Abstract.
Mosaicing of real-time three-dimensional echocardiography (RT3-DE) images aims at extending the field-of-view of overlapping images. Currently available methods discard most of the temporal information available in the time series. We investigate the added value of simultaneous registration of multiple temporal frames using common similarity metrics. We combine RT3-DE images of the left and right ventricles by registration and fusion. The standard approach of registering single frames, either end-diastolic (ED) or end-systolic (ES), is compared with simultaneous registration of multiple time frames, to evaluate the effect of using the information from all images in the metric. A transformation estimating the protocol-specific misalignment is used to initialize the registration. It is shown that multiframe registration can be as accurate as alignment of the images based on manual annotations. Multiframe registration using normalized cross-correlation outperforms any of the single-frame methods. As opposed to expectations, extending the multiframe registration beyond simultaneous use of ED and ES frames does not further improve registration results.
Keywords: image mosaicing, intensity-based registration, multiframe registration, real-time three-dimensional echocardiography
1. Introduction
Real-time three-dimensional (3-D) ultrasound is a safe, portable, and cost-efficient method routinely used by clinicians to visualize the inner organs of the body. In cardiology, it has many applications, like quantification of chamber volumes and wall motion.1 Disadvantageously, this technique is restricted by its rather narrow field-of-view (FOV) that prohibits direct visualization of large organs. Additionally, some anatomic regions can suffer from poor image quality due to an unfavourable direction of the ultrasound beam. Both limitations may be overcome by image mosaicing, where multiple differently oriented ultrasound volumes are registered and fused. This creates the possibility of size and volume measurements of large organs and simplifies their interpretability.2
Apart from extension of the FOV, a secondary advantage of image mosaicing is the improved visibility of anatomical structures in the overlapping part of the images. By fusion of multiple real-time 3-D echocardiography (RT3-DE) images, an improvement in image quality in terms of the visibility of endocardial borders as well as an increase in signal-to-noise and contrast-to-noise ratios was found.3,4 Additionally, it was shown that image-driven segmentation of the left ventricle (LV) performed better on multiview fused images than on single-view images.5
Accurate image registration is a prerequisite to obtain a fused image of high quality. Manual registration is time-consuming, which hampers wide adoption of mosaicing in clinical practice and explains the need for automatic registration methods.
The registration of RT3-DE data involves a time series of 3-D volumes, which makes the choice and number of temporal frames that are registered very important. Current techniques mostly employ single-frame registration of only end-diastolic (ED) or end-systolic (ES) time frames, as these frames can be easily identified. Grau et al.6 used the information available in both ED and ES frames to register echocardiography images in a multiframe registration strategy, in which the metric is optimized for both time frames simultaneously. Our study explores the effect of the included time frames on registration performance.
Besides the temporal aspect of the registration task, the choice of the similarity measure is of great importance. Multiple image-based methods are used for ultrasound image registration. Some of these methods are specifically designed for ultrasound registration, like ultrasound characteristics based methods 7 and phase-based image registration. In phase-based registration, the metric is based on the local phase and orientation of the images. Since phase is invariant to changes in both image brightness and contrast, it is theoretically suited for the registration of ultrasound images that are acquired from differing transducer positions.6 Next to this, more established metrics like normalized cross-correlation (NCC)3–5 and mutual information (MI)8 are used. Occasionally, global alignment of the images is achieved by tracking of the ultrasound probe, where the alignment can be refined using image-based registration techniques.9,10 For sparse registration of prestress to poststress echocardiography images, the superiority of NCC over sum-of-absolute-differences, sum-of-squared-differences, and normalized MI was reported.11
We assess multiview RT3-DE image registration by comparing several registration approaches, using RT3-DE images of the LV and right ventricle (RV). This work is an extension and deepening of our earlier findings12 including more data sets and a more elaborate evaluation. First, we examine the influence of incorporating information from multiple time frames in the metric by performing simultaneous multiframe registrations. Second, we evaluate the performance of the intensity-based metrics NCC and MI that are adopted in the field of ultrasound registration. Both the accuracy of the registration and the robustness of the different methods are assessed.
