Abstract
We have analyzed metagenomic fosmid clones from the deep chlorophyll maximum (DCM), which, by genomic parameters, correspond to the 16S ribosomal RNA (rRNA)-defined marine Euryarchaeota group IIB (MGIIB). The fosmid collections associated with this group add up to 4 Mb and correspond to at least two species within this group. From the proposed essential genes contained in the collections, we infer that large sections of the conserved regions of the genomes of these microbes have been recovered. The genomes indicate a photoheterotrophic lifestyle, similar to that of the available genome of MGIIA (assembled from an estuarine metagenome in Puget Sound, Washington Pacific coast), with a proton-pumping rhodopsin of the same kind. Several genomic features support an aerobic metabolism with diversified substrate degradation capabilities that include xenobiotics and agar. On the other hand, these MGIIB representatives are non-motile and possess similar genome size to the MGIIA-assembled genome, but with a lower GC content. The large phylogenomic gap with other known archaea indicates that this is a new class of marine Euryarchaeota for which we suggest the name Thalassoarchaea. The analysis of recruitment from available metagenomes indicates that the representatives of group IIB described here are largely found at the DCM (ca. 50 m deep), in which they are abundant (up to 0.5% of the reads), and at the surface mostly during the winter mixing, which explains formerly described 16S rRNA distribution patterns. Their uneven representation in environmental samples that are close in space and time might indicate sporadic blooms.
Introduction
Marine Euryarchaeota group II (MGII) are widely distributed in the global ocean (Massana et al., 2000) and are the dominant archaeal community within the euphotic zone of temperate waters (DeLong, 1992; Massana et al., 1997, 2000; Karner et al., 2001; Herndl et al., 2005; DeLong et al., 2006; Belmar et al., 2011). They have been classified into two major groups, MGIIA and MGIIB, by their 16S ribosomal RNA (rRNA) (Massana et al., 2000; Martin-Cuadrado et al., 2008; Galand et al., 2009; Belmar et al., 2011). Two other groups, MGIIC and D, related to hydrothermal and deep samples have also been proposed (Belmar et al., 2011). In addition, several subclusters have been identified in each of these general groups (Massana et al., 2000; Galand et al., 2010). For example, Euryarchaeota MGIIA is separated in subclusters K, L and M, and MGIIB is separated into subclusters O and N. Subcluster O comprises the WHARM subcluster (formerly phylotype II-CC) (Massana et al., 2000), widely distributed in surface waters (Galand et al., 2010; Hugoni et al., 2013). MGII representatives have a cosmopolitan distribution and are relatively abundant during summer months in temperate and tropical waters (Pernthaler et al., 2002; Herfort et al., 2007). In Mediterranean surface waters, there are reports of different seasonal prevalence, with MGIIB predominating in winter and MGIIA in summer (Galand et al., 2010; Hugoni et al., 2013). There is also evidence that indicates MGIIA is found more often at surface samples (Frigaard et al., 2006), while MGIIB is more frequently found in deeper waters (Massana et al., 2000; Galand et al., 2010; Hugoni et al., 2013).
To date, there are no cultivated representatives of MGII and little is known about their physiology and ecological role in the oceans. For years, the only information available came from a handful of metagenomic clones containing phylogenetic markers (16S and 23S rDNA) (Beja et al., 2000; Moreira et al., 2004; Frigaard et al., 2006; Martin-Cuadrado et al., 2008). In 2012, by using the de novo assembly from a metagenome from superficial estuarine waters, a composite genome sequence grouping 4–6 strains of Group II archaea (MG2-GG3) was published (Iverson et al., 2012). Recently, two single amplified genomes (SAGs) classified as MGII were released to the Genbank database (SCGC-AAA-288-C18, isolated from the North-Pacific Subtropical Gyre at 700 m deep, and SCGC-AB-629-J06 from Lake Washington, USA). However, both SAGs lack the ribosomal operon and their similarity to the MG2-GG3-assembled genome is very low (the euryarchaeal affiliation could be ascertained only for SCGC-AAA-288-C18). MG2-GG3, similar to many other marine prokaryotes, seems to be a photoheterotroph, possessing a rhodopsin to collect light energy but with the hallmarks of a heterotrophic lifestyle. The existence of many large peptidases, similar to the ubiquitous thermoacidophilic archaeon Aciduliprofundum boonei, a known protein degrader from deep-sea hydrothermal vents (Reysenbach et al., 2006), suggested that proteins may be important substrates for this microbe. Moreover, the presence of a complete fatty acid degradation pathway, together with proteins with a variety of adhesion domains and a type II/IV secretion systems to transport such proteins to the cell surface, led the authors to suggest a particle-associated lifestyle. The analysis of the MGII transcripts from coastal surface samples in northern California (Ottesen et al., 2013) also support the ability to metabolize large and complex polymers.
In most of the global ocean when the photic zone becomes thermally stratified (in summer, at temperate latitudes and permanently at tropical ones), phytoplankton biomass concentrates at the deep chlorophyll maximum (DCM) (Estrada et al., 1993; Huisman et al., 2006). The DCM is a layer, usually between 50 and 150 m deep, which is free from the strong hydrodynamism and damaging UV light of the surface and, on the other hand, is connected to the nutrient-rich deep waters. Thus, the DCM has a good compromise of available light and inorganic nutrients availability for planktonic photosynthetic microbes. There is some evidence in the literature, indicating that MGII could be relatively abundant at DCM depths. The maximum abundance of MGII in the Hawaii Ocean Time-Series permanent DCM by 16S rRNA gene amplification from a metagenomic fosmid library (DeLong et al., 2006) was found at 130 m. Ghai et al. (2010) found rRNA fragments of MGII in proportions close to 4% in the Mediterranean DCM using raw metagenomic reads. In addition, in a metagenomic fosmid library from the same sample, up to 22 fosmids (from a total of 197 larger than 10 kb) were classified as belonging to MGII.
Sequencing large metagenomic clones provides a powerful strategy for obtaining valuable information about the structure and ecology of uncultured microorganisms (Martin-Cuadrado et al., 2008; Ghai et al., 2013; Deschamps et al., 2014). From a metagenomic library constructed from the biomass recovered at the DCM in the Mediterranean, we sequenced about 7000 fosmid clones (Ghai et al., 2010, 2013; Mizuno et al., 2013) and we have analyzed the MGII genome fragments present in these fosmids. MGII contigs assembled from different DCM metagenomes obtained from the same location at different times were also included in this study. Here we present the first genomic data about a new class of abundant low GC MGIIB, which has been named Thalassoarchaea, a distant relative to the class represented by the MG2-GG3 assembly (Iverson et al., 2012).
Materials and methods
Sampling and sequencing
A metagenomic fosmid library (∼13 000 clones) was constructed with biomass recovered from the Mediterranean DCM (50 m, 38°4'6.64''N 0°13'55.18''W) on 15 October 2007. A metagenome (MedDCM-OCT2007) directly sequenced by 454 from the same filter and the sequencing results of nearly 1000 fosmids have been described previously in Ghai et al. (2010). The sequencing of additional metagenomic fosmids (∼6000) substantially extended these data sets and have been partially published in Ghai et al. (2013) and Mizuno et al. (2013). Three more samples were collected at the same location and at the chlorophyll maximum depth (as determined by a Seabird SBE 19, Sea-Bird Electronics, Bellevue, WA, USA, multiprobe profiler including fluorometers) in different years: June 2009 (65 m), July 2012 (75 m) and September 2013 (55 m). On each of these dates, sea water was collected at DCM depth and sequentially filtered on board using a positive pressure system. Nylon filters of 20 μm were used as prefilters, followed by 5 μm polycarbonate and finally 0.22 μm Sterivex filters (Durapore; Millipore, Billerica, MA, USA). Filters retaining the 20–5 μm (large fraction (LF), enriched in particle attached bacteria) and the 5–0.2 μm (small fraction, enriched in free-living planktonic cells) were conserved in lysis buffer (40 mM EDTA, 50 mM Tris/HCl and 0.75 M sucrose) at −20 °C until DNA extraction. Filters were thawed on ice and then treated with 1 mg ml−1 lysozyme and 0.2 mg ml−1 proteinase K (final concentrations). Nucleic acids were extracted with phenol–chloroform–isoamyl alcohol and chloroform–isoamyl alcohol, and then concentrated using a microconcentrator (Centricon 100; Amicon, Millipore). DNA integrity was checked by agarose gel electrophoresis and quantified with Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA).
Sequencing was done using Illumina HiSeq 2000 with 100 bp paired ends (Macrogen, Seoul, South Korea). For the 2009 sample, a total of 6.8 Gb of sequence data was produced for the large-size fraction (metagenome MedDCM-JUN2009-LF). A total of 13.6 Gb was obtained for the free-living fraction of the 2012 sample (MedDCM-JUL2012). In addition to the 2012 sample, a mate-paired library of 3 kb insert size was constructed from DNA previously amplified using the Illustra GenomiPhi V2 DNA amplification kit (GE Healthcare, Piscataway, NJ, USA) (50 ng to a final amount of 5.2 μg). As a result, an extra 6.3 Gb of sequence was obtained (metagenome MedDCM-JUL2012-3 kb). For the sample of September 2013, the sequencing of both size-fraction filters, 20–5 and 5–0.22 μm, generated 7.7 and 10.5 Gb, respectively (metagenomes MedDCM-SEP2013-LF and MedDCM-SEP2013, respectively).
Assembly and annotation
A total of 146 genomic fragments (>5 kb) with a GC content of 34–40% could be classified as Thalassoarchaea by annotation and tetranucleotide frequencies. Their origin and other characteristics are summarized in Supplementary Table 1.
The assembly of the DCM metagenomic fosmids from October 2007 were previously described in (Ghai et al., 2010, 2013; Mizuno et al., 2013). Briefly, 7152 fosmids were sequenced using 454-pyrosequencing and Illumina in two different rounds, each divided in 12/24 different batches containing 96 to ∼250 fosmids each. Using the contigs (>5 kb) assembled from this sample, a second assembly was performed using Geneious v.6.1.7 (http://www.geneious.com) with strict parameters (overlapping of >100 bp at 99% identity with no gaps). A total of 69 DNA fragments classified as Thalassoarchaea were obtained from this sample.
