Abstract
The genetic organization of the gene involved in the capsular polysaccharide (CPS) biosynthesis of Actinobacillus pleuropneumoniae serotype 14 has been determined. The DNA region for the CPS biosynthesis of serotype 14 (cps14) comprised 9 open reading frames, designated as cps14AB1B2B3CDEFG genes, encoding Cps14A to Cps14G protein, respectively. Cps14A was similar to CpsA of A. pleuropneumoniae serotypes 1, 4 and 12; the Cps14B1 and Cps14B2 were similar to CpsB of A. pleuropneumoniae serotypes 1, 4 and 12, suggesting that CPS structure of A. pleuropneumoniae serotype 14 would belong to Group I including A. pleuropneumoniae serotypes 1, 4, 12 and 15. Surprisingly, the overall nucleotide sequence, deduced amino acid sequence, and the genetic organization of the cps14 were nearly identical to those of Actinobacillus suis. This study will provide the molecular basic knowledge for development of diagnostics and vaccine of A. pleuropneumoniae serotype 14.
Keywords: Actinobacillus pleuropneumoniae serotype 14, Actinobacillus suis, capsular polysaccharide biosynthesis, genetic organization
Actinobacillus pleuropneumoniae is the causative agent of porcine pleuropneumonia [8]. To date, two biotypes and at least 15 serotypes are present in A. pleuropneumoniae on the basis of β-nicotinamide adenine dinucleotide (NAD) requirement for growth and the capsular polysaccharide (CPS) structure [4, 24]. A. pleuropneumoniae biotype 1 (NAD-dependent) has been divided into 14 serotypes (serotypes 1 to 13 and 15) and biotype 2 (NAD-independent) into 7 serotypes (serotypes 2, 4, 7, 9, 11, 13 and 14) [3, 19, 20, 23]. Isolation of biotype 2 strains from pleuropneumonic lungs of pigs is thought to be sporadic, and biotype 2 strains have been considered less virulent than biotype 1 [6, 8, 20]. The low rate of isolation of biotype 2 strains has been considered due to an underestimation by laboratories that are not familiar with this group, because of its NAD-independent growth [19]. Sporadic isolation of A. pleuropneumoniae biotype 2 serotype 14 strains has been reported only in Denmark [7, 20, 27]. However, Maldonado and other researchers have reported that biotype 2 serotype 14 could be implicated in non-sporadic outbreaks of fatal swine pleuropneumonia in Spain [18] as well as biotype 2 serotypes 2, 4, 7, 11 [19] and 13 [18], indicating that biotype 2 strains may have a potential to emerge, because of the current global movement of pigs.
The author reports here the nucleotide sequence and genetic organization of the genes involved in the capsular polysaccharide (CPS) biosynthesis of A. pleuropneumoniae biotype 2 serotype 14 (cps14). The first aim of this study is to obtain a molecular basic knowledge for development of the cps14 gene-based diagnostic tools for A. pleuropneumoniae serotype 14, such as PCR typing methods [1, 5, 12, 13, 15, 25, 27]. The second aim of this study is to obtain a molecular basic knowledge for development of vaccines, such as a genetically modified capsule-deficient mutant vaccine for A. pleuropneumoniae serotype 14 [9].
A. pleuropneumoniae biotype 2 serotype 14 strain 3906, which was kindly provided by Dr. L. O. Andresen, National Veterinary Institute, Technical University of Denmark [20], was used in the present study. The strain was grown on tryptic soy agar (Difco Laboratories, Detroit, MI, U.S.A.) supplemented with 5% horse blood and 100 µg/ml β-NAD at 37°C and 5% CO2.