2. Materials and Methods
2.1. Data
Apical RT3-DE images of 28 healthy volunteers were acquired with an iE33 ultrasound system (Philips Healthcare, Best, The Netherlands), equipped with an X3-1 matrix array transducer. Data acquisition was approved by the medical ethical committee and written informed consent was obtained from all volunteers. Image acquisition was done while the subject was lying in the left lateral decubitus position during a single end-expiratory breath-hold. For each volunteer, two RT3-DE scans were obtained in harmonic mode from 7 R-wave gated subvolumes. Acquisition of the LV was done from a standard apical view. The second scan was focused on the RV and was acquired from a modified apical view. The depth and angle of the ultrasound pyramid were adjusted to the minimal level encompassing the RV. A set of LV and RV images is displayed in Fig. 1. Since the images were acquired with electrocardiographic triggering (ECG)-triggering, the first and last frames (LFs) of the time sequences reflect corresponding moments in the heart phase, where the first frame is the ED frame. Frames corresponding to the ES phase, which was defined as the phase prior to opening of the mitral valve, were detected by visual inspection.
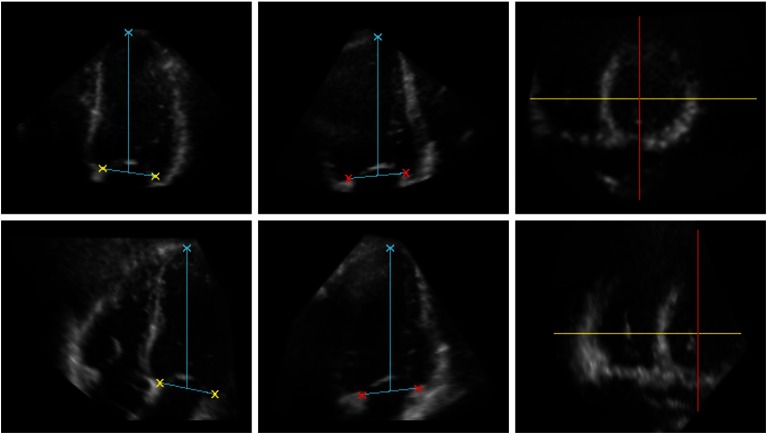
Fig. 1.
The annotated left ventricle (LV) data (upper row) and right ventricle (RV) data (lower row) at end diastole (ED); two-dimensional (2-D) slices from a RT3-DE data set. From left to right: Four-chamber view. Two-chamber view. Short-axis view, the directions of the four- and two-chamber views are depicted in yellow and red, respectively. The crosses indicate the position of the apex and the four points on the mitral valve annulus that were used for evaluation.
One subject was excluded from analysis because of inferior image quality of the RV data set. The remainder of the LV data (mean voxel size: ) consisted on average of 32 frames per heart cycle (range: 22 to 40 time frames). The RV data (mean voxel size: ) contained on average 29 frames per heart cycle (range: 24 to 35 frames).
2.2. Annotation
To evaluate the registration accuracy a total of five landmarks was used. Both the LV and RV data sets were independently annotated by two observers. The junction of the mitral valve leaflets and the mitral valve ring was indicated at four distinct positions in the two- and four-chamber views. The optimal two- and four-chamber views were found by the observers by rotation of the respective planes in a mid-ventrical short-axis view. The direction of the four-chamber view was always perpendicular to the two-chamber view. Additionally, the apex of the LV which was defined as the endocardial point most distant from the center of the mitral valve, was marked. In eight subjects, the LV apex was not visible and therefore its position was derived from the curvature and location of the LV endocardial borders. Before annotating the data, the observers agreed upon the placement of the landmarks and the selection of the views. Selection of the correct views and annotation of the data were performed following the method described by Nemes et al.13 ED and ES time frames, where the heart is in its two extreme states, as well as the LF of the time series were annotated using “3DStressView” (Biomedical Engineering, Thoraxcenter, Erasmus MC, Rotterdam, The Netherlands),13 while viewing the LV and RV data sets side-by-side. The position of the annotations is illustrated in Fig. 1.