Sequences from metagenomes MedDCM-JUN2009-LF, MedDCM-SEP2013-LF and MedDCM-SEP2013 were quality trimmed and assembled independently using meta-iterative de Bruijn de novo assembler (Peng et al., 2012). A combined assembly of the MedDCM-JUL2012 metagenome plus the sequences from the 3-kb library was also performed using the iterative de Bruijn de novo assembler (Peng et al., 2012). A total of 68 DNA fragments were classified as Thalassoarchaea from MedDCM-JUL2012 and 9 more from the metagenome MedDCM-SEP2013. From the LF metagenomes (MedDCM-SEP2009-LF and MedDCM-SEP2013-LF), no Euryarchaeota contigs >5 kb were obtained, probably due to the low coverage of this taxa present and the high percentage of eukaryotic reads obtained (based on the 18S to 16S rRNAs ratios found, only ∼1% of the sequences belonged to prokaryotes).
Gene predictions of the assembled fosmids were done using Prodigal (Hyatt et al., 2010). Ribosomal genes were identified using ssu-align (Nawrocki 2009) and meta_rna (Huang et al., 2009), and transfer RNAs were predicted using tRNAscan-SE (Lowe and Eddy 1997). Functional annotation was performed by comparison of predicted protein sequences against the NCBI NR database, Pfam (Bateman et al., 2004), COGs (Tatusov et al., 2001) and TIGRfams. Local BLAST searches against local databases were performed whenever necessary. The identification of MGII contigs was based on the condition that >50% of the open reading frames (ORFs) contained in the DNA fragment had their best hit to Euryarchaeota, and for this manual examination of each contig was performed. The contigs were named according to the time of sampling, that is, MedDCM-OCT2007-CX, MedDCM-JUL2012-CX and MedDCM-SEP2013-CX, where X is the number of the contig.
Oligonucleotide frequency analysis
The tetranucleotide and pentanucleotide frequencies of the contigs were computed using the wordfreq program from the EMBOSS package (Rice et al., 2000). Principal components analysis was performed using the FactoMineR package in ‘R' (Lê et al., 2008). The genomes of A. boonei T469, MG2-GG3, Nitrosopumilus maritimus and the single-cell genome of SCGC-AAA288-C18 were included in the analysis as references of marine archaea. The frequencies of randomly generated 30 kb fragments of these reference genomes were also included in the analysis. From the initial 154 genomic fragments, a total of 146 (69 from the fosmid collection MedDCM-OCT2007, 68 from MedDCM-JUL2012 and 9 from MedDCM-SEP2013) were clustered together and are described here.
Phylogenetic analysis
Archaeal 16S rRNA and 23S rRNA gene sequences were detected in the genomic fragments and aligned using MUSCLE (Edgar 2004) with their closest relatives as identified by BLAST (Altschul et al., 1990), those from available Euryarchaeota genomes and selected genome fragments present in Genbank and those from the Global Ocean Time Series (GOS) data set (available from http://camera.calit2.net/). Assembled site-specific GOS scaffolds were screened for the presence of ribosomal genes using a stringent cutoff of 98% identity in at least 97% of their length, ensuring that they belonged to the same lineage as the Mediterranean thalassoarchaeal 16S and 23S rRNA sequences assembled from the fosmids and metagenomes. Using the same stringent criteria, the entire Ribosomal Database Project (Cole et al., 2009) was also searched to identify related sequences. 16S rRNA sequences were screened and trimmed using ssu-align (Nawrocki, 2009). Phylogenetic reconstructions were conducted by maximum-likelihood and neighbor-joining methods using MEGA5 (v.5.2, GTR 1 CAT model and a gamma approximation with 1000 bootstraps) (www.megasoftware.net) (Hall, 2013), first using those sequences longer than 700 bp as reference and later including smaller metagenomic reads. In both cases, tree topologies were consistent.
For the rhodopsin tree and the geranylgeranylglyceryl phosphate synthase proteins, sequences were selected based on existing literature, PFAM domain searches and BLAST searches against NCBI-NR and the GOS data set metagenomic reads. Sequences were aligned using MUSCLE (Edgar, 2004) and a maximum-likelihood tree was constructed using MEGA5 (v.5.2, GTR 1 CAT model and a gamma approximation with 1000 bootstraps) (Hall, 2013).
Fluorescence in situ hybridization
For fluorescence in situ hybridization (FISH) detection of the Thalassoarchaea cells, a DCM water sample was recovered in June 2009 and fixed with a 1% paraformaldehyde solution. Subsamples of 15 ml were filtered at low pressure (100 mbar) through 0.2 μm pore size filters (Nucleopore, Whatman, Piscataway, NJ, USA) and kept at −20 °C. A new probe was used, specifically targeting low GC Thalassoarchaea (ThaMar 5′-GTAGTGAAACTATGGATCATTA-3′). The general euryarchaeal probe EURY806 (5′-CACAGCGTTTACACCTAG-3′) was used as a control. Oligonucleotide probes for FISH analysis were purchased from Isogen (Utretch, The Netherlands) and 5′ monolabeled with the indocarbocyanine dye, Cy3. The same filters were cut in quarters for hybridization with the different probes. FISH analysis was carried out by the standard protocol (Glockner et al., 1999) using 5 ng μl−1 of probe. The hybridization solution was prepared as described in DeLong (1992) and DeLong et al. (1999), with the following variations: 50% (vol/vol) formamide, 10% (wt/vol) dextran sulfate in 5 × SET (1 × SET is 150 mM NaCl, 1 mM Na2EDTA, 20 mM Tris-HCl, pH 7.8). Hybridization mixtures were incubated overnight in a hybridization oven at 65 °C. The filters were washed for 2 h at 45 °C in a solution containing 0.2 × SET and 50% (vol/vol) formamide. After incubation in a pre-warmed washing solution, filter sections were dipped in 80% ethanol, dried on Whatman 3 M paper (Whatman Ltd, Jenbach, Austria) and placed on a glass slide. Subsequently, they were mounted with a drop of DAPI (4′,6-diaminodino-2-phenylindole) at a final concentration of 1 μg ml−1.
Metagenomic recruitment
Recruitment plots of the virtual genome reconstructed with the thalassoarchaeal contigs, the MG2-GG3 and SAG SCGC-AAA-288-C18 were performed using BLASTN (a hit was considered only when the alignment was at least 50 nucleotides long). For estimating the abundance of these MGII cells in the metagenomes studied, only the best hits with an identity over 95% in at least 50 bp were considered. To compare the results among the different data sets, the number of reads was normalized considering the size of the contig and of the collection (number of reads per kilobase per gigabase of the collection). The meso and bathypelagic metagenome collections screened were: Marmara Sea-1000 m (Quaiser et al., 2011), Matapan-Vavilov Deep in the Mediterranean Sea-4900 m (Smedile et al., 2012), North Pacific subtropical gyre-ALOHA station-4000 m (Konstantinidis et al., 2009), Guaymas Basin hydrothermal vent in the Gulf of California (Lesniewski et al., 2012) and a metagenome obtained at 800 m in the South Atlantic Gyre (Swan et al., 2014).
Genome and proteome comparisons
For the genomic comparison showed in Figure 4, the thalassoarchaeal contigs were aligned with the genomes of MG2-GG3 and A. boonei T469 by TBLASTX, and only those alignments longer than 200 amino-acids where shown. Contigs with no similarities are not shown. A total of 4070 ORFs were detected within the 146 marine thalassoarchaeal contigs identified. Due to the occurrence of several overlaps among the DNA fragments, some genes were represented more than once. The proteins were clustered using CD-HIT at 50% similarity and 50% of coverage, resulting in a non-redundant proteome of 2435 ORFS. This set of proteins was compared separately to the proteins of the MG2-GG3 (1698), A. boonei T469 (1544) and SCGC_AAA288-C18 (398) genomes using a reciprocal best blast hit analysis to identify putative homologs. The same set was also compared with the proteins detected in the bathypelagic fosmids from the KM3 (9946 proteins), AD1000 (4215) and SAT1000 (1301) collections (Deschamps et al., 2014). Previously, all the sequences used in the comparison were annotated using the same methodology as the thalassoarchaeal contigs to avoid methodological discrepancies.
Accession numbers
The assembled contigs and the metagenomic data have been submitted to NCBI SRA and are accessible under the BioProject identifier PRJNA257723 (SRA run identifiers: MedDCM-SEP2013-LF SRR1539385; MedDCM-SEP2013 SRR1539645; MedDCM-JUL2012-3kb SRR1539382; MedDCM-JUL2012 SRR1539383; MedDCM-JUN2009-LF SRR1539203).