Genomic DNA for PCR templates was extracted as described previously [10]. PCR was done in a total volume of 50µl. The primers 5′-TCT AAR AYC GCA STA TGG CTA GGR CCT GAR GT-3′ and 5′-AAY GCT TTA TCA AAA GCG TGC CAA TGR CGC T-3′, located in cpxD gene encoding capsule export protein and lysA gene encoding diaminopimelate decarboxylase, respectively, were designed from previously published sequences [26], since the two genes are conserved in A. pleuropneumoniae and serotype specific CPS biosynthesis region is usually flanked by cpxD gene in Pasteurellaceae [14, 26]. A touchdown PCR with DNA polymerase KOD-FX Neo (Toyobo, Osaka, Japan) was done for amplification of the cps region as recommended by the supplier. The PCR conditions were as follows: step 1: 94 for 2 min (1 cycle); step 2: 98°C for 10 sec and 74°C for 15 min (5 cycles); step 3: 98°C for 10 sec and 72°C for 15 min (5 cycles); step 4: 98°C for 10 sec and 70°C for 15 min (5 cycles); step 5: 98°C for 10 sec and 68°C for 15 min (20 cycles); final step: 68°C for 10 min (1 cycle). Amplified PCR products were analyzed by agarose gel electrophoresis, stained with ethidium bromide (10 µg/ml) and visualized under ultraviolet light.
The PCR products were purified and sequenced as described previously [11]. Homology searches of the DDBJ /EMBL/Genbank databases were performed using the BLAST server at National Institute of Genetics, Japan. The nucleotide sequence of the CPS biosynthesis genes of A. pleuropneumoniae serotype 14 strain 3906 has been deposited in DDBJ/EMBL/Genbank under accession number AB810251.
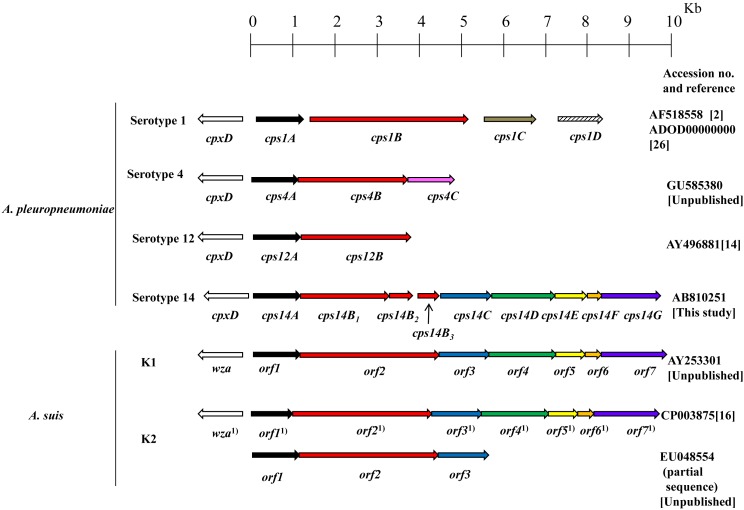
Comparison of overall nucleotide sequence determined in this study (11,497 nucleotides (nt)) revealed the highest similarity to the cps region of Actinobacillus suis (98% identity over 10,588 nt) [16]. Nine open reading frames (ORFs) were located between cpx14D and lysA genes (Fig. 1). The ORFs were designated as cps14AB1B2B3CDEFG genes (Fig. 1), encoding Cps14A to Cps14G protein, respectively. Cps14B1 showed homology to the N-terminal region of Cps1B, Cps4B and Cps12B from A. pleuropneumoniae serotypes 1, 4 and 12 and ORF 2 of A. suis, respectively; Cps14B2 showed homology to the central or C-terminal region of Cps1B, Cps4B and Cps12B from A. pleuropneumoniae serotypes 1, 4 and 12 and ORF2 from A. suis, respectively; Cps14B3 showed homology to C-terminal region of A. suis, indicating that three ORFs, cpsB1B2B3, seem to be generated due to the existence of the stop codons in A. pleuropneumoniae serotype 14 which are absent in the cpsB of A. pleuropneumoniae serotypes 1, 4, 12 and ORF2 of A. suis. The G+C content of the cps14AB1B2B3CDEFG ranged from 25% (in cps14B3) to 33% (in cps14G) (Table 1), which is lower than the 41 and 40% (overall G+C content of the A. pleuropneumoniae [26] and A. suis [16] genome, respectively), indicating that the region may be acquired by horizontal gene transfer.
Fig. 1.