2.3. Registration
Apical RT3-DE images focused on the LV of the heart were registered to RT3-DE images focused on the RV. Since the images were obtained at corresponding cardiac phases in the same subject, only translational and rotational differences were expected. For this reason, a rigid transformation was chosen to transform the images. The center of the image was used as center of rotation. The performance of two similarity measures, NCC and MI, was examined. NCC was calculated according to:
| (1) |
The metric was calculated for , which is the overlapping part of the fixed image and the transformed moving image , and it will depend on the transformation parameters that are stored in the vector . Since a rigid transformation model was chosen, contains three translation and three rotation parameters. The vector contains the coordinates for which the metric is evaluated. and are the mean intensities of the overlapping part of the fixed and transformed moving images, respectively.
| (2) |
B-spline Parzen windowing was used to construct the probability density function (PDF). and are the sets of regularly spaced intensity bins of the histogram for the fixed and moving image, respectively. is the marginal discrete PDF of the fixed image, is the marginal PDF of the moving image, and is the joint PDF.
High values of NCC or MI are associated with good image alignment. To find the transformation corresponding to the maximal metric, the metric was optimized using the adaptive stochastic gradient descent method.16 This optimizer applies a gradient descent optimization scheme to arrive at the minimum value of a cost function , which is the negative of the metric, in this case NCC or MI. The search direction is defined by the negative gradient of , as is stated in:
| (3) |
The step size is determined by the decreasing function at each iteration . A stochastic subsampling technique is used to accelerate optimization and the step size is adapted during the registration process.16
All data sets consist of multiple time frames covering the whole heart cycle. Since the probe position remains the same during acquisition, it was assumed that the transformation parameters do not change over the heart cycle. In addition to single-frame registration, which ignores the information available in other time frames, multiframe registration was applied to make use of the information in different time frames. To optimize the metric for several image pairs simultaneously a cost function was defined that is the average of the metric of all separate image pairs included in the registration process. This cost function is given by
| (4) |
and are collections of fixed and moving images, respectively, and is the number of time frames.
The influence of the number of image pairs involved in the registration process was examined by performing registration based on one or more selected temporal frames (ED or ES frames), as well as on a sequence of time frames. Since an equal number of LV and RV frames was required for multiframe registration, the RV data was interpolated between end diastole and end systole as well as between end systole and the last time frame of the data set.6 The new time frames were approximated by linear interpolation.
Data was registered using the open source registration toolbox elastix (Image Sciences Institute, UMC Utrecht, Utrecht, The Netherlands)17 in a multiresolution strategy with three resolutions (each resolution data was downsampled by a factor two; 750 iterations per resolution). 2048 samples were randomly selected per resolution, at the highest level 4096 samples were used. The number of bins for MI was 32, apart from the highest resolution level, where 64 bins were used. in Eq. (3) equals . All methods were implemented in “MeVisLab” (MeVis Medical Solutions, Bremen, Germany), an image processing environment that was also used for visualization of the data.
2.4. Initialization
Automatic registration was performed in two scenarios: (1) without prior knowledge, by initialization based on the image coordinate system, and (2) by initialization of the registration using a single initial transformation based on prior knowledge from five independent cases. Since the relative position of the LV and RV will be similar for all subjects, it was assumed that the difference in orientation of the LV and RV data was comparable for all subjects. The transformation that describes this difference was approximated by averaging the transformation parameters obtained by manual alignment of the LV and RV frames (for both end diastole and end systole) of five arbitrary data sets. The resulting transformation was applied to reduce the chance that optimization ends in a local optimum, thereby increasing the robustness of the registration. Manual alignment was done using an overlay representation of the LV and RV images. The relative position of the images was adapted till optimal overlap of corresponding structures was achieved.
2.5. Evaluation
Evaluation of the different automatic registration methods was based on the manual annotations made by the observers and in both the LV and RV data. The two point sets and , or and , were averaged over the observers to obtain the ground truth annotations and for the LV and RV images, respectively.
The LV and RV annotations of the observers as well as the ground truth annotations were rigidly registered by a closed-form least squares optimization algorithm18 to obtain the transformation parameters that map the RV image to the LV image. This resulted in and for the two observers and for the ground truth annotations. was used as ground truth transformation to compare with the transformation that was obtained by automatic registration of the LV and RV images.
