Abstract
The ‘bacterial switch' is a proposed regulatory point in the global sulfur cycle that routes dimethylsulfoniopropionate (DMSP) to two fundamentally different fates in seawater through genes encoding either the cleavage or demethylation pathway, and affects the flux of volatile sulfur from ocean surface waters to the atmosphere. Yet which ecological or physiological factors might control the bacterial switch remains a topic of considerable debate. Here we report the first field observations of dynamic changes in expression of DMSP pathway genes by a single marine bacterial species in its natural environment. Detection of taxon-specific gene expression in Roseobacter species HTCC2255 during a month-long deployment of an autonomous ocean sensor in Monterey Bay, CA captured in situ regulation of the first gene in each DMSP pathway (dddP and dmdA) that corresponded with shifts in the taxonomy of the phytoplankton community. Expression of the cleavage pathway was relatively greater during a high-DMSP-producing dinoflagellate bloom, and expression of the demethylation pathway was greater in the presence of a mixed diatom and dinoflagellate community. These field data fit the prevailing hypothesis for bacterial DMSP gene regulation based on bacterial sulfur demand, but also suggest a modification involving oxidative stress response, evidenced as upregulation of catalase via katG, when DMSP is demethylated.
Introduction
Dimethylsulfoniopropionate (DMSP) released from phytoplankton into seawater is consumed by bacteria within hours to days (Kiene and Linn, 2000) using one of two primary degradation pathways. While both pathways provide carbon and energy to bacterial cells, the cleavage pathway produces dimethylsulfide (DMS), a volatile precursor of atmospheric aerosols and potential condensation nuclei (Andreae, 1990), and the demethylation pathway provides reduced sulfur for biosynthesis of sulfur-containing amino acids (Kiene et al., 2000). The regulatory point between these two pathways determines the fate of DMSP-sulfur in the ocean and atmosphere and has been termed the ‘bacterial switch' (Simó, 2001). Factors proposed to influence this regulatory juncture include bacterial sulfur demand (González et al.,1999; Kiene et al., 2000; Simó, 2001), ultraviolet light stress (Slezak et al., 2007; Levine et al., 2012) and osmolyte requirements (Kiene et al., 2000; Reisch et al., 2008), but thus far none has satisfactorily explained the large variations in DMSP transformation patterns observed over time and space in the ocean. Although DMS emission to the atmosphere accounts for only a small fraction of the DMSP synthesized annually by marine phytoplankton (<2% of the >2000 Tg DMSP-S y−1; Moran et al., 2012), it contributes ∼40% of the atmospheric S burden (Simó, 2001) and impacts atmospheric chemistry both directly and through its degradation products (Kirkby et al., 2011; Quinn and Bates, 2011; Chen and Jang, 2012). Future changes in the environmental conditions that regulate these pathways (Six et al., 2013) have the potential to alter the radiation status of Earth.
In the coastal upwelling system of Monterey Bay, CA, previous metagenomic and metatranscriptomic surveys indicated consistent and abundant populations of a bacterial taxon with high genetic relatedness to Roseobacter strain HTCC2255 (Ottesen et al., 2011), a bacterium capable of both DMSP cleavage and demethylation (Newton et al., 2010). We took advantage of the predictable presence of this bacterium to design HTC2255-specific PCR primers for in situ observations for the first committed step in each of the two DMSP degradation pathways. The primer sets were employed on an Environmental Sample Processor (ESP), an autonomous instrument capable of performing qPCR analyses at sea (Preston et al., 2011) that was moored in Monterey Bay during a month-long period in October 2010, obtaining the first data on temporal DMSP regulation patterns in a single organism embedded in its natural environment.
Materials and methods
Metagenomic-based primer design
Marine metagenomic sequence data from Monterey Bay, CA (CAMERA accession CAM_PROJ_MontereyBay; 3 samples collected in October 2000, April 2001 and May 2001; http://camera.calit2.net/#; Rich et al., 2011) were mined for DMSP gene sequences by tBLASTn queries using full-length protein sequences with an E-value cutoff of 10−4. Hits were verified for the target annotation using BLASTx and a bit score cutoff of ⩾40 against NCBI RefSeq or to a >3000 member in-house database for dmdA (previously demonstrated to accurately distinguish dmdA from paralogs (Varaljay et al., 2010)). These sequences were used to design qPCR primer sets and 5′ nuclease probes (Supplementary Table S1) for the sequences with high similarity to Roseobacter strain HTCC2255 genes. The HTCC2255 dmdA gene falls outside the five defined clades based on Varaljay et al., 2010 and Howard et al., 2008, while the dddP gene falls into the Group A sequences of Peng et al., 2012. Archived DNA samples collected from Monterey Bay in 2006 and 2007 and during this study were used to confirm specificity of the probe/primer sets. An in silico specificity check was also carried out to determine if any primer/probe set would bind with non-target dmdA or dddP genes or with paralogs from the NCBI nucleotide (nt) database using NCBI Primer blast (http://www.ncbi.nlm.nih.gov/tools/primer-blast/).
Pre-deployment testing
Annealing temperature gradients and primer concentration matrices were used to determine optimal qPCR assay conditions when the HTCC2255-like dmdA and dddP genes were assayed on the ESP (Supplementary Table S1). qPCR assays consisted of 1 × Accuprime Supermix I (Invitrogen, Carlsbad, CA, USA), 300 nM hydrolysis probe labeled with FAM/BHQ-1 and 2.5 mM magnesium chloride final concentrations in 30 μl volumes. Reagents were loaded into the ESP and the qPCR assays were carried out as described previously (Preston et al., 2011; Robidart et al., 2011). Cycling conditions were as follows: 95 °C for 2.0 min, 42 cycles of 95 °C for 15 s and the specified annealing temperature for 1.0 min (see Supplementary Table S1). Pre-deployment standard curve efficiencies were 87–98%. Standards for qPCR assays were linearized clones with PCR product inserts. Cross-reactivity of standards at 107, 106 and 105 gene copies per reaction between primer/probe sets was minimal (⩽0.01% for any primer-standard pair). qPCR tests using surface seawater from the Coastal Data Information Program (CDIP) Station 156 in Monterey Bay and from Monterey Bay Wharf verified that quantification by all primer sets was comparable between the ESP module and a bench-top assay. A more detailed description of ESP procedures can be found elsewhere (Preston et al., 2009, 2011).