Schematic diagram of the genetic organization of the DNA region involved in the CPS of Actinobacillus pleuropneumoniae serotypes 1 (Accession number (no.) AF518558 [2] and ADOD00000000 [26]), 4 (Accsession no. GU585380 [unpublished]), 12 (Accession no. AY496881 [14]), 14 (AB810251 [this study]) and A. suis serotypes K1 (Accession no. AY253301 [unpublished]) and K2 (Accession no. CP003875 [16] and EU048554 [unpublished]). The arows in the same colors indicate open reading frames, of which encoding proteins show amino acid sequence homology. 1)Since the gene names are not designated by the author (Accession no. CP003875 [16]), the eight genes are named as described in A. suis serotype K1 (Accession no. AY253301) and serotype K2 (Accession no. EU048554). Length of orf1 under accession no. CP003875 was slightly smaller than that under accession no. EU048554, althogh the same strain H91-380 was used. It is unknown why identical or nearly identical nucleotide sequence of genes for CPS synthesis region and the common genetic organization are observed between A. suis serotpes K1 and K2 [16].
Table 1. Identity of Cps proteins of Actinobacillus pleuropneumoniae seotype 14 (Cps14) to that of A. pleuropneumoniae and Actinobacillus suis serotypes.
| Cps14 protein | Lengh of aaa of Cps14 |
G+C% of cps14 gene |
Bacterial species |
Serotype | Homologous protein |
Accession number |
Reference | % identity | Length over homologous aa |
|---|---|---|---|---|---|---|---|---|---|
| Cps14A | 370 | 30 | Apb) | 1 | Cps1A | AF518558 | [2] | 98.8 | 339 |
| Ap | 4 | Cps4A | GU585380 | Unpublished | 98.1 | 370 | |||
| Asc) | K1 | ORF1 | AY253301 | Unpublished | 95.9 | 368 | |||
| As | K2 | ORF1 | EU048554 | Unpublished | 95.9 | 368 | |||
| As | K2 | ORF1d) | CP003875 | [16] | 95.5 | 332 | |||
| Ap | 12 | Cps12A | AY496881 | [14] | 93.2 | 367 | |||
| Cps14B1 | 709 | 28 | As | K1 | ORF2 | AY253301 | Unpublished | 99.3 | 706 |
| As | K2 | ORF2d) | CP003875 | [16] | 99.3 | 706 | |||
| As | K2 | ORF2 | EU048554 | Unpublished | 99.3 | 706 | |||
| Ap | 4 | Cps4B | GU585380 | Unpublished | 52.5 | 713 | |||
| Ap | 1 | Cps1B | AF518558 | [2] | 49.9 | 717 | |||
| Ap | 12 | Cps12B | AY496881 | [14] | 35.5 | 713 | |||
| Cps14B2 | 190 | 28 | As | K1 | ORF2 | AY253301 | Unpublished | 99.5 | 189 |
| As | K2 | ORF2d) | CP003875 | [16] | 99.5 | 189 | |||
| As | K2 | ORF2 | EU048554 | Unpublished | 99.5 | 189 | |||
| Ap | 4 | Cps4B | GU585380 | Unpublished | 55.8 | 147 | |||
| Ap | 1 | Cps1B | AF518558 | [2] | 58.7 | 75 | |||
| Ap | 12 | Cps12B | AY496881 | [14] | 33.1 | 151 | |||
| Cps14B3 | 152 | 25 | As | K1 | ORF2 | AY253301 | Unpublished | 100 | 152 |
| As | K2 | ORF2d) | CP003875 | [16] | 100 | 152 | |||
| As | K2 | ORF2 | EU048554 | Unpublished | 100 | 152 | |||
| Cps14C | 410 | 27 | As | K1 | ORF3 | AY253301 | Unpublished | 99.5 | 410 |
| As | K2 | ORF3d) | CP003875 | [16] | 99.5 | 410 | |||
| As | K2 | ORF3 | EU048554 | Unpublished | 99.5 | 410 | |||
| Cps14D | 549 | 29 | As | K1 | ORF4 | AY253301 | Unpublished | 99.8 | 549 |
| As | K2 | ORF4d) | CP003875 | [16] | 99.8 | 549 | |||
| Cps14E | 238 | 29 | As | K1 | ORF5 | AY253301 | Unpublished | 99.6 | 238 |
| As | K2 | ORF5d) | CP003875 | [16] | 99.6 | 238 | |||
| Cps14F | 128 | 30 | As | K1 | ORF6 | AY253301 | Unpublished | 97.7 | 128 |
| As | K2 | ORF6d) | CP003875 | [16] | 97.7 | 128 | |||
| Cps14G | 522 | 33 | As | K1 | ORF7 | AY253301 | Unpublished | 99.4 | 522 |
| As | K2 | ORF7d) | CP003875 | [16] | 99.4 | 522 | |||
a) aa=Amino acid; b) A. pleuropneumoniae; c) A. suis; d) Protein name was designated in this study as named in A. suis serotype K1 [Accession number AY253301].