To compare the different transformations, the misalignment between different point sets was calculated. It was expressed as the average of the Euclidean distances between corresponding points as is stated in Eq. (5).
| (5) |
where and are two sets of points.
Table 1 organizes all distance measures used for evaluation. The annotation inconsistencies cover both the unreliability in the locations of the landmarks and the possible limitations of the chosen transformation model. The interobserver distances measure the disagreement between the two observers. The interobserver annotation distance (IAD) is the difference in the actual position of the landmarks. The interobserver transformation distance (ITD) is the discrepancy between the transformations found by the observers. The misalignment of the LV and RV images was calculated from the ground truth annotations in two occasions: (1) after alignment of the image axes and (2) after initialization, where the LV data was transformed with the initial transformation. The accuracy of the different automatic registration methods was expressed by the registration error and was assessed by comparing the automatic transformation with the manual transformation.
Table 1.
Distance measures used for evaluation. , , , , and are the vectors containing the transformation parameters that map the right ventricle (RV) image to the left ventricle (LV) image found by the observers and , the ground truth transformation, the automatic transformation and the transformation used to initialize the registration. and are the ground truth annotations and are the LV annotations transformed with the transformation described by .
| Annotation inconsistency | Observer |
| Manual | |
| Interobserver distances | Interobserver annotation distance (IAD) |
| Interobserver transformation distance (ITD) | |
| Misalignment LV and RV image | (1) Alignment of image axes |
| (2) After initialization | |
| Registration performance | Registration error |
All distance measures were calculated for ED and ES time frames. Additionally, the last time frame was used as independent reference frame not involved in the registration process. The registration errors were compared with each other and with the ITD. Based on the outcomes of the Shapiro–Wilk test, to test the differences for normality, statistical significance of the differences was assessed using a Wilcoxon-signed ranks test. The robustness of the methods was evaluated by means of the number of successfully registered data sets. For each time point, registration was considered to be successful if the registration error was smaller than the maximum of the ITD.
3. Results
The characteristics of the annotations and the amount of misalignment after alignment of the image axes and after initialization are given in Table 2. Misalignment was significantly reduced by initialization ().
Table 2.
The median and maximum inconsistency of the annotations of the observers as well as the ground truth annotations, the interobserver annotation distance (IAD) and the amount of misalignment of the LV and RV data before and after initialization, calculated for end-diastolic (ED), end-systolic (ES), and the last frame (LF) of the time series (27 data sets).
| ED | ES | LF | ||||
|---|---|---|---|---|---|---|
| Median (mm) | Max (mm) | Median (mm) | Max (mm) | Median (mm) | Max (mm) | |
| Annotation inconsistency | 1.7 | 4.2 | 1.8 | 3.5 | 1.8 | 3.5 |
| Annotation inconsistency | 2.1 | 5.0 | 1.5 | 4.0 | 2.3 | 4.4 |
| Annotation inconsistency manual | 1.4 | 4.5 | 1.3 | 3.3 | 1.6 | 3.7 |
| IAD LV | 4.1 | 8.4 | 4.2 | 6.4 | 3.9 | 8.6 |
| IAD RV | 4.7 | 8.4 | 4.7 | 8.0 | 4.2 | 8.0 |
| (1) Alignment of image axes | 36.4 | 59.1 | 30.5 | 49.7 | 35.1 | 58.2 |
| (2) After initialization | 16.8 | 29.6 | 14.7 | 33.2 | 16.2 | 29.6 |
The results of the experiments with initialization as well as the ITD are given in Table 3. Results without initialization are not shown due to low performance. The performance of NCC was generally better than the performance of MI based on the success rates that were achieved. Registration errors of the automatic methods were significantly smaller than the amount of misalignment after initialization ().
Table 3.