Results and discussion
Retrieval of rRNA-containing fosmids belonging to MGIIB
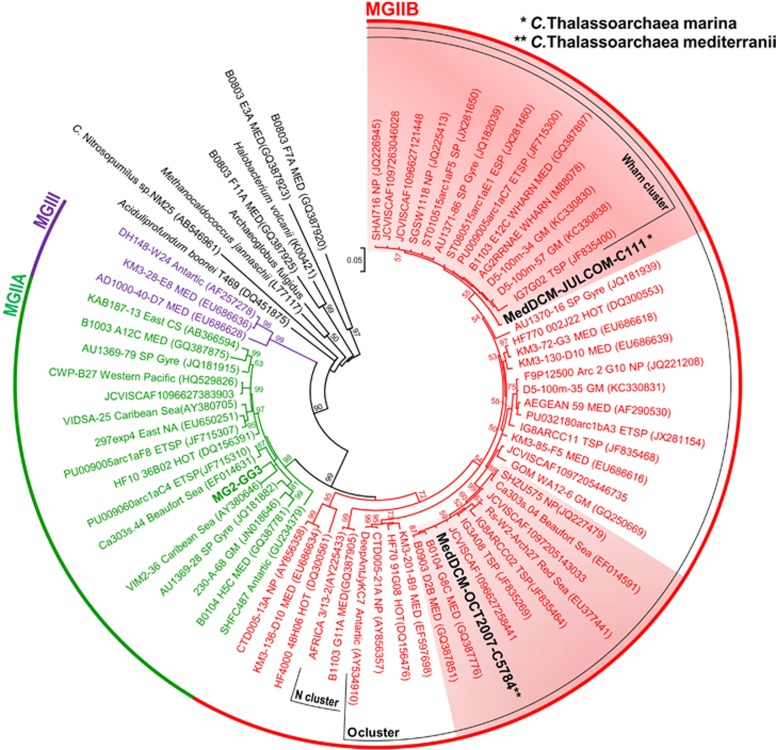
Metagenomic fosmid clones provide discrete natural contigs that can be efficiently assembled to obtain genomic fragments from all members in the community, even from those that are less prevalent and hence less accessible by direct sequencing and assembly (Ghai et al., 2013). During a systematic search of a Mediterranean DCM metagenomic fosmid library (Ghai et al., 2010) for rRNA genes, we identified four fosmids (6.2 to 36 to kb in size) containing rRNA sequences classified as MGII, two of them containing 16S rRNA and the other two 23S rRNA genes (Supplementary Figure 1). Interestingly, they had an atypical low GC content (34–40%) compared with the 52% of the assembled MG2-GG3 genome (Iverson et al., 2012) or the MGII fosmids previously described (DeLong et al., 1999; Moreira et al., 2004; Frigaard et al., 2006; Martin-Cuadrado et al., 2008; Rich et al., 2008; Ghai et al., 2010; Deschamps et al., 2014). MGII have the two rRNA genes separated in the genome (Moreira et al., 2004; Martin-Cuadrado et al., 2008), a feature observed also in the Thermoplasmatales and in A. boonei (Ruepp et al., 2000; Moreira et al., 2004). Hence, it was not surprising that the 16S and 23S genes were not contained in one single contig. The 16S rRNA genes detected in the DCM fosmids belonged to group IIB, hitherto known only by 16S RNA sequences. The two sequences were only 94% similar (well below the species threshold) but both clustered in the large subcluster O of MGIIB (Figure 1). One of them was affiliated with the surface WHARN subcluster (Massana et al., 2000). Fortunately, one large ribosomal protein cluster (spectinomycin or spc operon) is found in the proximity of the 16S rRNA gene (Moreira et al., 2004; Martin-Cuadrado et al., 2008), providing a very robust phylogenetic anchor. We generated maximum-likelihood trees for the alignments of the 16S rRNA genes (Figure 1) and also for a concatenated alignment of the 22 ribosomal proteins of the spc operon (Supplementary Figure 2). All analyses produced consistent results and unambiguously placed the sequences in a group that corresponds to the 16S rRNA group IIB (MGIIB). It is important to emphasize that the only genome available for MGII (Iverson et al., 2012) belongs to the 16S rRNA clade MGIIA. The two fosmids containing 23S rRNA genes were also clearly euryarchaeal. Although the number of 23S rRNA genes available for MGII is much smaller, a maximum-likelihood tree also clustered them separately from the 23S rRNA of the MG2-GG3 (Supplementary Figure 3), and belonging to a separate euryarchaeal clade. By GC content and other genomic parametric approaches, the 23S rRNA containing fosmids belonged to the same group as the 16S rRNA containing ones (see below). Confirming these results, the phylogenetic relationship of the typically archaeal geranylgeranylglyceryl phosphate synthase (GGGP) genes identified in fosmids from the same group, clustered together with A. boonei T469 and the Thermoplasmatales single cell genome SCGC AB-539-N05, isolated from marine sediments and clearly separated from the cluster of MG2-GG3 (Supplementary Figure 4). The affiliation of this gene also suggests that MGIIB may share the tetraether membrane lipids characteristic of the Thermoplasmatales (DeLong, 2006).
Figure 1.
16S rRNA phylogeny. Maximum-likelihood 16S rRNA gene tree (739 unambiguously aligned nucleotides) showing the relationship of the Thalassoarchaea (bold and black) with other MGII (MG2-GG3-assembled genome in bold and green). The clusters where thalassoarchaeal sequences appear are red shadowed. Numbers at nodes in major branches indicate bootstrap support (shown as percentages and only those >50%) by ML in MEGA 5.10. Scale bar represents the estimated number of substitutions per site. The different clusters are named following Galand et al., 2010. Sampling locations: MED, Mediterranean Sea; HOT, Hawaii Ocean Time-Series, North Pacific Gyre (ALOHA station); SP, South Pacific; ETSP, Eastern Tropical South Pacific; ESP, Eastern South Pacific; WP, western Pacific; SA, South Atlantic; NA, North Atlantic; GM, Gulf of Mexico; NP, North Pacific; CS, China Sea; NAEC, North American East Coast; GI, Galapagos Islands; CS, Caribbean Sea; TSP, Tropical South Pacific. The GOS data set identifiers are shown next to each GOS scaffold.
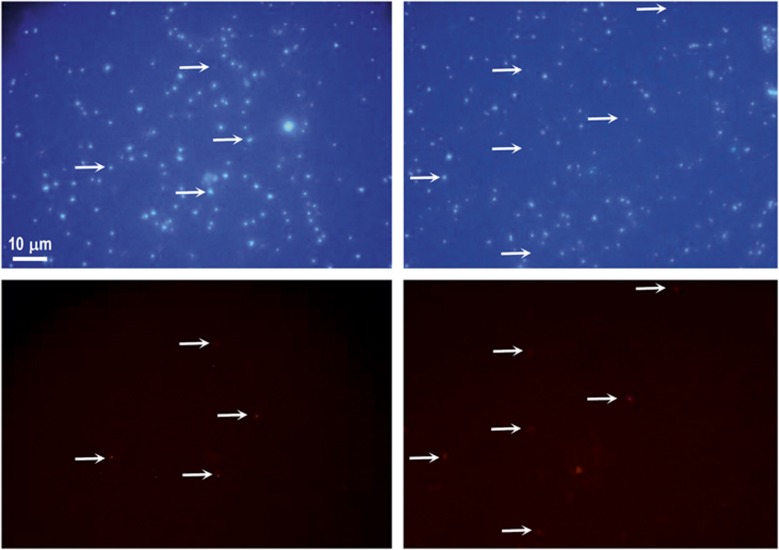
We examined whether similar sequences had been assembled before from other metagenomic data sets by searching the 16S rRNA and 23S rRNA genes in the entire collection of assembled scaffolds from the GOS data set (Venter et al., 2004; Rusch et al., 2007). Five GOS scaffolds were retrieved with a 16S rRNA 100% identical over 97% of the gene to either of our MGIIB 16S rRNA genes (species threshold level) (Supplementary Figure 1), and an additional five at 95% identity at 95% coverage. Therefore, it seems that the microbes represented by the MGIIB fosmids described here are geographically widespread and not an artifact of assembly. The GOS scaffolds were short and contained mostly only the rRNA sequence. However, when they also presented a few more genes they were syntenic to our fosmids. It is interesting to note that most of them were from temperate or tropical regions. When a similar search was performed using the MGIIA 16S rRNA found in the assembled MG2-GG3 (Iverson et al., 2012), no 16S rRNA sequence was found at >97% identity, suggesting less prevalence of this clade in open ocean or coastal metagenomes. In an effort to visualize the abundance of these MGIIB cells at the DCM we carried out FISH with a lineage-specific probe designed with the sequence of the 16S rRNA present in our fosmids. The plankton sample used came from the DCM of June 2009 (see Methods). A specific probe was designed to detect MGIIB and we could see a significant number of DAPI stained cells that hybridized with the probe (1.8%, n=10). Cells were very small, less than a 1 μm (Figure 2). Considering that we are providing an image for these microbes and on the grounds of the phylogenetic distance we would like to propose the taxonomic rank of class for the members of this group that overlaps with the 16S rRNA defined group IIB. We suggest the name Candidatus Thalassoarchaea marina and Candidatus Thalassoarchaea mediterranei, from the greek name for the sea ‘thalassa', for the microbes represented by the 16S rRNA sequences described here and stained by the probe (Figure 1).
Figure 2.
Microscopy of MGII cells. DCM seawater sample from Mediterranean Sea hybridized with fluorescein-labeled 16S rRNA group II archaeal probes (left panel, ‘ThaMar' probe and right panel, ‘Eury 806' probe). Images of the same field were captured using the fluorescein filter set (lower panels) and the DAPI filter set (upper panels). Scale bars, 10 μm.
Genomic reconstruction of MGIIB representatives
Reconstructing genomes from metagenomes is very challenging. First of all, there is the problem of the concurrent diversity. There are always several clones coexisting at any given time that although similar are by no means identical (Rodriguez-Valera et al., 2009; Gonzaga et al., 2012; Lopez-Perez et al., 2013; Kashtan et al., 2014). Secondly, some regions of these genomes are hypervariable, i.e. they vary sharply from one clone to another and are very poorly covered even in high coverage metagenomes. All these caveats prevent the reconstruction of complete and closed genomes from environments in which a significant diversity of microbes is present. Using metagenomic fosmids help in the process by providing long natural contigs that can be compared and joined together by genomic binning (Ghai et al., 2013).