ESP data collection
The ESP was fitted with an SBE 16plus CTD (Sea-Bird, Bellevue, WA, USA) and a Turner Cyclops 7 fluorometer (Turner Designs, Sunnyvale, CA, USA) for depth, temperature, salinity and chlorophyll (Chl a) measurements and deployed near Station M0 (36.835 N, 121.901 W) at a depth of 8.1 m (±0.7). A mooring at station M0 provided water column data from SBE 37 MicroCAT CTD sensors at depths of 1, 15, 20, 40 and 55 m. Between September 28 and October 28, 2010, 15 discrete samples were collected for in situ DNA extraction and qPCR, and 19 samples were collected for archiving (Supplementary Table S2). For in situ extraction and real-time qPCR in the ESP, cells from 1 l of seawater were collected onto 0.2 μm pore size 25 mm diameter filters. Filtration times were <50 min, and pressure across the membrane was between 25 and 28 psi. Filters were extracted as previously described (Preston et al., 2011). Primer sets were run in single reactions for each time point using 6.0 μl of the extracted DNA as a template. Negative (no template) controls were run once during the deployment and showed no amplification. Negative control lysates (to check for residual contamination in the ESP DNA extraction and amplification system) were run before, after and at three other times during the deployment, and the amplification signal was always <5% of the environmental samples. An internal positive control reaction (template included in the primer/probe reagent) that was run 14 times showed that the control cycle threshold (Ct) value was consistent and did not indicate inhibition (average Ct value=29.01 (±0.35)). As it was not possible to run standard curves on the ESP during deployment, these were conducted post-deployment with the same reagents. For ESP archive samples, approximately 1–2 h after in situ DNA extraction, a second 1.0 l seawater sample was filtered in-line through 5.0 and 0.2 μm pore size 25 mm diameter filters, preserved with two 20-min sequential incubations of 2.0 ml of RNA later (Ambion Life Technologies, Carlsbad, CA, USA) and stored on board the ESP until recovery. Following the deployment, the archived filters were removed, flash frozen in liquid nitrogen and stored at −80 °C.
Niskin sample collection
Ship casts using an SBE 19plus SEACAT CTD (Sea-Bird) with 5.0 l Niskin bottles were used to collect water samples at an average depth of 9.2 m (±0.7) within 1 km of the ESP for supplementary molecular and chemical measurements. Water was returned to the lab within 2 h of collection and filtered for DNA extraction. Triplicate 200–500 ml volumes of seawater were filtered by vacuum filtration and filters stored at −80 °C.
Biochemical measurements were carried out in triplicate with subsamples from the same Niskin bottle. Chl a was measured from 200 ml of seawater filtered onto Whatman GF/F filters extracted in 5 ml 90% acetone at −20 °C and quantified by fluorometry (Pennington and Chavez, 2000). Samples for total DMSP (DMSPt) analysis were collected as whole seawater and preserved with HCl (1.5% final concentrations), while samples for dissolved DMSP (DMSPd) analysis were collected by small volume gravity drip filtration through a GF/F filter and then immediately vacuum filtered through a 0.2 μm nylon filter and preserved in H2SO4 (1% final concentration) (Kiene and Slezak, 2006; Slezak et al., 2007). The 0.2 filtration step removed any DMSP-containing bacteria from the filtrate. Particulate DMSP (DMSPp) concentrations were calculated as the difference between DMSPt and DMSPd concentrations and represent the amount of DMSP in phytoplankton cells. All DMSP measurements were made by cleaving DMSP into DMS with strong alkali and quantifying DMS by gas chromatography.
Slides for phytoplankton taxonomic analysis were made on eight dates during the deployment by filtering 10–25 ml of whole seawater onto 0.2 μM black polycarbonate filters, preserving with 0.5% glutaraldehyde and freezing at −20 °C. Cells were counted under epifluorescence microscopy, and cell size, shape and volume were used to calculate microgram phytoplankton carbon per liter (see below for calculation details). For heterotrophic bacterial counts, 1.8 ml of whole seawater was preserved with a final concentration of 0.4% paraformaldehyde and flash frozen in liquid nitrogen. Samples were stored at −80 °C until analysis on a Beckman Coulter (Brea, CA, USA) Altra flow cytometer for simultaneous detection of DNA (Hoechst-stained cells, 1 μg ml−1 final concentration), pigments (to exclude cyanobacteria), and forward and 90° light scatter (Monger and Landry, 1993).
Post-deployment extractions, qPCR and RT-qPCR
DNA was extracted from filters obtained by Niskin bottle sampling (Supplementary Table S2) using a modified version of the Qiagen DNAeasy protocol that mimics the method used in the ESP (Preston et al., 2011). RNA was extracted from the RNAlater-preserved ESP archive filters using a modification of the AllPrep DNA/RNA mini extraction kit (Qiagen, Valencia, CA, USA) using bead-beating. RNA extracts were DNase digested using the Turbo DNA-free kit (Ambion) with double the enzyme volume to ensure no DNA contamination. Laboratory qPCR assays of DMSP genes and transcripts used similar conditions and primer/probe concentrations as for the ESP (Supplementary Table S1). Primers were designed for five additional HTCC2255 genes: downstream demethylation pathway gene dmdB (methylymercaptopropionate CoA-ligase; Reisch et al., 2011), downstream cleavage pathway genes prpE and acuI (propionyl-CoA synthetase and acrylate utilization protein; Todd et al., 2012), and reactive oxygen species genes sodD (superoxide dismutase) and katG (catalase) (Supplementary Table S1), and qPCR was carried out for all genes on an iCycler iQ or iCycler iQ5 (Bio-Rad, Hercules, CA, USA). For reverse-transcription (RT)-qPCR, the Invitrogen OneStep Express kit (SuperScript III) or SYBR One-step supermix for specific priming of cDNA synthesis was used in 25 μl final volumes. DNA template was added in a 1:10 dilution and RNA template in 1:10 or 1:20 dilutions. Tenfold serially diluted standard curves representing 101 to 107 copies per reaction were included on every plate, along with triplicate no-template control reactions. Reactions without reverse transcriptase confirmed the absence of genomic DNA contamination. Laboratory standard curve efficiencies were 90%–103% and R2 values were >0.99. Size and specificity of the qPCR and RT-qPCR products were verified by agarose gel electrophoresis or melt curve analysis, and the limit of detection for quantification was ∼10 gene copies per reaction for both dmdA and dddP. One of the last ESP archive filters collected on October 28 when battery power was low had poor RNA yield and was removed from further analysis.
DMSP-carbon calculations
The percent of phytoplankton carbon present in the form of DMSP was calculated from the DMSPp values and chlorophyll a (Chl a) data. Chl a data were converted to carbon by assuming 80 μg C per microgram Chl a (Banse, 1977). The Chl a-based carbon estimates were compared with those made on selected dates by direct microscopic counting and sizing of phytoplankton cells, and found to agree well; Chl a-based data averaged 60% of microscopy data (range: 36–85%) and were significantly correlated (R=0.95, n=8).