At the amino acid level, Cps14A to Cps14G showed a homology to proteins of A. pleuropneumoniae serotypes 1, 4, 12 and A. suis [2, 14, 16] and did not show any significant homology to CPS proteins of A. pleuropneumoniae other serotypes. Their identities are shown in Table 1. Serotype-specific enzymes that are involved in the CPS biosynthesis are probably responsible for the dissimilarities of the CPS chemical structures [26]. However, A. pleuropneumoniae serotypes 1 to 13 and 15 can be divided into three groups based on basic differences of their chemical compositions and the structures of the CPS: Group I (serotypes 1, 4, 12 and 15), with CPS composed solely of repeating oligosaccharide units joined through phosphate linkages; Group II (serotypes 5 and 10), with CPS composed of repeating oligosaccharide units; Group III (serotypes 2, 3, 6, 7, 8, 9, 11 and 13), with CPS composed of teichoic acid polymers joined through phosphate diester linkages [14, 17, 21, 22]. The genetic organization of the cps genes provided molecular evidence to support the grouping of A. pleuropneumoniae serotypes described above [26]. A. pleuropneumoniae serotype 14 carried the cps14A gene encoding putative CPS phosphotransferase (Table 1). This enzyme shared by serotypes 1, 4 and 12 may be involved in the chemical linkage of phosphate in the linear CPS backbone [26]. Therefore, it is suggested that the CPS of serotype 14 belongs to Group I including serotypes 1, 4 and 12 [14, 26], although the CPS chemical structure of serotypes 14 should be determined by the chemical structural characterization. Surprisingly, the overall nucleotide sequence, deduced amino acid sequence and the genetic organization of the cps14 were nearly identical to those of A. suis (Table 1 and Fig. 1). High similarities in the nucleotide sequence of the cps genes of some A. pleuropneumoniae serotypes may be suggestive evidences for chemical structural similarity, but not for antigenic similarity of CPS [14, 26]. Therefore, the antigenic similarity of the CPS between A. pleuropneumoniae serotype 14 and A. suis should be determined for development of serological diagnostics and vaccine for the 2 organisms.
In conclusion, the author believes that this study will provide the molecular basic knowledge for diagnostics and vaccine development of A. pleuropneumoniae serotype 14.
Acknowledgments
The author thanks to Ms. K. Miyata for help of drawing of Fig. 1 and Dr. H. Kobayashi for his kind critical reading of the manuscript.