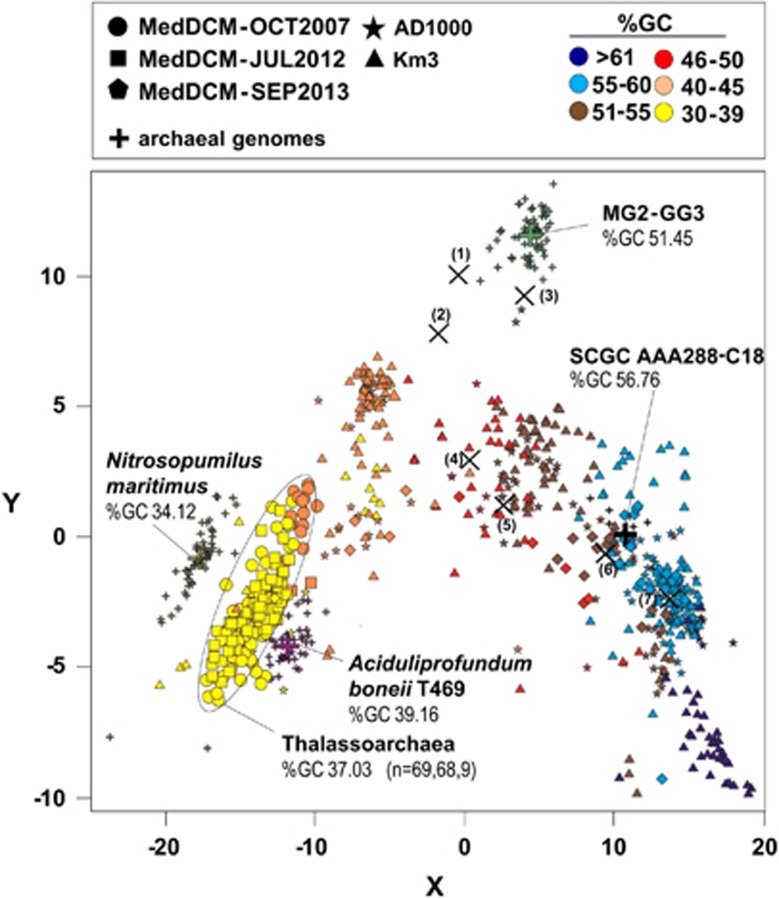
We identified a total of 146 fosmids (total sequence ∼4 Mb) that were affiliated to MGIIB by binning the fosmids containing similar tetranucleotide frequencies to the MGIIB 16S and 23S rRNA containing fosmids described above (Figure 3) (the main features of the contigs can be seen in Supplementary Table 2). They represent the first extensive collection of genomic fragments from this elusive marine euryarchaeal group. All the fosmids were manually curated and we checked that most hits of the ORFs belonged to Euryarchaeota. The comparison with the assembled genome MG2-GG3 (Iverson et al., 2012) or to the cultivated euryarchaeon A. boonei T469 indicated only a distant relationship to either, and very little synteny (Figure 4). The clustered fosmids had lower and homogeneous GC content (range 34.1–40.4%, average 37.03%), much lower than the one described for the MG2-GG3-assembled genome (52%) (Iverson et al., 2012) and closer to A. boonei T469 (34%). The MGII fosmids described up to now in the literature have also higher GC content (DeLong 1992; DeLong et al., 1999; Moreira et al., 2004; Frigaard et al., 2006; Martin-Cuadrado et al., 2008; Rich et al., 2008; Ghai et al., 2010). The low similarity to other available genomes concurs with the rRNA and the ribosomal protein concatenate phylogenetic trees (see above) supporting that these microbes belong to a new class.
Figure 3.
Comparison of MGII fosmids and contigs with known archaeal genomes. Principal component analysis of tetranucleotide frequencies of the low-GC MGII MedDCM-OCT2007 fosmids and the assembled low-GC MGII contigs from the MedDCM-JUL2012 and MedDCM-SEP2013 data sets. Reference genomes are shown as crosses (N. maritimus, A. boonei T469, MG2-GG3 and the SAG SCGC_AAA288-C18). Smaller crosses around the larger ones represent the tetranucleotide frequencies of 35 kb fragments from the same genome. Colors correspond to the %GC accordingly with the legend. The contigs classified as Thalassoarchaea are marked by a circle and the number of contigs from each of the data sets (MedDCM-OCT2007, MedDCM-JUL2012 and MedDCM-SEP2013) is indicated. The length of the assembled DNA fragments and %GC average are also shown. Numbers in brackets indicate MGII fosmids containing 16S rRNA sequences previously described: (1) HF10-29C11, %GC 45.56; (2) HF10-3D09, %GC 46.84; (3) HF70-59C08, %GC 51.36; (4) HF70-19B12, %GC 48.75; (5) EF100-57A08, %GC 50.66; (6) DeepAnt-15E7, %GC 56.03; (7) HF4000-APKG2H5, %GC 56.08).
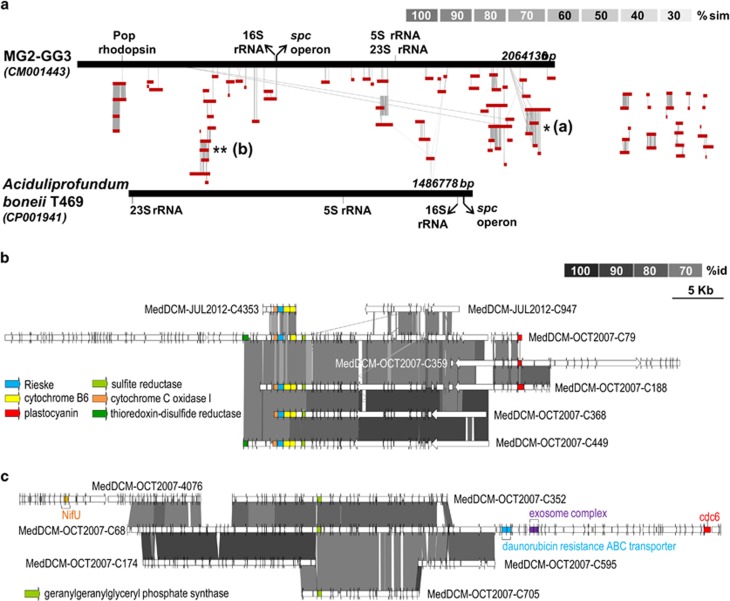
Figure 4.
Genomic comparisons. (a) Linear representation of the synteny of thalassoarchaeal contigs with MG2-GG3-assembled genome and A. boonei T469. (b and c) Comparative genomic organization of thalassoarchaeal contigs. Conserved genomic regions between contigs are indicated by gray shaded areas, gray intensity being a function of sequence similarity by BLASTN. Specific ORFs mentioned in the text are highlighted by a graphic code.
The identity of the two thalassoarchaeal 16S rRNAs (94%) indicates that more than one species were present simultaneously in the fosmid collection. In addition, we found several contigs from overlapping genome regions that were largely syntenic but which average nucleotide identity (ANI) ranged from 98% to 80% (Figure 4), i.e. consistent with more than one species being present. To establish how many we searched among the DNA fragments for different versions of 36 housekeeping genes (Supplementary Table 3). The results indicate that although there are clearly more than one abundant species, their number is likely not very high, probably two, and diverging less than 20% ANI, i.e. within a single genus. The use of other sequence parameters such as codon usage, pentanucleotide frequencies, %GC or the coverage in the metagenomes did not permit to separate them.
We analyzed the contigs for the presence of 35 (Raes et al., 2007) or 100 (Albertsen et al., 2013) previously defined orthologous markers to estimate the completeness of the assembled genomes. Using these criteria, the thalassoarchaeal genomic fragments retrieved represent between 78 and 100% of a complete genome. By the same extrapolation, the estimated genome length of an individual genome of this group would be ∼2 Mb, very similar to the estimated size of the MG2-GG3 genome (Iverson et al., 2012). A total of 4074 ORFs could be identified in the thalassoarchaeal fosmids. Most hits (61%) against the nr database (Genbank) were to Euryarchaeota, 16% to Bacteria and 2% to eukaryotes (the remaining 21% were unclassified). The corresponding proteins were clustered using CD-HIT at 50% similarity and coverage resulting in a smaller data set of 2435 non-redundant proteins. This set was compared to the 1698 proteins of the MG2-GG3 genome (using a reciprocal best blast hit analysis) and 1427 proteins were found to share more than 50% similarity. However, the average similarity was low (65%). Only 639 homologs were found within the 1544 proteins of A. boonei T469 (average similarity 60%). The SAG SCGC-AAA-288-C18 from 700 m deep in the central North Pacific also had a low similarity (average 70%). The highest similarities were found with the proteins of the large set of MGII fosmids retrieved from bathypelagic Mediterranean samples (Ionian Sea 3000 m, 324 fosmids; and Adriatic Sea 1000 m, 139 fosmids) (Deschamps et al., 2014). Of the non-redundant thalassoarchaeal proteins 1599 had a hit with Adriatic (38% of their total) and 1850 with the Ionian fosmids. However, synteny was seldom conserved and average similarity was of 74 and 73% respectively, suggesting that the deep MGII belong to very different taxa.
Inferred metabolic and ecological features
We found genes coding for enzymes for glycolysis, the tricarboxylic acid cycle and oxidative phosphorylation. However, due perhaps to the incomplete nature of these genomes, not all genes could be found, e.g. Thalassoarchaea cells appear to contain many of the enzymes of the Embden–Meyerhof–Parnas (EMP) pathway for the metabolism of hexose sugars, with the exception of the first (glucokinase) and the last (pyruvate kinase). For the first, several carbohydrate kinases of unknown specificity found could serve as alternatives. For the second, a pyruvate phosphate dikinase was found that could operate in the catabolic direction, as have been described in some archaea (Tjaden et al., 2006). Similar proteins were also found among the fosmids from the deep Mediterranean libraries (Deschamps et al., 2014) but they were absent in the MG2-GG3 or any other MGII genomic fragment. Therefore, further investigations must be performed in this direction to clarify if the EMP functions in the gluconeogenic direction rather than the glycolytic pathway, as has been proposed for other Archaea (Hutchins et al., 2001; Hallam et al., 2006). Along these lines, typical gluconeogenesis enzymes such as pyruvate carboxylase (subunits A and B) and a gene coding for a phosphoenolpyruvate carboxykinase, both typical gluconeogenic enzymes, were found. As in other Euryarchaeota (Makarova et al., 1999; Makarova and Koonin 2003; Hallam et al., 2006) glucose 1-dehydrogenase, gluconolactonase and 2-keto-3-deoxy gluconate aldolase homologues are absent, suggesting that the Entner–Duodoroff hexose catabolic pathway is not present. In addition, we identified a complete non-oxidative pentose phosphate pathway, but the irreversible oxidative branch was missing. The reactions of the oxidative branch are important for generating NADPH, which is a source of reducing energy required by many enzymes in central biosynthetic pathways. However, we found genes for enzymes such as a 2,5-didehydrogluconate reductase, or a malate dehydrogenase that could act as alternatives for reducing NADP+ to NADPH.