16S rRNA sequencing and analysis
DNA samples from ESP archived filters were used in triplicate PCR amplifications with bacterial 16S rRNA primers (Bakt_341F and Bakt_805R) over 25 cycles as per Herlemann et al., 2011 to quantify the relative abundance of Roseobacter HTCC2255 16S rRNA genes during the deployment. The primers were modified with 454 Titanium adaptors and sample-specific 5-bp barcodes. PCR assays used the Phusion high-fidelity DNA polymerase (Thermo Fisher Scientific, Waltham, MA, USA) and 0.5 μM final concentrations of each primer. Following PCR, amplicons were purified with Agencourt Ampure XP (Beckman Coulter) using a 1:1 volume of PCR product to Ampure XP beads, quantified with Quant-iT PicoGreen (Invitrogen), and pooled in equal concentration for Roche/454 Titanium sequencing at the Georgia Genomics Facility (University of Georgia).
Sequences of 16S rRNA were analyzed using the QIIME pipeline (Caporaso et al., 2010) downloaded from http://www.qiime.org/. Sequences without perfect matches to primer and barcode sequences were removed, and remaining sequences were separated by barcode ID and denoised using AmpliconNoise (Quince et al., 2011). Sequences of 16S rRNA were clustered into operational taxonomic units (OTUs) based on a 97% sequence similarity, and taxonomy was assigned using the Greengenes classifier using the latest build (gg_otus_4feb2011). Reference sequences from each OTU were also compared with a marine 16S rRNA sequence custom database according to Biers et al., 2009 using Smith-Waterman pairwise alignments (Smith and Waterman, 1981) and requiring sequence overlaps of ⩾80%. HTCC2255 sequences were identified based on an alignment with the full-length 16S rRNA sequence available in the HTCC2255 metagenomic assembly of Iverson et al., 2012 because only a partial sequence was assembled in the genome of the original HTCC2255 culture. If possible, sequences were assigned to species level taxa with ⩾97% identity across the overlap. Archaea, chloroplast and unassigned (those not classified to the kingdom level; <2% of the total) sequences were removed before analyses of taxonomic structure.
Results and Discussion
Roseobacter HTCC2255 DMSP genes in Monterey Bay
The abundance of DMSP genes in populations related to Roseobacter strain HTCC2255 was measured autonomously over a month-long period in the upwelling system of Monterey Bay, CA, USA. Several previous metagenomic and metatranscriptomic studies had indicated that bacterial populations with high genetic relatedness to HTCC2255 (⩾95% average amino acid identity) were consistently present at this site (DeLong, 2005; Ottesen et al., 2011; Rich et al., 2011). We therefore retrieved DMSP gene sequences from the available Monterey Bay metagenomic data sets (Rich et al., 2011) and designed species-specific PCR assays that could be employed on the ESP to characterize gene dynamics over time. For dmdA, the gene encoding the first step in the demethylation pathway (Howard et al., 2006), 21% of the Monterey Bay hits had highest homology to the HTCC2255 gene with an average identity of 97%. For dddP, the gene encoding the first step in the cleavage pathway (Todd et al., 2009), 43% of the hits had highest homology to HTCC2255, also with an average identity of 97%. qPCR primer sets and corresponding 5′ nuclease probes specific to the Monterey Bay populations of HTCC2255 were designed from these sequences (Supplementary Figure S1, Supplementary Table S1). Pre-deployment tests of specificity using previously archived DNA from Monterey Bay surface water showed that amplicons from dmdA and dddP primer sets (n=12) matched the HTCC2255 reference sequences (Supplementary Figure S1). When the primer/probe sets were subjected to an in silico specificity check against the NCBI nucleotide (nt) database, no unintended matches were found, either for paralogs or for the correct homologs of dmdA and dddP in non-target bacterial groups.
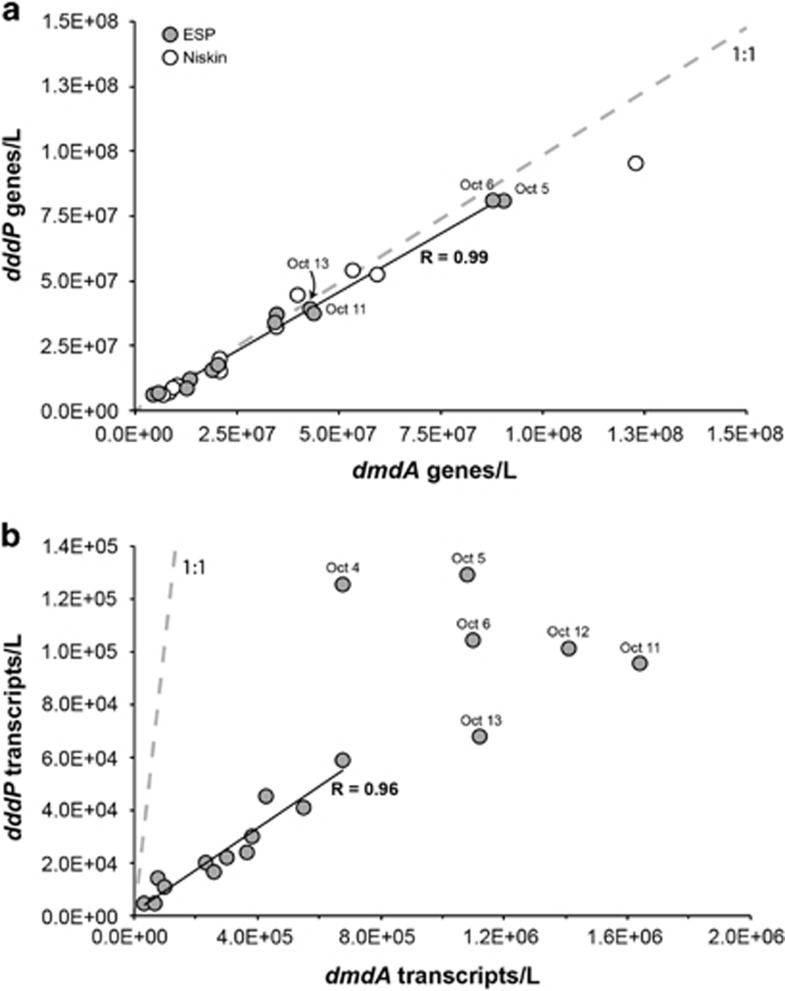
Following primer design and testing, the in situ qPCR analysis was carried out on the ESP between September 28 and October 28, 2010 while the instrument was moored at Station M0 at a depth of ∼8 m. Analysis of DNA extracted onboard the instrument from ∼1 × 109 bacterial cells per sample collected by filtration from ∼1 l seawater showed that dmdA and dddP were present in M0 surface water in a 1:1 stoichiometery, and that they varied in a tightly coordinated manner throughout the deployment (Figure 1a; R=0.99). This was the expected outcome for genes present in single copy in the same bacterial genome. Because battery power and reagent reservoir constraints on the 2010 version of the ESP limited the in situ gene quantification to 15 non-replicated time points over the 34-day deployment, the autonomous sampling was augmented on 11 occasions with discrete Niskin grab samples collected manually at the M0 mooring and processed in the laboratory. Equal abundances of dmdA and dddP genes were also found in the Niskin water samples, and these matched the ESP-based abundances well (Figure 1a). Using DMSP gene counts as proxies for genome counts and assuming one genome per cell, abundance of HTCC2255 at the deployment site averaged 3.4 × 107 cells l−1 but varied 16-fold over the course of the deployment. Comparing this number to the flow cytometric counts of total heterotrophic bacterioplankton at the site, we calculate that HTCC2255-like cells comprised between 0.2% and 4% of the bacterial community during the deployment (Supplementary Figure S2A). Other taxa dominant at the site were typical of coastal bacterioplankton communities and included Alphaproteobacteria in the SAR11 and SAR116 lineages and members of the Gammaproteobacteria and Bacteroidetes (Supplementary Figure S2B).