Median and maximum interobserver transformation distance, ITD. Median registration errors and success rates for the experiments with initialization. Registration performance was evaluated on ED, ES, and the last time frame and was considered to be successful if the registration error was smaller than the maximum ITD. Single (ED): single-frame registration of ED time frames; Single (ES): single-frame registration of ES time frames; Multi (ED and ES): multiframe registration including only ED and ES time frames; Multi (ED to ES): multiframe registration of ED up to ES time frames inclusive; Multi (ES to LF): multiframe registration of ES frames up to LF inclusive; Multi (ED to LF): multiframe registration including all time frames (27 data sets).
| ED | ES | LF | |||||
|---|---|---|---|---|---|---|---|
| Median | Max | Median | Max | Median | Max | ||
| (mm) | (mm) | (mm) | (mm) | (mm) | (mm) | ||
| ITD |
3.7 | 6.6 | 2.8 | 6.2 | 3.1 | 6.6 | |
| Median (mm) | Success rate (%) | Median (mm) | Success rate (%) | Median (mm) | Success rate (%) | ||
| NCC | Single (ED) | 3.7 | 85 | 3.0 | 85 | 2.4 | 85 |
| Single (ES) | 3.7 | 93 | 2.6 | 93 | 3.1 | 93 | |
| Multi (ED and ES) | 3.6 | 93 | 2.6 | 93 | 2.8 | 93 | |
| Multi (ED to ES) | 3.9 | 93 | 2.9 | 89 | 3.1 | 93 | |
| Multi (ES to LF) | 3.7 | 85 | 3.0 | 85 | 3.0 | 85 | |
| Multi (ED to LF) | 3.8 | 89 | 2.9 | 89 | 2.9 | 89 | |
| MI | Single (ED) | 3.9 | 78 | 3.5 | 74 | 2.9 | 78 |
| Single (ES) | 4.2 | 81 | 2.7 | 81 | 3.1 | 81 | |
| Multi (ED and ES) | 3.8 | 81 | 2.6 | 81 | 2.7 | 81 | |
| Multi (ED to ES) | 3.8 | 81 | 2.8 | 81 | 2.7 | 81 | |
| Multi (ES to LF) | 3.7 | 85 | 2.9 | 85 | 2.8 | 85 | |
| Multi (ED to LF) | 3.8 | 85 | 2.9 | 85 | 2.6 | 85 | |
Highest success rates, i.e., the percentage of cases where the registration error is smaller than the maximum ITD, were achieved by two different registration strategies, namely single-frame registration of the ES frame, and multiframe registration including ED and ES time frames, using NCC as metric. However, the latter strategy returned the smallest registration errors, based on the last time frame that was not involved in the registration process. No statistically significant differences between the registration accuracy of the different approaches were found. The boxplot in Fig. 2 presents the range of the registration errors when NCC is used as metric.
Fig. 2.
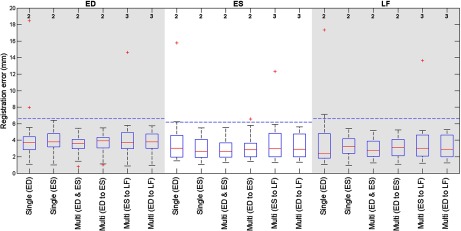
Boxplot showing the registration errors at end diastole (ED), end systole (ES) and the last time frame (LF) for the different registration strategies. NCC was used as metric. Multiframe registration with ED and ES frames results in the smallest deviation in the registration errors. At the top the number of data sets with a registration error over 20 mm is given. The horizontal (dashed) lines give the maximal interobserver transformation distance, or ITD, for the different temporal frames.
Figure 3 shows an example of the LV and RV data before and after registration. It shows that by combining the RV and LV images, the FOV is extended and a high-quality image covering both ventricles is obtained.
Fig. 3.
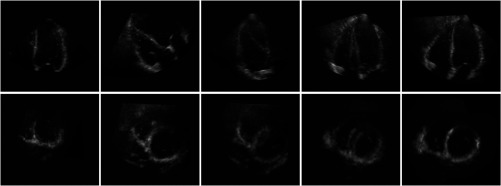
The four-chamber view (top row) and short-axis view (lower row) at ED; 2-D slices of a 3-D volume data set are shown. From left to right: LV image. RV image. Fused LV and RV images (alignment of image axes). Fused LV and RV images (LV data is transformed by the initial transformation). Fused LV and RV images after successful registration (NCC, multiframe registration including ED and ES frames). Mean-intensity fusion was used for image fusion.
4. Discussion
This study compares different intensity-based registration approaches for the alignment of RT3-DE images. For this purpose, RT3-DE images focused on the LV and RV were used.