One of the metabolic features that we can infer is that these representatives of group IIB are facultative photoheterotrophs as seems to be the case of MG2-GG3. Three different rhodopsin genes were found among our contigs, sharing a similarity between 76 and 92% (Figure 5). Rhodopsin genes are widespread in the open oceans (Beja et al., 2000) and are used as back up for heterotrophic energy generation using sunlight. These rhodopsin genes were neither adjacent to the 16S rRNA nor to the archaeal geranylgeranylglyceryl phosphate synthase genes (Figures 4 and 5) as found previously for other MGII fosmids (Frigaard et al., 2006), including those assembled previously from the Mediterranean DCM (Ghai et al., 2010). Two different genomic contexts were found for the thalassoarchaeal rhodopsins, likely corresponding to different species (Figure 5). Several GOS scaffolds were found to be syntenic to these rhodopsin containing fosmids, but none larger than 5 kb. The phylogenetic tree (Supplementary Figure 5) shows the rhodopsin of the thalassoarchaeal representatives to be closely related to the one found in MG2-GG3. Both are in a cluster (clade B) separate from other rhodopsins from bacterial or eukaryotic origin (clade A). We would like to suggest the term thalassorhodopsin to designate this group. Their key residues (listed herein with EBAC31A08 numbering), indicate some important differences with other related rhodopsins (Supplementary Figure 6). Residue 105, involved in spectral tuning, was a methionine, characteristic of green absorbing rhodopsins from shallow depths (Fuhrman et al., 2008) of the Bacteriodetes proteorhodopsin isoform, such as in the marine flavobacterium Dokdonia donghaensis MED134 (Gomez-Consarnau et al., 2007; Riedel et al., 2010). It is known that different amino acid substitutions in residues aspartic-97 (D) and glutamic-108 (E), which function as Schiff base proton acceptor and donor in many proteorhodopsins, provide different capacities to pump outward different ions, varying from H+ (Gushchin et al., 2013), Na+ (Inoue et al., 2013) and even lithium (Inoue et al., 2013). These residues are partially conserved in the thalassorhodopsins and instead of the E, the basic aminoacid lysine (K) is found, a very unusual residue in proton pumping rhodopsins found previously only in Exiguobacterium sibiricum, a permafrost soil Gram-positive (Balashov et al., 2013; Gushchin et al., 2013). This suggests that thalassorhodopsins are proton pumps and add even more diversity to these ubiquitous photoreceptors.
Figure 5.
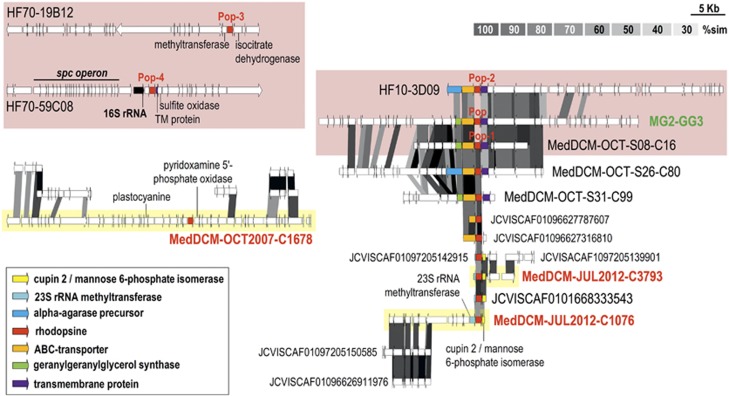
Comparison of rhodopsin genes. Comparative genomic organization of thalassoarchaeal contigs containing rhodopsin genes (yellow-shadowed rectangles) in context with other genomic fragments containing the MGII Pop, Pop-1, Pop-2, Pop-3 and Pop-4 rhodopsins (pink-shadowed rectangles) (Iverson et al., 2012). Scaffolds from the GOS collections similar to the thalassoarchaeal sequences where also included in the comparison.
In one of the contigs containing the rhodopsin gene we found a gene for a plastocyanin (69% similar to its homolog in MG2-GG3). A manual inspection of the predicted proteins showed their best similarities to be with halocyanins (Supplementary Figure 10). These are archaeal blue copper redox proteins (type I) found in halophilic archaea such as Natronomonas pharaonis (Mattar et al., 1994) that serve as mobile electron carrier at a peripheral membrane protein. The presence of the typical copper binding domain in our thalassoarchaeal subunits was confirmed. Close to the halocyanin, we found in two contigs a gene cluster coding for a Rieske/cytochrome b (Rieske/cytb) complex (Figure 4) also with their best similarities with haloarchaeal subunits. This complex was found in four more contigs (Figure 2), suggesting that this is widespread electron transport chain among these archaea. The Rieske/cytb complexes are energy-converting enzymes that operate between the initial electron donors and the terminal electron acceptors of respiratory chains. In halophiles, two distinct clusters of Rieske/cytb encoding genes have been described, (i) the Rieske/cytb complex cluster along with the NAR-encoding genes implied in the denitrifying chain (Martinez-Espinosa et al., 2007) and (ii) the Rieske/cytb complex cluster with a gene coding for an halocyanin followed by a cytochrome b (Nitschke et al., 2010). The implication of this second type of gene cluster in aerobic respiration seems the most likely function, where the halocyanin will work as an electron acceptor of the Rieske/cytb complex and the donor to a cytochrome oxidase (Baymann et al., 2012). The MGIIB Rieske/cytb complexes found have the canonical order as in the haloarchaeal ones, but the halocyanin is found in a separate gene cluster nearby (Figure 4). Two extra genes not present in haloarchaea were conserved among the thalassoarchaeal contigs, a cysteine rich protein and a gene for a NADPH sulfite reductase. Other difference is that the haloarchaeal Rieske subunits are predicted to be of high potential, while these thalassoachaeal homologs have the same residues than the actinobacterial medium potential subunits (Lebrun et al., 2006; Baymann et al., 2012). Previously, it has been suggested that Haloarchaea acquired their Rieske/cytb complexes by different lateral gene transfer events from the Actinobacteria or Thermus/Deinococcus group (Baymann et al., 2012). The similarity in the thalassoarchaeal subunits to residues of the Actinobacterial ones, together with the delocalization of halocyanin also supports this hypothesis. We can speculate that the presence of the Rieske/cytb complex in MGIIB might optimize the energy yield of respiratory metabolism due to the efficient proton pumping capacity of the complex. This could reflect the necessity to answer to an extra demand of energy in conditions of fast growth, typical of bloomers or r-strategists (see below).
The assembly mechanism of many archaeal surface structures is related to the one of bacterial type IV pili (Lassak et al., 2012). The genome of MG2-GG3 contains these genes together with a complete cluster of flagellum biosynthesis (Iverson et al., 2012). In our case, neither genes related with type IV secretion systems nor with flagellum assembly were identified. Therefore, thalassoarchaeal cells are likely to be non motile.
Five percent (137 genes) of the total number of ORFs identified in the contigs were transporters. Unfortunately, 37% of them could not be assigned a substrate. We found four contigs containing a complete set of genes for the uptake of phosphonate, indicating the capacity to utilize this refractory form of phosphorus abundant in marine habitats (Martinez et al., 2010). Additionally, two clusters of genes were found coding for high affinity inorganic phosphate ABC transporters. Transporters for branched chain amino acids, peptides and oligopeptides were 10.2% of the total. Interestingly, 26.3% of the transporters were related with drug/multidrug transport systems, e.g. a daunorubicin resistance ABC transporter (Figure 4). These are ATP-dependent transport proteins which efflux a variety of compounds as a defense against toxic compounds or just transporting endogenous metabolites out of cells. Also a bleomycin hydrolase was found. This abundance of resistance mechanisms may indicate a defensive lifestyle, typical of organisms exposed to high natural toxin concentrations, such as blooms of cyanobacteria producers of toxins.
Although no sugar transporters could be identified, a putative alpha-agarase like protein was detected (34% similarity to the experimentally tested agarase of Thalassomonas agarivorans) (Hatada et al., 2006) (Figure 5). Other agarases were also detected near the 16S rRNA gene in other MGII fosmids already published (Figure 5). Breaking into oligosaccharides agarose or agaropectin present in the cell wall of many algae, the presence of this enzyme would make accessible an abundant food source in the ocean.
Recently, the metatranscriptomes of a serial samples from a coastal upwelling in the North California coast has been published (Ottesen et al., 2013). About 5% of the transcripts of each collection were classified as MGII archaea, which synchronized their transcription with Pelagibacter and SAR86 marine groups. We confirmed the presence of Thalassoarchaea in this metatranscriptomic collection finding a similar number of transcripts than for P. ubique HTCC7211 and SAR86 (Supplementary Figure 7). In order to find out which genes were metabolically active at the sampling time, we calculated the reads per kilobase per gigabase of the collection values for the thalassoarchaeal ORFs (cutoff 95% identity) for each of the metatranscriptomes. Only 316 ORFs (<8% of the total) had transcripts, indicative of a low coverage. In addition to the expected hits to ribosomal proteins, elongation factors and translation initiation factors (that is, SUI1), which were abundant across all the data sets, we also found a putative collagen-like cell surface protein in all the collections. It has been described that in blooming bacterioplankton, genes predicted to increase cell surface adhesiveness are highly transcribed, having an important role in microbial aggregation (Rinta-Kanto et al., 2012). Along these lines, we found a putative thalassoarchaeal flotillin being highly expressed (61% similar to its homolog in MG2-GG3). Flotillins are proteins that form membrane microdomains implicated in signal transduction, vesicle trafficking and cytoskeleton rearrangements (Bach and Bramkamp, 2013). They trigger changes in cell surface properties in response to environmental signals inducing the formation of polysaccharides (D'Argenio and Miller, 2004). Therefore, the high frequency of this flotillin-like protein suggests that aggregation was happening at the time of sampling. Other highly expressed transcripts were rhodopsins, cold shock proteins, amino acids ABC transporters and the previously mentioned bleomycin hydrolase.