Figure 1.
(a) Gene counts for in situ ESP (gray circles) and Niskin (white circles) samples and (b) transcript counts for ESP archived filters. In situ ESP data are missing in panel a for October 4 (neither gene was measured), October 12 (dmdA was not measured) and October 28–30 (ESP battery drain). The correlation coefficient in panel a is for in situ ESP data only. Including the Niskin data also resulted in a correlation coefficient of 0.99. The correlation in panel b is for samples outside the October 4–14 period only. Including all samples resulted in a correlation coefficient of 0.85. Gray dashed lines in both panels indicate the 1:1 line.
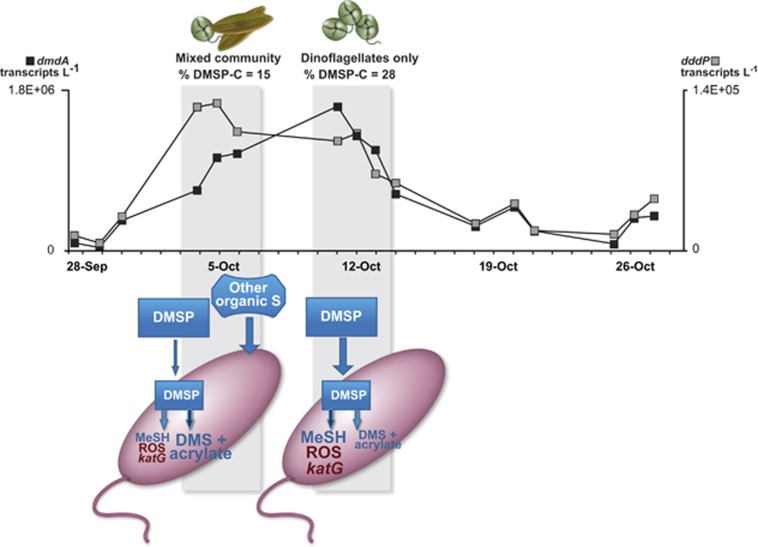
Additional filters archived by the ESP at the majority of the in situ qPCR time points were extracted for RNA post-deployment and used in reverse-transcription qPCR to track DMSP gene expression over time. dmdA transcripts were ∼14-fold more abundant than dddP transcripts (5.8 × 105 versus 5.1 × 104 l−1; Figure 1), a difference that may indicate higher inventories of DmdA compared with DddP in the protein pool of HTCC2255 cells, or may reflect different half-lives of the transcripts or proteins. In any case, it is consistent with earlier comparisons of DMSP gene transcripts in seawater (Levine et al., 2012; Varaljay et al., 2012). Although different in absolute numbers, transcript abundances (Figure 1b; R=0.96) and expression ratios (Supplementary Figure S3A) showed similar patterns for the two genes and were significantly correlated at the beginning (September 28–30) and end of the deployment (October 20–27). However, transcripts from the two genes varied independently from October 4 to 14, when dddP transcript numbers peaked first (October 4–6) and dmdA transcript numbers peaked later (October 11–14) (Figure 2). This period of decoupled expression was also the time of greatest abundance of HTCC2255 at the M0 mooring, with cell numbers estimated at 108 l−1 and accounting for up to 4% of the heterotrophic bacteria.
Figure 2.
Modified version of the bacterial sulfur demand conceptual model applied to Monterey Bay, CA, USA populations of Roseobacter HTCC2255 showing differential regulation of DMSP demethylation and cleavage pathways. Gray shading indicates the periods of decoupled gene expression, dominated first by a mixed diatom (Pseudo-nitzchia australis) and dinoflagellate (Prorocentrum micans) community, and then by a P. micans only community. Black squares indicate dmdA transcript counts (left axis); gray squares indicate dddP transcript counts (right axis). The size of the lettering in the cartoon shows hypothesized relative internal concentrations of degradation product methanethiol (MeSH) and reactive oxygen species (ROS) and the relative level of katG expression. Darker arrow indicates an upregulated pathway. The modified model hypothesizes that when DMSP accounts for a large percentage of phytoplankton production (October 11–14), dmdA is upregulated to bias metabolism towards the demethylation pathway, satisfying the cells' sulfur requirements but producing H2O2; katG upregulation then furnishes ROS protection to the cell. When DMSP accounts for a small percentage of phytoplankton C (October 4–6) and alternate organic S compounds are present, dddP is upregulated to bias metabolism towards the cleavage pathway, satisfying the cells' requirements with non-DMSP organic sulfur and avoiding ROS stress generated during demethylation.
Because evidence of the decoupled expression period was based on only six samples that were unreplicated due to ESP constraints, we also checked expression levels of HTCC2255 genes metabolically downstream of dmdA and dddP to evaluate the robustness of the observation. Demethylation pathway gene dmdB (methylymercaptopropionate CoA-ligase; Reisch et al., 2011) and cleavage pathway genes prpE and acuI (propionyl-CoA synthetase and acrylate utilization protein; Todd et al., 2012) were present in abundances equal to dmdA and dddP (Supplementary Figure S4), consistent with expectations for single-copy genes in the same genomes. The transcription of these genes during the October 4–14 decoupled expression period for dmdA and dddP was correlated within a pathway but not between competing pathways. That is, statistically significant correlations were found between transcript numbers for dmdB and dmdA (R=0.73) and between prpE and dddP (R=0.92) and acuI and dddP (R=0.81), but not between transcript numbers for genes in different pathways (Table 1). Transcript patterns for all genes in the two pathways were well coordinated outside of this decoupled period, and correlations calculated over the full deployment were all statistically significant (Table 1).
Table 1. Pearson correlation coefficients for transcript copy numbers of HTCC2255 genes during the full deployment (September 28–October 27, 2010) and during the period of uncoupled regulation only (October 4–14).
|
Full deployment |
Uncoupled regulation period |
|||
|---|---|---|---|---|
| dmdA | dddP | dmdA | dddP | |
| dmdB | 0.95* | 0.82* | 0.73* | 0.17 |
| prpE | 0.80* | 0.95* | 0.08 | 0.92* |
| acuI | 0.89* | 0.96* | 0.51 | 0.81* |
| sodD | 0.90* | 0.88* | 0.49 | 0.24 |
| katG | 0.90* | 0.81* | 0.73* | 0.26 |
Asterisks indicate significant positive correlations (P<0.05).