REFERENCES
- 1.Angen Ø., Ahrens A., Jessing S. G.2008. Development of a multiplex PCR test for identification of Actinobacillus pleuropneumoniae serovar 1, 7, and 12. Vet. Microbiol. 132: 312–318. doi: 10.1016/j.vetmic.2008.05.010 [DOI] [PubMed] [Google Scholar]
- 2.Bandara A. B., Lawrence M. L., Veit H. P., Inzana T. J.2003. Association of Actinobacillus pleuropneumoniae capsular polysaccharide with virulence in pigs. Infect. Immun. 71: 3320–3328. doi: 10.1128/IAI.71.6.3320-3328.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck M., van den Bosch J. F., Jongenelen I. M. C. A., Loeffen P. L. W., Nielsen R., Nicolet J., Frey J.1994. RTX toxin genotypes and phenotypes in Actinobacillus pleuropneumoniae field strains. J. Clin. Microbiol. 32: 2749–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackall P. J., Klaasen H. L. B. M., van den Bosch H., Kuhnert P., Frey J.2002. Proposal of a new serovar of Actinobacillus pleuropneumoniae: serovar 15. Vet. Microbiol. 84: 47–52. doi: 10.1016/S0378-1135(01)00428-X [DOI] [PubMed] [Google Scholar]
- 5.Bossé J. T., Li Y., Angen Ø, Weinert L. A., Chaudhuri R. R., Holden M. T., Williamson S. M., Maskell D. J., Tucker A. W., Wren B. W., Rycroft A. N., Langford P. R. and BRaDP1T consortium. 2014. Multiplex PCR assay for unequivocal differentiation of Actinobacillus pleuropneumoniae serovars 1 to 3, 5 to 8, 10, and 12. J. Clin. Microbiol. 52: 2380–2385. doi: 10.1128/JCM.00685-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dom P., Hommez J., Castryck F., Devriese L. A., Haesebrouck F.1994. Serotyping and quantitative determination of in vitro antibiotic susceptibility of Actinobacillus pleuropneumoniae strains isolated in Belgium (July 1991-August 1992). Vet. Q. 16: 10–13. doi: 10.1080/01652176.1994.9694407 [DOI] [PubMed] [Google Scholar]
- 7.Fussing V., Barford K., Nielsen R., Møller K., Nielsen J. P., Wegener H. C., Bisgaard M.1998. Evaluation and application of ribotyping for epidemiological studies of Actinobacillus pleuropneumoniae in Denmark. Vet. Microbiol. 62: 145–162. doi: 10.1016/S0378-1135(98)00205-3 [DOI] [PubMed] [Google Scholar]
- 8.Gottschalk M.2012. Actinobacillosis. pp. 653–669. In Diseases of Swine, 10th ed. (Zimmerman, J.J., Karriker, L.A., Ramirez, A., Schwarz, K.J. and Stevenson, G.W. eds.), Wiley-Blackwell, Oxford. [Google Scholar]
- 9.Inzana T. J., Glindemann G., Fenwick B., Longstreth J., Ward D.2004. Risk assessment of transmission of capsule-deficient, recombinant Actinobacillus pleuropneumoniae. Vet. Microbiol. 104: 63–71. doi: 10.1016/j.vetmic.2004.08.004 [DOI] [PubMed] [Google Scholar]
- 10.Ito H., Uchida I., Sekizaki T., Ooishi E., Kawai T., Okabe T., Taneno A., Terakado N.1995. Molecular cloning of an Actinobacillus pleuropneumoniae outer membrane lipoprotein (OmlA) from serotype 5a. Microb. Pathog. 18: 29–36. [PubMed] [Google Scholar]
- 11.Ito H., Ishii H., Akiba M.2004. Analysis of the complete nucleotide sequence of an Actinobacillus pleuropneumoniae streptomycin-sulfonamide resisitance plasmid, pMS260. Plasmid 51: 41–47. doi: 10.1016/j.plasmid.2003.10.001 [DOI] [PubMed] [Google Scholar]
- 12.Ito H.2010. Development of a cps-based multiplex PCR for typing of Actinobacillus pleuropenumoniae serotypes 1, 2 and 5. J. Vet. Med. Sci. 72: 653–655. doi: 10.1292/jvms.09-0529 [DOI] [PubMed] [Google Scholar]
- 13.Jessing S. G., Angen Ø., Inzana T. J.2003. Evaluation of a multiplex PCR test for simultaneous identification and serotyping of Actinobacillus pleuropneumoniae serotypes 2, 5 and 6. J. Clin. Microbiol. 41: 4095–4100. doi: 10.1128/JCM.41.9.4095-4100.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jessing S. G., Ahrens P., Inzana T. J., Angen Ø.2008. The genetic organization of the capsule biosynthesis region of Actinobacillus pleuropneumoniae serotypes 1, 6, 7 and 12. Vet. Microbiol. 129: 350–359. doi: 10.1016/j.vetmic.2007.12.003 [DOI] [PubMed] [Google Scholar]
- 15.Lo T. M., Ward C. K., Inzana T. J.1998. Detection and identification of Actinobacillus pleuropneumoniae serotype 5 by multiplex PCR. J. Clin. Microbiol. 36: 1704–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacInnes J. I., Mackinnon J., Bujold A. R., Ziebell K., Kropinski A. M., Nash J. H.2012. Complete genome sequence of Actinobacillus suis H91-0380, a virulent serotype O2 strain. J. Bacteriol. 194: 6686–6687. doi: 10.1128/JB.01633-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacLean L. L., Perry M. B., Vinogradoc E.2004. Characterization of the antigenic lipopolysaccharide O chain and the capsular polysaccharide produced by Actinobacillus pleuropneumoniae serotype 13. Infect. Immun. 72: 5925–5930. doi: 10.1128/IAI.72.10.5925-5930.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maldonado J., Martinez E., Blanco M., Riera P., Gottschalk M., Navas J.2006. Typing of NAD-independent Actinobacillus pleuropneumoniae by PCR and RFLP analysis. Abstr. Proc. 19th IPVS Congr. 238.