Presence of thalassoarchaeal reads in metagenomic collections
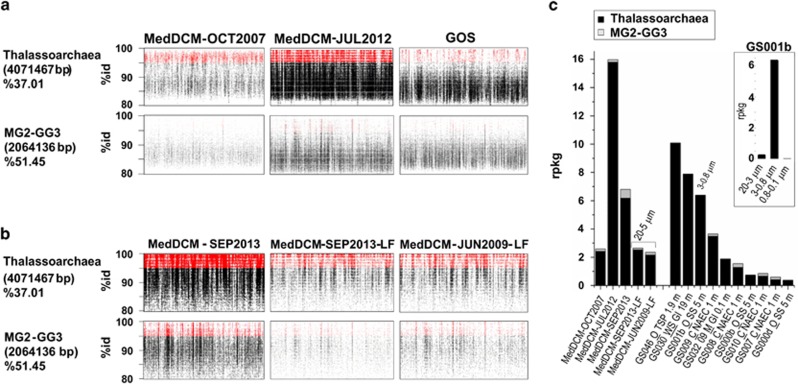
In the MedDCM-OCT2007 454-metagenome (Ghai et al., 2010), a total of 4% of the 16S rRNA sequences were attributed to Euryarchaeota. However, when recruitment assays were performed using a virtual genome reconstructed with the thalassoarchaeal contigs, only 0.5% of the reads in MedDCM-OCT2007 and MedDCM-JUL2012 metagenomes were recruited over 95% of identity (species cutoff) (Figure 6). However, the MG2-GG3 genome recruits one order of magnitude less (0.019% and 0.002%, respectively) and no hits were detected for SAG SCGC-AAA-288-C18. We examined the geographic distribution of the thalassoarchaeal genomes, analyzing their presence in the GOS database (Venter et al., 2004; Rusch et al., 2007) that contains mainly surface samples. In general, the collections with more hits came from the tropics (for example, tropical Pacific, Galapagos Island and Sargasso Sea), with high temperature (22.9–26.9 °C range in the metadata provided). However, they were also detected in GS009, Block Island, and in GS008, Newport Harbor, where temperatures decrease to 11–9.4 °C (Figure 6). They were very abundant at GS046, 300 miles from Polynesia, but not in other collections recovered in consecutive days at similar depths in the same transect (every 100/200 miles). In summary, the abundance of this group appears to be very uneven including in warm waters. One possible explanation is the heterogeneity of surface waters, which are submitted to a strong hydrodynamism. Microbial communities coming from deeper waters may upwell eventually. A different but non-exclusive explanation is that the thalassoarchaeal cells produce localized blooms. In this sense, one of the collections where they were better represented was a warm seep from the Galapagos Island (GS030; 19 m, 26.9 °C). Such a high temperature could promote the growth of bloomers.
Figure 6.
Metagenome recruitments. (a) Metagenomic reads recruited (BLASTN) by the thalassoarchaeal contigs and MG2-GG3 genome in metagenomes from the Mediterranean deep chlorophyll maximum MedDCM-OCT2007, MedDCM-JUL2012 and from the GOS. (b) Metagenomic reads recruited (BLASTN) by the thalassoarchaeal contigs and MG2-GG3 genome in metagenomes from the Mediterranean DCM recovered from different pore-size filters, small fraction (5–0.22 μm) MedDCM-SEP2013 and LF (20–5 μm) MedDCM-JUN2009-LF and MedDCM-SEP2013-LF. (c) Recruitment comparison of the Mediterranean data sets and GOS collections. Metagenomes obtained with large-fraction filter size are indicated.
Analyzing the few available metagenomic depth profile collections (subtropical gyres of North Atlantic (Bermuda Atlantic Time Series) and north Pacific (Hawaii Ocean Time-Series, Station ALOHA); (Coleman and Chisholm 2010; DeLong et al., 2006)), we found evidence of the preference of thalassoarchaeal cells for the photic zone (above 100 m) (Supplementary Figure 8). However, their abundance was one or two orders of magnitude lower than in the Mediterranean DCM (Supplementary Figure 8). Thalassoarchaeal sequences were also scarce along the colder waters of the coast of the northeast subarctic Pacific NESAP collections (Allers et al., 2013) and also in the metagenomes from the eastern tropical south Pacific (Stewart et al., 2012), where reads matching thalassoarchaeal sequences peaked in abundance in the small-size fraction (1.6–0.22 μm) upper water column samples (70 m). These authors reported that 50% of the sequences matching Euryarchaeota peaked in abundance at this depth on prefilters (3–1.6 μm). Of these sequences, those matching MGII constituted 40% of the total, indicating that still an important fraction of MGII remains in the smaller fraction.
A high prevalence of group IIA in particle-rich coastal waters has been described (Galand et al., 2010). In this respect, the presence of genes for the degradation of proteins and lipids suggested that the MG2-GG3-assembled genome may have a particle-attached lifestyle (Iverson et al., 2012), although no direct evidence was provided. To clarify this issue, three more metagenomes were sequenced from Mediterranean DCM samples recovered in 2009 and 2013 (same location the DCM-2007 and 2012) but from the large-size filter. MedDCM-JUN2009-LF and MedDCM-SEP2013-LF were obtained from biomass recovered from the 20 to 5μm fraction. MedDCM-SEP2013 was obtained from the smaller 5- to 0.22-μm fraction. When the corresponding recruitments were done using the thalassoarchaeal genomic fragments, homologous sequences were preferentially found (three to eight times more) in the small-fraction data sets than in the large ones (Figure 6), suggesting that these microbes are free living. The MG2-GG3-assembled genome was not significantly present in either fraction. However, for the sample from the Sargasso Sea GS001, most of the homologous sequences to Thalassoarchaea where found in the 3- to 0.8-μm filter (GS001b) instead of the smaller fraction (inset in Figure 6). Recently, some data sets have been published that come from river estuaries or plumes (similar to the environment from which MG2-GG3 metagenome was obtained) (Smith et al., 2013; Satinsky et al., 2014). These metagenomes have large numbers of reads homologous to both MG2-GG3 and Thalassoarchaea. In the Columbia River plume (very close to the site of origin of MG2-GG3), this assembled genome was found almost equally in the small (0.8–0.1 μm) than in the larger fraction (3–0.8 μm) (Supplementary Figure 9), although Thalassoarchaea recruited scarcely. In the metagenomes of the Amazon plume, both microbes were found in one LF collection (156–2 μm) (Supplementary Figure 8). Interestingly, Thalassoarchaea did not recruit from any of the other collections from the same study, including the smaller size fraction retrieved at the same location. In this case, dramatic differences were found even in collections taken only 1 h apart, that is, ACM25 and ACM1. One plausible explanation is the blooming of both Euryarchaea in the sample.
Several meso and bathypelagic metagenome collections were also screened with negative results (see Materials and Methods). They were also absent in marine collections from polar regions (Alonso-Saez et al., 2012) and from the Baltic collections (Larsson et al., 2014). The Red Sea DCM metagenome (365 Mb) (Thompson et al., 2013) was also analyzed, and although the sample was described as recovered at the DCM (50 m), no significant recruitment was found.
Conclusions
Owing to cultivation bias, Archaea were considered extremophilic or anaerobic microbes for many years. Still, the number of isolates of non-extremophilic archaea is very small. The reasons for this reluctance to grow in pure culture are still obscure but, in any case, our knowledge remains restricted to culture-independent approaches such as metagenomic assemblies. In fact, the number of marine planktonic microbes brought into culture is small for any group and the difficulties of cultivating members of the Pelagibacterales illustrate this point. Here, by sequencing metagenomic fosmids and assembling high-coverage metagenomes, we are providing information about a group of Euryarchaeota hitherto known only from 16S rRNA sequences. Using specific FISH probes, we could visualize the cells and assess their abundance at ca. 2% of the DAPI-stained cells. The genomic information indicates that (at least some representatives of this group) are planktonic photoheterotrophic microbes similar to other known components of the open ocean planktonic microbiota such as the Pelagibacterales or the Actinomarinales. They are all non-motile small cells, with streamlined genomes, and with diverse heterotrophic capabilities, as should be expected from the diverse composition of dissolved organic matter in the open ocean. However, Thalassoarchaea appear very unevenly in metagenomic data sets. This is true also of the available genome of MGIIA, MG2-GG3, which seems to be more abundant in estuarine systems or river plumes. Thalassoarchaea are present in all the available Mediterranean DCM samples and other temperate and tropical seas. They seem to be also detectable at the DCMs at Bermuda Atlantic Time Series (Sargasso) and Hawaii Ocean Time-Series (Central Pacific), and therefore the DCM seems to be its main habitat. In addition, they appear at the surface during sporadic blooms. For example, MGIIB 16S rRNA representatives have been found before at the surface during winter mixing, and many of them cluster together with Thalassoarchaea. Their preference for the DCM can explain this apparently paradoxical finding. They could bloom after hydrodynamic mixing with deeper nutrient-rich waters as happens in temperate latitudes during winter, or more sporadic phenomena due to currents, river plume fertilization and so on. They might use light as an additional energy source and start growing as soon as nutrient concentrations increase after water column mixing.
This bloomer strategy is more often found in particle-associated microbes. However, the data presented seem inconclusive regarding the known representatives of MGIIA or B. Although MG2-GG3 genome indicated a particle-attached lifestyle, it recruits in different size fractions depending on the sample and mostly appears in the small filter sizes, like the Thalassoarchaea. This might be an artifact of the filtration method (larger size filters collect smaller microbes as they become clogged) but it is also possible that MGII representatives are present in both habitats. These hypotheses remain open to be tested in the future with metagenomes obtained with different filter sizes or alternative methodologies. However, it is clear that Thalassoarchaea are an abundant component of microbial plankton in the Mediterranean DCM and also in other oceans. Further work of genome reconstruction coupled to single-cell genomics or development of cultivation methods to obtain laboratory cultures will allow a better understanding of this widespread and important marine microbial group.