Phytoplankton dynamics in Monterey Bay
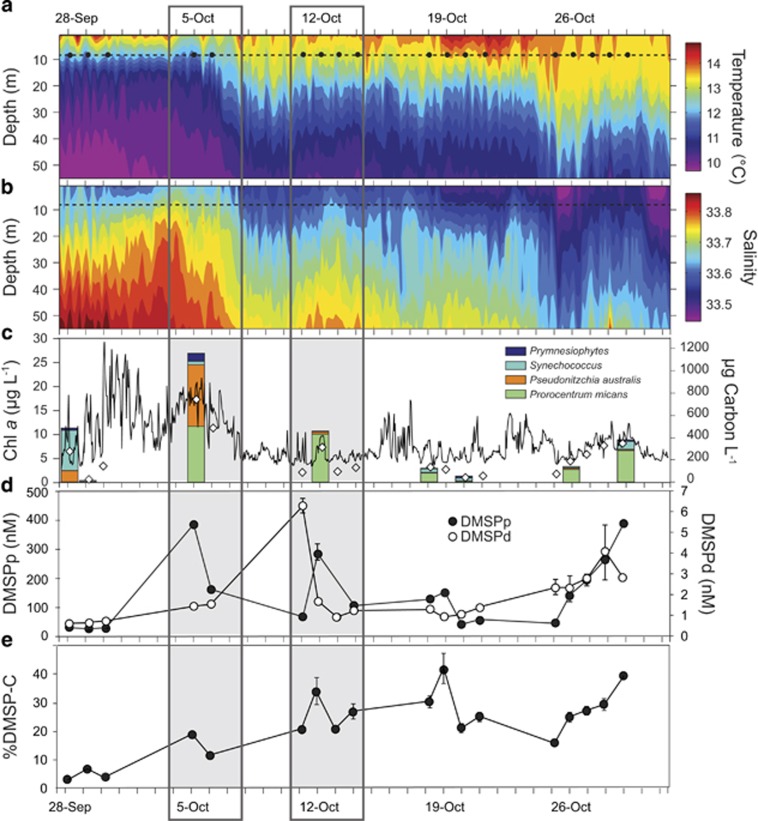
Phytoplankton composition changed through the deployment, shifting between communities dominated by cyanobacteria, diatoms and dinoflagellates in a manner consistent with known responses to changing hydrography in the region (Ryan et al., 2014) (Figure 3). A Synechococcus-dominated phase from September 28 to 30 was characterized by low concentrations of chlorophyll (Chl a), DMSPp (particulate DMSP within phytoplankton cells) and DMSPd (dissolved DMSP in seawater) (Figure 3). The October 4–14 time period of uncoupled DMSP gene expression was marked first by a mixed phytoplankton community consisting of the diatom Pseudo-nitzchia australis (48% of phytoplankton C) and the dinoflagellate Prorocentrum micans (43% of phytoplankton C), the former being a poor DMSP producer and the latter a strong DMSP producer (Keller et al., 1989); this period coincided with a signal of upwelled waters and was marked by high Chl a and DMSPp concentrations (October 4–7). Following a shift to warmer and fresher waters later in this period (October 11–14) (Figure 3), the phytoplankton community consisted nearly exclusively of P. micans (94% of phytoplankton C). Near the end of the deployment (October 26–29), a further freshening and destratification of the water column was associated with slight increases in Chl a, DMSPp and DMSPd concentrations in another P. micans-dominated community (Figure 3); few gene abundance or expression measurements were available during this last phase due to low battery power of the ESP.
Figure 3.
(a, b) Time series of water column temperature and salinity collected by CTD sensors on the M0 mooring. The dotted lines indicate the deployment depth of the ESP and black dots in panel a indicate molecular sampling events. (c) Concentration of Chlorophyll a (Chl a) collected by a CTD instrument deployed on the ESP (solid line), and discrete measurements of Chl a (gray diamonds) and phytoplankton biomass (stacked bars, color coded by major species) obtained from Niskin bottle collections near the ESP mooring. (d, e) Concentrations of particulate DMSP (DMSPp), dissolved DMSP (DMSPd) and DMSP carbon as a percent of phytoplankton carbon obtained from the Niskin collections. Dark gray boxes identify periods of uncoupled gene expression.
Does the sulfur demand hypothesis fit?
Wide variation in the percent of dissolved DMSP converted to DMS in ocean surface waters (3% to 30% Kiene and Linn, 2000) has most frequently been attributed to the energetic benefit to bacteria of acquiring pre-reduced sulfur rather than expending three ATP equivalents to reduce seawater sulfate (Kiene et al., 2000; Pinhassi et al., 2005; Moran et al., 2012). This ‘bacterial sulfur demand' hypothesis proposes that demethylation is favored by marine bacteria when reduced sulfur is limiting because the sulfur from DMSP can be assimilated into biomass through the demethylation pathway (Kiene et al., 1999) but not the cleavage pathway (Todd et al., 2009). However, sulfur may also be available to bacteria from organic compounds other than DMSP (Weinitschke et al., 2006; Reisch et al., 2011; Durham et al., 2014). Thus, the hypothesis proposes that when DMSP is the predominant source of available organic sulfur (relative to other organic sulfur compounds, not based on absolute concentrations; Pinhassi et al., 2005), bacteria preferentially use the demethylation pathway to meet their biosynthetic requirements; but when alternate organic sulfur compounds are more available, the cleavage pathway is favored because DMSP-sulfur is no longer necessary for biosynthesis (Pinhassi et al., 2005; Merzouk et al., 2006, 2008; Lizotte et al., 2009; Rinta-Kanto et al., 2011).
The availability of DMSP to marine bacteria is influenced by the taxonomy of the dominant phytoplankton because the percent contribution of DMSP to carbon in phytoplankton (%DMSP-C; Kiene and Linn, 2000) varies among taxonomic groups. By major group, cultured cyanobacterial cells average <0.001%DMSP-C, diatoms average 0.4%, coccolithophorids average 5%, and dinoflagellates average 11% (Stefels et al., 2007), although within-group variation is quite high. Although this is not a direct measure of DMSP concentrations as a proportion of total bioavailable organic sulfur in phytoplankton cells, it serves as a proxy for that value. For example, diatom cells typically contain low %DMSP-C but synthesize sulfonates and other alternate organic sulfur compounds (Benson and Lee, 1972; Weinitschke et al., 2006; Merzouk et al., 2008; Boroujerdi et al., 2012; Durham et al., 2014). Accordingly, DMSP-sulfur typically makes up only ∼6% of the sulfur in diatom cells, but ∼40% in dinoflagellate cells (Park et al., 2014).