For evaluation, two observers manually annotated the data. These annotations were averaged to obtain the ground truth annotations. The IAD is the difference in the actual position of the annotations made by the observers. Generally, the image quality of the LV is higher in the LV data than in the RV data resulting in an improved visibility of the structures that are used as landmark. This explains the lower IAD for the LV images. Based on the annotation of the apex and mitral valve, the IAD appears to be comparable to the interobserver variability found by Leung et al.,11 and we refer to this study for statistics on the intraobserver variability.
The inconsistency in the ground truth annotations is smaller than the inconsistencies in the annotations of the individual observers. This denotes the increase in reliability that is achieved by averaging the annotations of the observers. The rigid transformation aligning the ground truth annotations, yields the ground truth transformation . Despite the small inconsistency due to difficulties for the observers in indicating the corresponding positions in both the LV and RV data sets, and the possibly too restricted rigid transformation model, it is the best achievable reference standard. Ground truth transformations are unavailable and evaluation on synthetic or phantom data would not resemble the variability in clinical ultrasound data.
The registration error measures the difference between the automatic and manual transformation and is compared with the difference in the transformations found by the two observers, the ITD. Registration is considered successful when the registration error is smaller than the maximum ITD. In these cases, the difference between the automatic and manual, ground truth, transformation is smaller than the difference between the transformations found by two different observers. This implies that the automated registration performs as accurate as an arbitrary additional experienced observer.
Initialization helps to improve registration results by decreasing the initial amount of misalignment. In this study, we use a single initial transformation for all data sets that is based on the protocol-specific difference between the LV and RV images. Table 2 shows that using a transformation that is derived from only five data sets, misalignment is decreased by half, indicating the effectiveness of this type of initialization. To test the robustness of the initialization, the best performing method was initialized using the average manual transformation of different collections of five data sets as well as the average of all data sets. Although registration results were not identical for all combinations, initialization always leads to higher success rates compared with no initialization.
Though the differences in performance between the examined registration strategies are not statistically significant, a finding we assign to the limited size of our data set, our results do reveal some trends that we will discuss here. To begin with, the use of NCC as metric yields better registration results than the use of MI. This behavior suggests that the additional degrees of freedom of MI are disadvantageous for our data. Focusing on NCC, it is shown that by single-frame registration of the ES frame a higher success rate is achieved than by registration solely based on the ED frame. The contracted state of the heart during end systole improves the visibility of the cardiac structures, which facilitates registration.
Of the examined multiframe registration strategies, inclusion of both the ED and ES frames results in the highest success rate. Remarkably, registration with a series of time frames degrades performance. Several factors might contribute to this. First, the ED and ES frames represent the heart in its two extreme states and will contain the most diverse information. Adding more frames will increase the risk of adding confounding signal. Second, the interpolation of the RV data can negatively affect image quality by the introduction of interpolation artifacts. Last, because the diastolic phase is longer than the systolic phase, a high number of frames of the time sequence will resemble the ED frame. Since single-frame registration shows that registration of ED frames performs worse than ES frames, inclusion of all time frames can negatively influence registration performance. This can explain the poorer performance of multiframe registration of the diastolic heart phase, in terms of registration success. These results are in line with the findings of Grau et al.,6 who touch upon this subject by noticing that the performance of their phase-based registration method did not improve by inclusion of other time frames besides the ED and ES frames.
Of all examined methods, best results are obtained using a multiframe registration strategy with ED and ES frames to optimize the metric NCC. It outperforms any single-frame method on success rate. Only single-frame ES registration is on par in terms of success rate. However, evaluation on the unbiased LF of the time sequence shows that the multiframe method is again superior. Figure 2 shows that the smallest deviation in the registration error is achieved by multiframe registration with ED and ES frames, supporting the conclusion that this is the best method within this experimental setup.
Figure 3 shows that by fusion of the LV and RV images, the FOV is extended and anatomical information from the individual images is combined to get an anatomically more complete view of the heart. Unfortunately, some parts of the heart, like the anterior RV wall, are not well visualized in the fused image either, due to insufficient quality of the RV image. This is a limitation of fusing no more than two images. Inclusion of more volumes that specifically aim at visualization of the parts of the heart that are not well imaged, may offer a solution for this.