Acknowledgments
This work was supported by projects MICROGEN (Programa CONSOLIDER-INGENIO 2010 CDS2009-00006), MEDIMAX BFPU2013-48007-P from the Spanish Ministerio de Economía y Competitividad, the French Agence Nationale de la Recherche (ANR-08-GENM-024-001, EVOLDEEP), MaCuMBA Project 311975 of the European Commission FP7 (FEDER funds supported this project), ACOMP/2014/024 and AORG 2014/032.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol. 2013;31:533–538. doi: 10.1038/nbt.2579. [DOI] [PubMed] [Google Scholar]
- Alonso-Saez L, Waller AS, Mende DR, Bakker K, Farnelid H, Yager PL, et al. Role for urea in nitrification by polar marine Archaea. Proc Natl Acad Sci USA. 2012;109:17989–17994. doi: 10.1073/pnas.1201914109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Allers E, Wright JJ, Konwar KM, Howes CG, Beneze E, Hallam SJ, et al. Diversity and population structure of Marine Group A bacteria in the Northeast subarctic Pacific Ocean. ISME J. 2013;7:256–268. doi: 10.1038/ismej.2012.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach JN, Bramkamp M. Flotillins functionally organize the bacterial membrane. Mol Microbiol. 2013;88:1205–1217. doi: 10.1111/mmi.12252. [DOI] [PubMed] [Google Scholar]
- Balashov SP, Petrovskaya LE, Imasheva ES, Lukashev EP, Dioumaev AK, Wang JM, et al. Breaking the carboxyl rule: lysine 96 facilitates reprotonation of the Schiff base in the photocycle of a retinal protein from Exiguobacterium sibiricum. J Biol Chem. 2013;288:21254–21265. doi: 10.1074/jbc.M113.465138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baymann F, Schoepp-Cothenet B, Lebrun E, van Lis R, Nitschke W. Phylogeny of Rieske/cytb complexes with a special focus on the Haloarchaeal enzymes. Genome Biol Evol. 2012;4:720–729. doi: 10.1093/gbe/evs056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beja O, Suzuki MT, Koonin EV, Aravind L, Hadd A, Nguyen LP, et al. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ Microbiol. 2000;2:516–529. doi: 10.1046/j.1462-2920.2000.00133.x. [DOI] [PubMed] [Google Scholar]
- Belmar L, Molina V, Ulloa O. Abundance and phylogenetic identity of archaeoplankton in the permanent oxygen minimum zone of the eastern tropical South Pacific. FEMS Microbiol Ecol. 2011;78:314–326. doi: 10.1111/j.1574-6941.2011.01159.x. [DOI] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman ML, Chisholm SW. Ecosystem-specific selection pressures revealed through comparative population genomics. Proc Natl Acad Sci USA. 2010;107:18634–18639. doi: 10.1073/pnas.1009480107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Argenio DA, Miller SI. Cyclic di-GMP as a bacterial second messenger. Microbiology. 2004;150:2497–2502. doi: 10.1099/mic.0.27099-0. [DOI] [PubMed] [Google Scholar]
- DeLong EF. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong EF, Taylor LT, Marsh TL, Preston CM. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl Environ Microbiol. 1999;65:5554–5563. doi: 10.1128/aem.65.12.5554-5563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong EF. Archaeal mysteries of the deep revealed. Proc Natl Acad Sci USA. 2006;103:6417–6418. doi: 10.1073/pnas.0602079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU, et al. Community genomics among stratified microbial assemblages in the ocean's interior. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- Deschamps P, Zivanovic Y, Moreira D, Rodriguez-Valera F, López-García P. Pangenome evidence for extensive inter-domain horizontal transfer affecting lineage-core and shell genes in uncultured planktonic Thaumarchaeota and Euryarchaeota. Genome Biol Evol. 2014;6:1549–1563. doi: 10.1093/gbe/evu127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada M, Marrasé C, Latasa M, Berdalet E, Delgado M, Riera T. Variability of deep chlorophyll maximum characteristics in the Northwestern Mediterranean. Marine Ecol Prog Ser. 1993;92:289–300. [Google Scholar]
- Frigaard NU, Martinez A, Mincer TJ, DeLong EF. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature. 2006;439:847–850. doi: 10.1038/nature04435. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, Schwalbach MS, Stingl U. Proteorhodopsins: an array of physiological roles. Nat Rev Microbiol. 2008;6:488–494. doi: 10.1038/nrmicro1893. [DOI] [PubMed] [Google Scholar]
- Galand PE, Casamayor EO, Kirchman DL, Potvin M, Lovejoy C. Unique archaeal assemblages in the Arctic Ocean unveiled by massively parallel tag sequencing. ISME J. 2009;3:860–869. doi: 10.1038/ismej.2009.23. [DOI] [PubMed] [Google Scholar]
- Galand PE, Gutiérrez-Provecho C, Massana R, Gasol JM, Casamayor EO. Inter-annual recurrence of archaeal assemblages in the coastal NW Mediterranean Sea (Blanes Bay Microbial Observatory) Limnol Oceanogr. 2010;55:2117–2125. [Google Scholar]
- Ghai R, Martin-Cuadrado A, Gonzaga A, Garcia-Heredia I, Cabrera R, Martin J, et al. Metagenome of the Mediterranenan deep chlorophyl maximum studied by direct and fosmid library 454 pyrosequencing. ISME J. 2010;4:1154–1166. doi: 10.1038/ismej.2010.44. [DOI] [PubMed] [Google Scholar]
- Ghai R, Mizuno CM, Picazo A, Camacho A, Rodriguez-Valera F. Metagenomics uncovers a new group of low GC and ultra-small marine Actinobacteria. Sci Rep. 2013;3:2471. doi: 10.1038/srep02471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glockner FO, Fuchs BM, Amann R. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Consarnau L, Gonzalez JM, Coll-Llado M, Gourdon P, Pascher T, Neutze R, et al. Light stimulates growth of proteorhodopsin-containing marine Flavobacteria. Nature. 2007;445:210–213. doi: 10.1038/nature05381. [DOI] [PubMed] [Google Scholar]
- Gonzaga A, Martin-Cuadrado AB, Lopez-Perez M, Mizuno CM, Garcia-Heredia I, Kimes NE, et al. Polyclonality of concurrent natural populations of Alteromonas macleodii. Genome Biol Evol. 2012;4:1360–1374. doi: 10.1093/gbe/evs112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gushchin I, Chervakov P, Kuzmichev P, Popov AN, Round E, Borshchevskiy V, et al. Structural insights into the proton pumping by unusual proteorhodopsin from nonmarine bacteria. Proc Natl Acad Sci USA. 2013;110:12631–12636. doi: 10.1073/pnas.1221629110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BG. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol. 2013;30:1229–1235. doi: 10.1093/molbev/mst012. [DOI] [PubMed] [Google Scholar]
- Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y, Sugahara J, et al. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci USA. 2006;103:18296–18301. doi: 10.1073/pnas.0608549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatada Y, Ohta Y, Horikoshi K. Hyperproduction and application of alpha-agarase to enzymatic enhancement of antioxidant activity of porphyran. J Agric Food Chem. 2006;54:9895–9900. doi: 10.1021/jf0613684. [DOI] [PubMed] [Google Scholar]
- Herfort L, Schouten S, Abbas B, Veldhuis MJ, Coolen MJ, Wuchter C, et al. Variations in spatial and temporal distribution of Archaea in the North Sea in relation to environmental variables. FEMS Microbiol Ecol. 2007;62:242–257. doi: 10.1111/j.1574-6941.2007.00397.x. [DOI] [PubMed] [Google Scholar]
- Herndl GJ, Reinthaler T, Teira E, van Aken H, Veth C, Pernthaler A, et al. Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl Environ Microbiol. 2005;71:2303–2309. doi: 10.1128/AEM.71.5.2303-2309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Gilna P, Li W. Identification of ribosomal RNA genes in metagenomic fragments. Bioinformatics. 2009;25:1338–1340. doi: 10.1093/bioinformatics/btp161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugoni M, Taib N, Debroas D, Domaizon I, Jouan Dufournel I, Bronner G, et al. Structure of the rare archaeal biosphere and seasonal dynamics of active ecotypes in surface coastal waters. Proc Natl Acad Sci USA. 2013;110:6004–6009. doi: 10.1073/pnas.1216863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman J, Pham Thi NN, Karl DM, Sommeijer B. Reduced mixing generates oscillations and chaos in the oceanic deep chlorophyll maximum. Nature. 2006;439:322–325. doi: 10.1038/nature04245. [DOI] [PubMed] [Google Scholar]
- Hutchins AM, Holden JF, Adams MW. Phosphoenolpyruvate synthetase from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 2001;183:709–715. doi: 10.1128/JB.183.2.709-715.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Ono H, Abe-Yoshizumi R, Yoshizawa S, Ito H, Kogure K, et al. A light-driven sodium ion pump in marine bacteria. Nat Commun. 2013;4:1678. doi: 10.1038/ncomms2689. [DOI] [PubMed] [Google Scholar]
- Iverson V, Morris RM, Frazar CD, Berthiaume CT, Morales RL, Armbrust EV. Untangling genomes from metagenomes: revealing an uncultured class of marine Euryarchaeota. Science. 2012;335:587–590. doi: 10.1126/science.1212665. [DOI] [PubMed] [Google Scholar]
- Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- Kashtan N, Roggensack SE, Rodrigue S, Thompson JW, Biller SJ, Coe A, et al. Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science. 2014;344:416–420. doi: 10.1126/science.1248575. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Braff J, Karl DM, DeLong EF. Comparative metagenomic analysis of a microbial community residing at a depth of 4,000 meters at station ALOHA in the North Pacific subtropical gyre. Appl Environ Microbiol. 2009;75:5345–5355. doi: 10.1128/AEM.00473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J, Celepli N, Ininbergs K, Dupont CL, Yooseph S, Bergman B, et al. Picocyanobacteria containing a novel pigment gene cluster dominate the brackish water Baltic Sea. ISME J. 2014;8:1892–1903. doi: 10.1038/ismej.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassak K, Ghosh A, Albers SV. Diversity, assembly and regulation of archaeal type IV pili-like and non-type-IV pili-like surface structures. Res Microbiol. 2012;163:630–644. doi: 10.1016/j.resmic.2012.10.024. [DOI] [PubMed] [Google Scholar]