The relative importance of DMSP as a source of sulfur in Monterey Bay phytoplankton was therefore predicted based on %DMSP-C calculations by assuming a carbon:Chl a ratio of 80 (Banse, 1977) (Figure 3). %DMSP-C in phytoplankton biomass averaged 18% at the time of peak DMSP cleavage pathway expression occurring during the mixed diatom/dinoflagellate phase (October 4–6), and nearly doubled to 34% at the time of peak DMSP demethylation pathway expression during the dinoflagellate-only phase (October 11–14; Figure 3). Across the full deployment period, there was a weak (non-significant) positive relationship between the ratio of expression for genes of the two DMSP pathways (dmdA:dddP) and the calculated %DMSP-C in phytoplankton biomass (R=0.38; P=0.14; Supplementary Figure S3B). Overall, these observations are consistent with the predictions of the bacterial sulfur demand hypothesis that relative availability of DMSP controls the balance between competing degradation pathways in HTCC2255 populations, and that expression of genes in the demethylation pathway is favored when % DMSP-carbon is higher and therefore DMSP is the predominant source of bioavailable organic sulfur.
Is there more to the story?
A puzzling aspect of the sulfur demand hypothesis is that the diffusional loss of DMS that occurs in the cleavage pathway represents not only the release of superfluous sulfur, but also loss of reduced carbon and energy that is typically in short supply in ocean environments. Thus, the regulatory scheme implicit in the S demand hypothesis in which reduced carbon taken up by marine bacteria is subsequently released rather than catabolized is difficult to fully reconcile regardless of the relative abundance of DMSP in the environment. We therefore evaluated whole-genome transcriptional changes in a model cultured Roseobacter to ask whether a broader analysis of gene expression patterns might suggest alternative regulation hypotheses. Because the original HTCC2255 strain was lost from culture soon after isolation, experiments were carried out with Ruegeria pomeroyi DSS-3, a roseobacter relative of HTCC2255 that similarly possesses genes for both DMSP degradation pathways. When R. pomeroyi degraded DMSP in laboratory cultures, we noticed strong upregulation of katG, the gene encoding catalase (Supplementary Figure S5), indicating a possibility that reactive oxygen species (ROS), and specifically H2O2, may be generated during DMSP degradation. Although these studies were carried out for a pure culture exposed to DMSP levels several orders above typical ocean concentrations (80 μM versus low nM), they suggested that ROS-related gene transcript expression should be investigated in HTCC2255 populations in Monterey Bay.
HTCC2255-specific qPCR primer sets were designed for sodD (superoxide dismutase; a ROS protection gene not upregulated in the laboratory cultures of R. pomeroyi) and katG (Supplementary Table S1) and used to analyze RNA extracted from the archived ESP filters. Consistent with laboratory-grown R. pomeroyi, there was no indication that HTCC2255 sodD expression shifted in relation to expression of DMSP degradation pathways. However, there was a positive correlation of katG expression with dmdA (R=0.73, n=7; P=0.03) but not dddP (R=0.26, n=7) during the period when regulation expression was uncoupled (Table 1). This synchronous expression of catalase and demethylation genes by HTCC2255 is suggestive of a possible link between H2O2 stress and demethylation reactions, a relationship also suggested in studies of the Alphaproteobacterium Hyphomicrobium sp. EG, which generates H2O2 when oxidizing methanethiol (Suylen et al., 1986), a final product of the demethylation pathway (Supplementary Figure S4A) (Reisch et al., 2011). Although these data are preliminary, a role for ROS stress is also consistent with previous observations that UV light, another source of ROS, may have a role in pathway preference (Slezak et al., 2007; Levine et al., 2012). Follow-up studies are needed to determine whether including ROS stress as a possible regulatory factor can provide new insights into why releasing reduced carbon and sulfur in the form of DMS might be advantageous for bacterial survival under certain circumstances (Figure 2).
Insights from taxon-specific field observations
Surface ocean bacterioplankton presides over a divergence point in the marine sulfur cycle where the fate of DMSP is determined yet the regulation is poorly understood. Evaluating how well current hypotheses explain DMSP transformations has been hampered by the small number of studies addressing DMSP gene expression in the ocean (Vila-Costa et al., 2010, 2014; Levine et al., 2012) and the fact that most oceanographic data amalgamate transcriptional responses from multiple community members. During this month-long study spanning substantial shifts in physical conditions, phytoplankton community structure and DMSP concentrations in Monterey Bay waters, an autonomous ocean sensor successfully interrogated the activity of a single DMSP-degrading bacterial species embedded in, yet distinguishable from, the larger microbial community. In addition, it provided an opportunity to evaluate hypotheses of pathway regulation in an ocean setting.
Of the five major taxa of marine bacteria known to participate in DMSP degradation (Moran et al., 2012), three lineages are currently thought to carry demethylation genes but not cleavage genes, and thus while they consume dissolved DMSP and eliminate it from possible conversion to DMS, they probably cannot produce DMS directly; these include the SAR11, marine gammaproteobacterium OM60, and SAR324 lineages. In contrast, DMSP-degrading members of the Roseobacter and SAR116 lineages can, like HTCC2255, carry genes for both demethylation and cleavage (Newton et al., 2010; Moran et al., 2012), and therefore more directly influence the fate of DMSP. Emerging technological capabilities for conducting taxon-specific observations of gene regulation in the ocean are allowing us to explore, for the first time, the diversity of gene regulation strategies among the key bacterial groups affecting partitioning of organic sulfur between the ocean and atmosphere.