- 19.Maldonado J., Valls L., Martinez, Riera P.2009. Isolation rates, serovars, and toxin genotypes of nicotinamide adenine dinucleotide-independent Actinobacillus pleuropneumoniae among pigs suffering from pleuropneumonia in Spain. J. Vet. Diagn. Invest. 21: 854–857. doi: 10.1177/104063870902100615 [DOI] [PubMed] [Google Scholar]
- 20.Nielsen R., Andresen L. O., Plambeck T., Nielsen J. P., Kamp E. M., Jorsal S. E.1997. Serological characterization of Actinobacillus pleuropneumoniae biotype 2 strains isolated from pigs in two Danish herds. Vet. Microbiol. 54: 35–46. doi: 10.1016/S0378-1135(96)01267-9 [DOI] [PubMed] [Google Scholar]
- 21.Perry M. B., Altman E., Brisson J. R., Beynon L. M., Richards J. C.1990. Structural characteristics of the antigenic capsular polysaccharides involved in the serological classification of Actinobacillus (Haemophilus) pleuropneumoniae strains. Serodiagn. Immunother. Infect. Dis 4: 299–308. doi: 10.1016/0888-0786(90)90018-J [DOI] [Google Scholar]
- 22.Perry M. B., MacLean L. L., Vinogradov E.2005. Structural characterization of the antigenic capsular polysaccharide and lipopolysaccharide O-chain produced by Actinobacillus pleuropneumoniae serotype 15. Biochem. Cell Biol. 83: 61–69. doi: 10.1139/o04-112 [DOI] [PubMed] [Google Scholar]
- 23.Perry M. B., Angen Ø., MacLean L. L., Lacouture S., Kokotovic B., Gottschalk M.2012. An atypical biotype I Actinobacillus pleuropneumoniae serotype 13 is present in North America. Vet. Microbiol. 156: 403–410. doi: 10.1016/j.vetmic.2011.11.024 [DOI] [PubMed] [Google Scholar]
- 24.Pohl S., Bertschinger H., Frederiksen W., Manheim W.1983. Transfer of Haemophilus pleuropneumoniae and the Pasteurella haemolytica–like organism causing porcine necrotic pleuropneumonia to the genus Actinobacillus (Actinobacillus pleuropneumoniae comb. Nov.) on the basis of phenotypic and deoxyribonucleic acid relatedness. Int. J. Syst. Bacteriol. 33: 510–514. doi: 10.1099/00207713-33-3-510 [DOI] [Google Scholar]
- 25.Schuchert J. A., Inzana T. J., Angen Ø., Jessing S.2004. Detection and identification of Actinobacillus pleuropneuminiae serotypes 1, 2, and 8 by multiplex PCR. J. Clin. Microbiol. 42: 4344–4348. doi: 10.1128/JCM.42.9.4344-4348.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Z., Chen X., Li L., Wang S., Chen H., Zhou R.2010. Comparative genomic characterization of Actinobacillus pleuropneumoniae. J. Bacteriol. 192: 5625–5636. doi: 10.1128/JB.00535-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou L., Jones S. C. P., Angen Ø., Bosse J. T., Nash J. H. E., Frey J., Zhou R., Chen H. C., Kroll J. S., Rycroft A. N., Langford P. R.2008. Multiplex PCR that can distinguish between immunologically cross-reactive serovar 3, 6, and 8 Actinobacillus pleuropneumoniae strains. J. Clin. Microbiol. 46: 800–803. doi: 10.1128/JCM.01787-07 [DOI] [PMC free article] [PubMed] [Google Scholar]