- Lê S, Josse J, Husson F. FactoMineR: an R package for multivariate analysis. J Stat Software. 2008;25:1–18. [Google Scholar]
- Lebrun E, Santini JM, Brugna M, Ducluzeau AL, Ouchane S, Schoepp-Cothenet B, et al. The Rieske protein: a case study on the pitfalls of multiple sequence alignments and phylogenetic reconstruction. Mol Biol Evol. 2006;23:1180–1191. doi: 10.1093/molbev/msk010. [DOI] [PubMed] [Google Scholar]
- Lesniewski RA, Jain S, Anantharaman K, Schloss PD, Dick GJ. The metatranscriptome of a deep-sea hydrothermal plume is dominated by water column methanotrophs and lithotrophs. ISME J. 2012;6:2257–2268. doi: 10.1038/ismej.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Perez M, Gonzaga A, Rodriguez-Valera F. Genomic diversity of ‘deep ecotype' Alteromonas macleodii isolates. Evidence for pan-Mediterranean clonal frames. Genome Biol Evol. 2013;5:1220–1232. doi: 10.1093/gbe/evt089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Galperin MY, Grishin NV, Tatusov RL, Wolf YI, et al. Comparative genomics of the Archaea (Euryarchaeota): evolution of conserved protein families, the stable core, and the variable shell. Genome Res. 1999;9:608–628. [PubMed] [Google Scholar]
- Makarova KS, Koonin EV. Comparative genomics of Archaea: how much have we learned in six years, and what's next. Genome Biol. 2003;4:115. doi: 10.1186/gb-2003-4-8-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Cuadrado AB, Rodriguez-Valera F, Moreira D, Alba JC, Ivars-Martinez E, Henn MR, et al. Hindsight in the relative abundance, metabolic potential and genome dynamics of uncultivated marine archaea from comparative metagenomic analyses of bathypelagic plankton of different oceanic regions. ISME J. 2008;2:865–886. doi: 10.1038/ismej.2008.40. [DOI] [PubMed] [Google Scholar]
- Martinez-Espinosa RM, Dridge EJ, Bonete MJ, Butt JN, Butler CS, Sargent F, et al. Look on the positive side! The orientation, identification and bioenergetics of 'Archaeal' membrane-bound nitrate reductases. FEMS Microbiol Lett. 2007;276:129–139. doi: 10.1111/j.1574-6968.2007.00887.x. [DOI] [PubMed] [Google Scholar]
- Martinez A, Tyson GW, Delong EF. Widespread known and novel phosphonate utilization pathways in marine bacteria revealed by functional screening and metagenomic analyses. Environ Microbiol. 2010;12:222–238. doi: 10.1111/j.1462-2920.2009.02062.x. [DOI] [PubMed] [Google Scholar]
- Massana R, Murray AE, Preston CM, DeLong EF. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl Environ Microbiol. 1997;63:50–56. doi: 10.1128/aem.63.1.50-56.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massana R, DeLong EF, Pedros-Alio C. A few cosmopolitan phylotypes dominate planktonic archaeal assemblages in widely different oceanic provinces. Appl Environ Microbiol. 2000;66:1777–1787. doi: 10.1128/aem.66.5.1777-1787.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattar S, Scharf B, Kent SB, Rodewald K, Oesterhelt D, Engelhard M. The primary structure of halocyanin, an archaeal blue copper protein, predicts a lipid anchor for membrane fixation. J Biol Chem. 1994;269:14939–14945. [PubMed] [Google Scholar]
- Mizuno CM, Rodriguez-Valera F, Kimes NE, Ghai R. Expanding the marine virosphere using metagenomics. PLoS Genet. 2013;9:e1003987. doi: 10.1371/journal.pgen.1003987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D, Rodriguez-Valera F, Lopez-Garcia P. Analysis of a genome fragment of a deep-sea uncultivated Group II euryarchaeote containing 16S rDNA, a spectinomycin-like operon and several energy metabolism genes. Environ Microbiol. 2004;6:959–969. doi: 10.1111/j.1462-2920.2004.00644.x. [DOI] [PubMed] [Google Scholar]
- Nawrocki EP. Structural RNA Homology Search and Alignment using Covariance ModelsPhD thesis, Washington University in Saint Louis, School of Medicine: St Louis, MO, USA 2009.
- Nitschke W, van Lis R, Schoepp-Cothenet B, Baymann F. The “green” phylogenetic clade of Rieske/cytb complexes. Photosynth Rese. 2010;104:347–355. doi: 10.1007/s11120-010-9532-1. [DOI] [PubMed] [Google Scholar]
- Ottesen EA, Young CR, Eppley JM, Ryan JP, Chavez FP, Scholin CA, et al. Pattern and synchrony of gene expression among sympatric marine microbial populations. Proc Natl Acad Sci USA. 2013;110:E488–E497. doi: 10.1073/pnas.1222099110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Leung HC, Yiu SM, Chin FY. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- Pernthaler A, Preston CM, Pernthaler J, DeLong EF, Amann R. Comparison of fluorescently labeled oligonucleotide and polynucleotide probes for the detection of pelagic marine bacteria and archaea. Appl Environ Microbiol. 2002;68:661–667. doi: 10.1128/AEM.68.2.661-667.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaiser A, Zivanovic Y, Moreira D, Lopez-Garcia P. Comparative metagenomics of bathypelagic plankton and bottom sediment from the Sea of Marmara. ISME J. 2011;5:285–304. doi: 10.1038/ismej.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J, Korbel JO, Lercher MJ, von Mering C, Bork P. Prediction of effective genome size in metagenomic samples. Genome Biol. 2007;8:R10. doi: 10.1186/gb-2007-8-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reysenbach AL, Liu Y, Banta AB, Beveridge TJ, Kirshtein JD, Schouten S, et al. A ubiquitous thermoacidophilic archaeon from deep-sea hydrothermal vents. Nature. 2006;442:444–447. doi: 10.1038/nature04921. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Rich VI, Konstantinidis K, DeLong EF. Design and testing of ‘genome-proxy' microarrays to profile marine microbial communities. Environ Microbiol. 2008;10:506–521. doi: 10.1111/j.1462-2920.2007.01471.x. [DOI] [PubMed] [Google Scholar]
- Riedel T, Tomasch J, Buchholz I, Jacobs J, Kollenberg M, Gerdts G, et al. Constitutive expression of the proteorhodopsin gene by a flavobacterium strain representative of the proteorhodopsin-producing microbial community in the North Sea. Appl Environ Microbiol. 2010;76:3187–3197. doi: 10.1128/AEM.02971-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinta-Kanto JM, Sun S, Sharma S, Kiene RP, Moran MA. Bacterial community transcription patterns during a marine phytoplankton bloom. Environ Microbiol. 2012;14:228–239. doi: 10.1111/j.1462-2920.2011.02602.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B, Pasic L, Thingstad TF, Rohwer F, et al. Explaining microbial population genomics through phage predation. Nat Rev Microbiol. 2009;7:828–836. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- Ruepp A, Graml W, Santos-Martinez ML, Koretke KK, Volker C, Mewes HW, et al. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature. 2000;407:508–513. doi: 10.1038/35035069. [DOI] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, et al. The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satinsky BM, Zielinski BL, Doherty M, Smith CB, Sharma S, Paul JH, et al. The Amazon continuum dataset: quantitative metagenomic and metatranscriptomic inventories of the Amazon River plume, June 2010. Microbiome. 2014;2:17. doi: 10.1186/2049-2618-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedile F, Messina E, La Cono V, Tsoy O, Monticelli LS, Borghini M, et al. Metagenomic analysis of hadopelagic microbial assemblages thriving at the deepest part of Mediterranean Sea, Matapan-Vavilov Deep. Environ Microbiol. 2012;3:1462–2920. doi: 10.1111/j.1462-2920.2012.02827.x. [DOI] [PubMed] [Google Scholar]
- Smith MW, Zeigler Allen L, Allen AE, Herfort L, Simon HM. Contrasting genomic properties of free-living and particle-attached microbial assemblages within a coastal ecosystem. Front Microbiol. 2013;4:120. doi: 10.3389/fmicb.2013.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart FJ, Ulloa O, DeLong EF. Microbial metatranscriptomics in a permanent marine oxygen minimum zone. Environ Microbiol. 2012;14:23–40. doi: 10.1111/j.1462-2920.2010.02400.x. [DOI] [PubMed] [Google Scholar]
- Swan BK, Chaffin MD, Martinez-Garcia M, Morrison HG, Field EK, Poulton NJ, et al. Genomic and metabolic diversity of Marine Group I Thaumarchaeota in the mesopelagic of two subtropical gyres. PLoS One. 2014;9:e95380. doi: 10.1371/journal.pone.0095380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, et al. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LR, Field C, Romanuk T, Ngugi D, Siam R, El Dorry H, et al. Patterns of ecological specialization among microbial populations in the Red Sea and diverse oligotrophic marine environments. Ecol Evol. 2013;3:1780–1797. doi: 10.1002/ece3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden B, Plagens A, Dorr C, Siebers B, Hensel R. Phosphoenolpyruvate synthetase and pyruvate, phosphate dikinase of Thermoproteus tenax: key pieces in the puzzle of archaeal carbohydrate metabolism. Mol Microbiol. 2006;60:287–298. doi: 10.1111/j.1365-2958.2006.05098.x. [DOI] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.