5. Conclusions
In this study, we investigated the role of temporal multiframe registration for multiview RT3-DE registration. Accurate registration is essential to obtain a high-quality extended FOV image by means of image mosaicing. We showed that multiframe registration with multiple time frames improves registration results compared with single-frame registration. The ED and ES frame are most suited for this. Registration of these two time frames in combination with the metric NCC performed best in terms of the number of successfully registered data sets and registration error. Furthermore, the method achieved similar accuracy as manual alignment of the data by experienced observers. Notably, extending the multiframe approach beyond simultaneous registration of ED and ES frames did not further improve automatic registration.
Acknowledgments
The authors would like to thank G. van Burken of the Department of Biomedical Engineering, Thoraxcenter, Erasmus Medical Center, Rotterdam, The Netherlands, for the required software. This research is supported by the Dutch Technology Foundation STW (project number 10847) which is the applied science division of NWO, and the Technology Program of the Ministry of Economic Affairs, The Netherlands.
Biographies
Harriët W. Mulder received her MSc degree in medical natural sciences (specialization medical physics) at the VU University, Amsterdam, The Netherlands, in 2010. Since March 2010, she has worked as a PhD student at the Image Sciences Institute, University Medical Center Utrecht, in Utrecht. Currently, she is working on the registration and fusion of transesophageal echocardiography (TEE) images.
Marijn van Stralen obtained his MSc in medical-technical computer science at Utrecht University. He received his PhD in medical image analysis on automated analysis of 3-D echocardiography from Leiden University (2009), working at the Leiden University Medical Center and the Erasmus Medical Center, Rotterdam. Since 2008, he is a postdoc on 4-D medical image analysis at the Image Sciences Institute, UMC Utrecht. His work focuses on MR imaging, specifically, abdominal perfusion imaging.
Heleen B. van der Zwaan did a PhD project on the clinical use of three-dimensional echocardiography for assessment of right ventricular function in patients with congenital heart disease. She completed this work in June 2011. She is currently working as a cardiology resident.
K. Y. Esther Leung received her MS degree in applied physics from the Delft University of Technology, The Netherlands. The subject of her thesis was image processing for ultrasound strain imaging. In 2009, she received her PhD degree (cum laude) on the subject of three-dimensional stress echocardiography, at the Erasmus Medical Center. She is currently a clinical physics resident at the Albert Schweitzer hospital. Her research interests include medical technology, image processing, and radiation safety.
Johan G. Bosch is an associate professor and staff member at the Department of Biomedical Engineering, Thoraxcenter, EMC Rotterdam. He specializes in 2-D/3-D echocardiographic image processing/analysis and (3-D) transducer development. His main research interests are optimal border detection, geometrical and statistical models, and anatomical and physical knowledge representations for border detection. He is the leader of projects on 3-D segmentation and 3-D ultrasound guidance in electrophysiology and involved in 2-D and 3-D carotid and 3-D TEE imaging.
Josien P. W. Pluim received her MSc degree in computer science (University of Groningen, NL) in 1996 and a PhD degree (Utrecht University, NL) in 2001. Her PhD research was performed at the Image Sciences Institute, University Medical Center Utrecht, on mutual information-based image registration. She is currently an associate professor at the Image Sciences Institute, Utrecht. Her research group works on various topics in medical image analysis, in particular for oncology and digital pathology.
