific pyridine nucleotide dehydrogenase complex mediates cyclic electron transport in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 1995;36:661–668. [Google Scholar]
- Mitschke J, Georg J, Scholz I, Sharma CM, Dienst D, Bantscheff J, et al. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci USA. 2011;108:2124–2129. doi: 10.1073/pnas.1015154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr W, Intermaggio MP, LaRoche J. Diel rhythm of nitrogen and carbon metabolism in the unicellular, diazotrophic cyanobacterium Crocosphaera watsonii WH8501. Environ Microbiol. 2010;12:412–421. doi: 10.1111/j.1462-2920.2009.02078.x. [DOI] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Mi H. Cyanobacterial NADPH dehydrogenase complexes. Photosynth Res. 2007;93:69–77. doi: 10.1007/s11120-006-9128-y. [DOI] [PubMed] [Google Scholar]
- Ottesen EA, Young CR, Eppley JM, Ryan JP, Chavez FP, Scholin CA, et al. Pattern and synchrony of gene expression among sympatric marine microbial populations. Proc Natl Acad Sci USA. 2013;110:E488–E497. doi: 10.1073/pnas.1222099110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottesen EA, Young CR, Gifford SM, Eppley JM, Marin R, Schuster SC, et al. Multispecies diel transcriptional oscillations in open ocean heterotrophic bacterial assemblages. Science. 2014;345:207–212. doi: 10.1126/science.1252476. [DOI] [PubMed] [Google Scholar]
- Peters G, Meeks J. The Azolla-Anabaena symbiosis — basic biology. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:193–210. [Google Scholar]
- Poretsky RS, Gifford S, Rinta-Kanto J, Vila-Costa M, Moran MA. Analyzing gene expression from marine microbial communities using environmental transcriptomics. J Vis Exp. 2009;24:1086. doi: 10.3791/1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poretsky RS, Hewson I, Sun S, Allen AE, Zehr JP, Moran MA. Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environ Microbiol. 2009;11:1358–1375. doi: 10.1111/j.1462-2920.2008.01863.x. [DOI] [PubMed] [Google Scholar]
- Rodrigue S, Materna AC, Timberlake SC, Blackburn MC, Malmstrom RR, Alm EJ, et al. Unlocking short read sequencing for metagenomics. PLoS One. 2010;5:e11840. doi: 10.1371/journal.pone.0011840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satinsky BM, Crump BC, Smith CB, Sharma S, Zielinski B, Doherty M, et al. Microspatial gene expression patterns in the Amazon River plume. Proc Natl Acad Sci USA. 2014;111:11085–11090. doi: 10.1073/pnas.1402782111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T, Ilikchyan I, Rabouille S, Zehr JP. Genome-wide analysis of diel gene expression in the unicellular N2-fixing cyanobacterium Crocosphaera watsonii WH 8501. ISME J. 2010;4:621–632. doi: 10.1038/ismej.2009.148. [DOI] [PubMed] [Google Scholar]
- Shi Y, Tyson GW, DeLong EF. Metatranscriptomics reveals unique microbial small RNAs in the ocean's water column. Nature. 2009;459:266–269. doi: 10.1038/nature08055. [DOI] [PubMed] [Google Scholar]
- Shi Y, Tyson GW, Eppley JM, DeLong EF. Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J. 2010;5:999–1013. doi: 10.1038/ismej.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipe RF, Carpenter EJ, Govil SR, Capone DG. Limitation of phytoplankton production by Si and N in the western Atlantic Ocean. Mar Ecol Prog Ser. 2007;338:33–45. [Google Scholar]
- Stewart FJ, Ottesen EA, DeLong EF. Development and quantitative analyses of a universal rRNA-subtraction protocol for microbial metatranscriptomics. ISME J. 2010;4:896–907. doi: 10.1038/ismej.2010.18. [DOI] [PubMed] [Google Scholar]
- Subramaniam A, Yager P, Carpenter E, Mahaffey C, Björkman K, Cooley S, et al. Amazon River enhances diazotrophy and carbon sequestration in the tropical North Atlantic Ocean. Proc Natl Acad Sci USA. 2008;105:10460–10465. doi: 10.1073/pnas.0710279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundström BG. Observations on Rhizosolenia clevei Ostenfeld (Bacillariophyceae) and Richelia intracellularis Schmidt (Cyanophyceae) Bot Mar. 1984;27:345–356. [Google Scholar]
- Turk-Kubo KA, Achilles KM, Serros TRC, Ochiai M, Montoya JP, Zehr JP. Nitrogenase (nifH) gene expression in diazotrophic cyanobacteria in the Tropical North Atlantic in response to nutrient amendments. Front Microbiol. 2012;3:386. doi: 10.3389/fmicb.2012.00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villareal TA. Nitrogen-fixation by the cyanobacterial symbiont of the diatom genus Hemiaulus. Mar Ecol Prog Ser. 1991;76:201–204. [Google Scholar]
- Villareal TA, Carpenter EJ. Buoyancy regulation and the potential for vertical migration in the oceanic cyanobacterium Trichodesmium. Microb Ecol. 2003;45:1–10. doi: 10.1007/s00248-002-1012-5. [DOI] [PubMed] [Google Scholar]
- Yeung LY, Berelson WM, Young ED, Prokopenko MG, Rollins N, Coles VJ, et al. Impact of diatom-diazotroph associations on carbon export in the Amazon River plume. Geophys Res Lett. 2012;39:L18609. [Google Scholar]
- Zehr JP, Kudela RM. Nitrogen cycle of the open ocean: from genes to ecosystems. Annu Rev Mar Sci. 2011;3:197–225. doi: 10.1146/annurev-marine-120709-142819. [DOI] [PubMed] [Google Scholar]
- Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.