Acknowledgments
We thank J Birch, B Roman and S Jenson for technical expertise; E Demir, K Buck, F Chavez, T Pennington, M Blum, J Harvey and C Wahl for help with protocols and sampling; the Captain and crew of the R/V Zephyr and the R/V John Martin; B Carter for assistance with MEGAMER facilities; C Smith, K Turk, M Hogan and B Higgins for advice on qPCR processing; L Oswald for DMSP measurements; S Tanner and R Michisaki for phytoplankton enumeration; K Selph for flow cytometry measurements; S Bertilsson, D Herlemann and P Yager for advice on 16S rRNA primers; S Sharma, A Rivers and W Sheldon for bioinformatics assistance; and C English for graphics assistance. This work was funded in part by NSF OCE-1342694 (to MAM and RPK) and by the Gordon and Betty Moore Foundation through grants GBMF#538.01 (to MAM) and GBMF#1761 (to JPZ).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Andreae MO. Ocean-atmosphere interactions in the global biogeochemical sulfur cycle. Mar Chem. 1990;30:1–29. [Google Scholar]
- Banse K. Determining the carbon-to-chlorophyll ratio of natural phytoplankton. Marine Biol. 1977;41:199–212. [Google Scholar]
- Benson AA, Lee RF. The sulphoglycolytic pathway in plants. Biochem J. 1972;128:29P–30P. doi: 10.1042/bj1280029pb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biers EJ, Sun S, Howard EC. Prokaryotic genomes and diversity in surface ocean waters: interrogating the global ocean sampling metagenome. Appl Environ Microbiol. 2009;75:2221–2229. doi: 10.1128/AEM.02118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroujerdi AFB, Lee PA, DiTullio GF, Janech MG, Vied SB, Bearden DW. Identification of isethionic acid and other small molecule metabolites of Fragilariopsis cylindrus with nuclear magnetic resonance. Anal Bioanal Chem. 2012;404:777–784. doi: 10.1007/s00216-012-6169-2. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Jang M. Secondary organic aerosol formation from photooxidation of a mixture of dimethyl sulfide and isoprene. Atmos Environ. 2012;46:271–278. [Google Scholar]
- DeLong EF. Microbial community genomics in the ocean. Nat Rev Microbiol. 2005;3:459–469. doi: 10.1038/nrmicro1158. [DOI] [PubMed] [Google Scholar]
- Durham BP, Sharma S, Luo H, Smith CB, Amin SA, Bender SJ, et al. Cryptic carbon and sulfur cycling between surface ocean plankton. Proc Natl Acad Sci USA. 2014;112:453–457. doi: 10.1073/pnas.1413137112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González JM, Kiene RP, Moran MA. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the α-subclass of the class Proteobacteria. Appl Environ Microbiol. 1999;65:3810–3819. doi: 10.1128/aem.65.9.3810-3819.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlemann DPR, Labrenz M, Jurgens K, Bertilsson S, Waniek JJ, Andersson AF. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011;5:1571–1579. doi: 10.1038/ismej.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard EC, Henriksen JR, Buchan A, Reisch CR, Bürgmann H, Welsh R, et al. Bacterial taxa that limit sulfur flux from the ocean. Science. 2006;314:649–651. doi: 10.1126/science.1130657. [DOI] [PubMed] [Google Scholar]
- Howard EC, Sun S, Biers EJ, Moran MA. Abundant and diverse bacteria involved in DMSP degradation in marine surface waters. Environ Microbiol. 2008;10:2397–2410. doi: 10.1111/j.1462-2920.2008.01665.x. [DOI] [PubMed] [Google Scholar]
- Iverson V, Morris RM, Frazar CD, Berthiaume CT, Morales RL, Armbrust EV. Untangling genomes from metagenomes: revealing an uncultured class of marine Euryarchaeota. Science. 2012;335:587–590. doi: 10.1126/science.1212665. [DOI] [PubMed] [Google Scholar]
- Keller MD, Bellows WK, Guillard RRL.1989Dimethyl sulfide production in marine phytoplanktonIn: Saltzman E, Cooper WJ, (eds)Biogenic Sulfur in the Environment American Chemical Society: Washington, DC, USA; 167–182. [Google Scholar]
- Kiene RP, Linn LJ, Bruton JA. New and important roles for DMSP in marine microbial communities. J Sea Res. 2000;43:209–224. [Google Scholar]
- Kiene RP, Linn LJ, González JM, Moran MA, Bruton JA. Dimethylsulfoniopropionate and methanethiol are important precursors of methionine and protein-sulfur in marine bacterioplankton. Appl Environ Microbiol. 1999;65:4549–4558. doi: 10.1128/aem.65.10.4549-4558.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiene RP, Linn LJ. Distribution and turnover of dissolved DMSP and its relationship with bacterial production and dimethylsulfide in the Gulf of Mexico. Limnol Oceanogr. 2000;45:849–861. [Google Scholar]
- Kiene RP, Slezak D. Low dissolved DMSP concentrations in seawater revealed by small-volume gravity filtration and dialysis sampling. Limnol Oceanogr Methods. 2006;4:80–95. [Google Scholar]
- Kirkby J, Curtius J, Almeida J, Dunne E, Duplissy J, Ehrhart S, et al. Role of sulphuric acid, ammonia and galactic cosmic rays in atmospheric aerosol nucleation. Nature. 2011;476:429–433. doi: 10.1038/nature10343. [DOI] [PubMed] [Google Scholar]
- Levine NM, Varaljay VA, Toole DA, Dacey JWH, Doney SC, Moran MA. Environmental, biochemical, and genetic drivers of DMSP degradation and DMS production in the Sargasso Sea. Environ Microbiol. 2012;14:1210–1223. doi: 10.1111/j.1462-2920.2012.02700.x. [DOI] [PubMed] [Google Scholar]
- Lizotte M, Levasseur M, Kudo I, Suzuki I, Tsuda A, Kiene RP, et al. Iron-induced alterations of bacterial DMSP metabolism during SEEDS II. Deep-Sea Res Pt II Top Stud Oceanogr. 2009;56:2889–2898. [Google Scholar]
- Merzouk A, Scarratt M, Michaud S, Lizotte M, et al. Bacterial DMSP metabolism during the senescence of the spring diatom bloom in the Northwest Atlantic. Mar Ecol Prog Ser. 2008;369:1–11. [Google Scholar]
- Merzouk A, Levasseur M, Scarratt M, Michaud S, Rivkin R, Hale M, et al. DMSP and DMS dynamics during a mesoscale iron fertilization experiment in the Northeast Pacific - Part II: Biological cycling. Deep-Sea Res Pt II Top Stud Oceanogr. 2006;53:2370–2383. [Google Scholar]
- Monger BC, Landry MR. Flow cytometric analysis of marine bacteria with hoechst 33342. Appl Environ Microbiol. 1993;59:905–911. doi: 10.1128/aem.59.3.905-911.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran MA, Reisch CR, Kiene RP, Whitman WB. Genomic insights into bacterial DMSP transformations. Ann Rev Mar Sci. 2012;4:523–542. doi: 10.1146/annurev-marine-120710-100827. [DOI] [PubMed] [Google Scholar]
- Newton RJ, Griffin LE, Bowles KM, Meile C, Gifford SM, Givens CE, et al. Genome characteristics of a generalist marine bacterial lineage. ISME J. 2010;4:784–798. doi: 10.1038/ismej.2009.150. [DOI] [PubMed] [Google Scholar]