References
- 1.Burri M. V., et al. , “Review of novel clinical applications of advanced, real-time, 3-dimensional echocardiography,” Transl. Res. 159(3), 149–164 (2012). 10.1016/j.trsl.2011.12.008 [DOI] [PubMed] [Google Scholar]
- 2.Wachinger C., Wein W., Navab N., “Registration strategies and similarity measures for three-dimensional ultrasound mosaicing,” Acad. Radiol. 15(11), 1404–1415 (2008). 10.1016/j.acra.2008.07.004 [DOI] [PubMed] [Google Scholar]
- 3.Szmigielski C., et al. , “Real-time 3D fusion echocardiography,” JACC Cardiovasc. Imaging 3(7), 682–690 (2010). 10.1016/j.jcmg.2010.03.010 [DOI] [PubMed] [Google Scholar]
- 4.Rajpoot K., et al. , “Multiview fusion 3-D echocardiography: improving the information and quality of real-time 3-D echocardiography,” Ultrasound Med. Biol. 37(7), 1056–1072 (2011). 10.1016/j.ultrasmedbio.2011.04.018 [DOI] [PubMed] [Google Scholar]
- 5.Rajpoot K., et al. , “The evaluation of single-view and multi-view fusion 3D echocardiography using image-driven segmentation and tracking,” Med. Image Anal. 15(4), 514–528 (2011). 10.1016/j.media.2011.02.007 [DOI] [PubMed] [Google Scholar]
- 6.Grau V., Becher H., Noble J. A., “Registration of multiview real-time 3-D echocardiographic sequences,” IEEE Trans. Med. Imaging 26(9), 1154–1165 (2007). 10.1109/TMI.2007.903568 [DOI] [PubMed] [Google Scholar]
- 7.Wachinger C., Klein T., Navab N., “Locally adaptive nakagami-based ultrasound similarity measures,” Ultrasonics 52(4), 547–554 (2012). 10.1016/j.ultras.2011.11.009 [DOI] [PubMed] [Google Scholar]
- 8.Shekhar R., et al. , “Registration of real-time 3-D ultrasound images of the heart for novel 3-D stress echocardiography,” IEEE Trans. Med. Imaging 23(9), 1141–1149 (2004). 10.1109/TMI.2004.830527 [DOI] [PubMed] [Google Scholar]
- 9.Poon T. C., Rohling R. N., “Three-dimensional extended field-of-view ultrasound,” Ultrasound Med. Biol. 32(3), 357–369 (2006). 10.1016/j.ultrasmedbio.2005.11.003 [DOI] [PubMed] [Google Scholar]
- 10.Housden R. J., et al. , “Extended-field-of-view three-dimensional transesophageal echocardiography using image-based x-ray probe tracking,” Ultrasound Med. Biol. 39(6), 993–1005 (2013). 10.1016/j.ultrasmedbio.2012.12.018 [DOI] [PubMed] [Google Scholar]
- 11.Leung K. Y. E., et al. , “Sparse registration for three-dimensional stress echocardiography,” IEEE Trans. Med. Imaging 27(11), 1568–1579 (2008). 10.1109/TMI.2008.922685 [DOI] [PubMed] [Google Scholar]
- 12.Mulder H. W., et al. , “Registration of multi-view apical 3D echocardiography images,” Proc. SPIE 7962, 79621W (2011). 10.1117/12.878042 [DOI] [Google Scholar]
- 13.Nemes A., et al. , “Side-by-side viewing of anatomically aligned left ventricular segments in three-dimensional stress echocardiography,” Echocardiogr.- J. Cardiovasc. Ultrasound Allied Tech. 26(2), 189–195 (2009). 10.1111/echo.2009.26.issue-2 [DOI] [PubMed] [Google Scholar]
- 14.Viola P., Wells W. M., “Alignment by maximization of mutual information,” Int. J. Comput. Vision 24(2), 137–154 (1997). 10.1023/A:1007958904918 [DOI] [Google Scholar]
- 15.Maes F., et al. , “Multimodality image registration by maximization of mutual information,” IEEE Trans. Med. Imaging 16(2), 187–198 (1997). 10.1109/42.563664 [DOI] [PubMed] [Google Scholar]
- 16.Klein S., et al. , “Adaptive stochastic gradient descent optimisation for image registration,” Int. J. Comput. Vision 81(3), 227–239 (2009). 10.1007/s11263-008-0168-y [DOI] [Google Scholar]
- 17.Klein S., et al. , “Elastix: a toolbox for intensity-based medical image registration,” IEEE Trans. Med. Imaging 29(1), 196–205 (2010). 10.1109/TMI.2009.2035616 [DOI] [PubMed] [Google Scholar]
- 18.Horn B. K. P., “Closed-form solution of absolute orientation using unit quaternions,” J. Opt. Soc. Am. A-Opt. Image Sci. Vision 4(4), 629–642 (1987). 10.1364/JOSAA.4.000629 [DOI] [Google Scholar]