- Ottesen EA, Marin R, III, Preston CM, Young CR, Ryan JP, Scholin CA, et al. Metatranscriptomic analysis of autonomously collected and preserved marine bacterioplankton. ISME J. 2011;5:1881–1895. doi: 10.1038/ismej.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K-T, Lee K, Shin K, Jeong HJ, Kim KY. Improved method for minimizing sulfur loss in analysis of particulate organic sulfur. Anal Chem. 2014;86:1352–1356. doi: 10.1021/ac403649m. [DOI] [PubMed] [Google Scholar]
- Peng M, Xie Q, Hu H, Hong K, Todd JD, Johnston AW, et al. Phylogenetic diversity of the dddP gene for dimethylsulfoniopropionate-dependent dimethyl sulfide synthesis in mangrove soils. Can J Microbiol. 2012;4:523–530. doi: 10.1139/w2012-019. [DOI] [PubMed] [Google Scholar]
- Pennington JT, Chavez FP. Seasonal fluctuations of temperature, salinity, nitrate, chlorophyll and primary production at station H3/M1 over 1989-1996 in Monterey Bay, California. Deep-Sea Res Pt II. 2000;47:947–973. [Google Scholar]
- Pinhassi J, Simó R, González JM, Vila M, Alonzo-Sáez L, Kiene RP, et al. Dimethylsulfoniopropionate turnover is linked to the composition and dynamics of the bacterioplankton assemblage during a microcosm phytoplankton bloom. Appl Environ Microbiol. 2005;71:7650–7660. doi: 10.1128/AEM.71.12.7650-7660.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston CM, Harris A, Ryan JP, Roman B, Marin R, 3rd, Jensen S, et al. Underwater application of quantitative PCR on an ocean mooring. PLoS one. 2011;6:e22522. doi: 10.1371/journal.pone.0022522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston CM, Marin R, III, Jensen SD, Feldman J, Birch JM, Massion EI, et al. Near real-time, autonomous detection of marine bacterioplankton on a coastal mooring in Monterey Bay, California, using rRNA-targeted DNA probes. Environ Microbiol. 2009;11:1168–1180. doi: 10.1111/j.1462-2920.2009.01848.x. [DOI] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Davenport R, Turnbaugh P. Removing noise from pyrosequenced amplicons. BMC Bioinformatics. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn PK, Bates TS. The case against climate regulation via oceanic phytoplankton sulphur emissions. Nature. 2011;480:51–56. doi: 10.1038/nature10580. [DOI] [PubMed] [Google Scholar]
- Reisch CR, Moran MA, Whitman WB. Dimethylsulfoniopropionate-dependent demethylase (DmdA) from Pelagibacter ubique and Silicibacter pomeroyi. J Bacteriol. 2008;190:8018–8024. doi: 10.1128/JB.00770-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisch CR, Stoudemayer MJ, Varaljay VA, Amster IJ, Moran MA, Whitman WB, et al. Novel pathway for assimilation of dimethylsulphoniopropionate widespread in marine bacteria. Nature. 2011;473:208–211. doi: 10.1038/nature10078. [DOI] [PubMed] [Google Scholar]
- Rich VI, Pham VD, Eppley J, Shi Y, DeLong EF. Time-series analyses of Monterey Bay coastal microbial picoplankton using a ‘genome proxy' microarray. Environ Microbiol. 2011;13:116–134. doi: 10.1111/j.1462-2920.2010.02314.x. [DOI] [PubMed] [Google Scholar]
- Rinta-Kanto JM, Bürgmann H, Gifford SM, Sun S, Sharma S, del Valle DA, et al. Analysis of sulfur-related transcription by Roseobacter communities using a taxon-specific functional gene microarray. Environ Microbiol. 2011;13:453–467. doi: 10.1111/j.1462-2920.2010.02350.x. [DOI] [PubMed] [Google Scholar]
- Robidart JC, Preston CM, Paerl RW, Turk KA, Mosier AC, Francis CA, et al. Seasonal Synechococcus and Thaumarchaeal population dynamics examined with high resolution with remote in situ instrumentation. ISME J. 2011;6:513–523. doi: 10.1038/ismej.2011.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan JP, McManus MA, Kudela RM, Artigas ML, Bellingham JG, et al. Boundary influences on HAB phytoplankton ecology in a stratification-enhanced upwelling shadow. Deep-Sea Res Pt II. 2014;101:63–79. [Google Scholar]
- Simó R. Production of atmospheric sulfur by oceanic plankton: biogeochemical, ecological and evolutionary links. Trends Ecol Evol. 2001;16:287–294. doi: 10.1016/s0169-5347(01)02152-8. [DOI] [PubMed] [Google Scholar]
- Six KD, Kloster S, Ilyina T, Archer SD, Zhang K, et al. Global warming amplified by reduced sulphur flyuxes as a result of ocean acidification. Nature Clim Change. 2013;3:975–978. [Google Scholar]
- Slezak D, Kiene R, Toole D, Simó R, Kieber D. Effects of solar radiation on the fate of dissolved DMSP and conversion to DMS in seawater. Aquat Sci. 2007;69:377–393. [Google Scholar]
- Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Stefels J, Steinke M, Turner S, Malin G, Belviso S. Environmental constraints on the production and removal of the climatically active gas dimethylsulphide (DMS) and implications for ecosystem modelling. Biogeochemistry. 2007;83:245–275. [Google Scholar]
- Suylen GMH, Stefess GC, Kuenen JG. Chemolithotrophic potential of a Hyphomicrobium species capable of growth on methylated sulfur-compounds. Arch Microbiol. 1986;146:192–198. [Google Scholar]
- Todd JD, Curson AR, Dupont CL, Nicholson P, Johnston AW. The dddP gene, encoding a novel enzyme that converts dimethylsulfoniopropionate into dimethyl sulfide, is widespread in ocean metagenomes and marine bacteria and also occurs in some Ascomycete fungi. Environ Microbiol. 2009;6:1376–1385. doi: 10.1111/j.1462-2920.2009.01864.x. [DOI] [PubMed] [Google Scholar]
- Todd JD, Curson AR, Sullivan MJ, Kirkwood M, Johnston AW. The Ruegeria pomeroyi acuI gene has a role in DMSP catabolism and resembles yhdH of E. coli and other bacteria in conferring resistance to acrylate. PLoS One. 2012;4:e35947. doi: 10.1371/journal.pone.0035947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varaljay VA, Gifford SM, Wilson ST, Sharma S, Karl DM, Moran MA. Bacterial dimethylsulfoniopropionate degradation genes in the oligotrophic North Pacific Subtropical Gyre. Appl Environ Microbiol. 2012;78:2775–2782. doi: 10.1128/AEM.07559-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varaljay VA, Howard EC, Sun S, Moran MA. Deep sequencing of a dimethylsulfoniopropionate-degrading gene (dmdA) by using PCR primer pairs designed on the basis of marine metagenomic data. Appl Environ Microbiol. 2010;76:609–617. doi: 10.1128/AEM.01258-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila-Costa M, Rinta-Kanto JM, Poretsky RS, Sun S, Kiene RP, Moran MA. Microbial controls on DMSP degradation and DMS formation in the Sargasso Sea. Biogeochemistry. 2014;120:295–305. [Google Scholar]
- Vila-Costa M, Rinta-Kanto JM, Sun S, Sharma S, Poretsky R, Moran MA. Transcriptomic analysis of a marine bacterial community enriched with dimethylsulfoniopropionate. ISME J. 2010;4:1410–1420. doi: 10.1038/ismej.2010.62. [DOI] [PubMed] [Google Scholar]
- Weinitschke S, Denger K, Smits THM, Hollemeyer K, Cook AM. The sulfonated osmolyte N-methyltaurine is dissimilated by Alcaligenes faecalis and by Paracoccus versutus with release of methylamine. Microbiology. 2006;152:1179–1186. doi: 10.1099/mic.0.28622-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.