Abstract
Genes that encode for enzymes that can degrade petroleum hydrocarbons (PHs) are critical for the ability of microorganisms to bioremediate soils contaminated with PHs. Distributions of two petroleum-degrading genes AlkB and Nah in soils collected from three zones of the Dagang Oilfield, Tianjin, China were investigated. Numbers of copies of AlkB ranged between 9.1 × 105 and 1.9 × 107 copies/g dry mass (dm) soil, and were positively correlated with total concentrations of PHs (TPH) (R2 = 0.573, p = 0.032) and alkanes (C33 ~ C40) (R2 = 0.914, p < 0.01). The Nah gene was distributed relatively evenly among sampling zones, ranging between 1.9 × 107 and 1.1 × 108 copies/g dm soil, and was negatively correlated with concentrations of total aromatic hydrocarbons (TAH) (R2 = −0.567, p = 0.035) and ∑16 PAHs (R2 = −0.599, p = 0.023). Results of a factor analysis showed that individual samples of soils were not ordinated as a function of the zones.
Contamination of aquatic and terrestrial environments with petroleum hydrocarbons (PHs) represents a serious problem worldwide1. A total mass of between 8 × 104 and 1 × 107 tons of PHs per year have been estimated to be released into ecosystems globally2. Contamination of the environment with PHs is not uniform with locations where PHs are extracted, refined, or shipped typically being at greater risk of contamination. In China, there are more than 10 high-production oilfields, of which the Dagang Oilfield has 1.87 × 104 square kilometers of exploration area with an annual production of 4.3 million tons of crude oil and 360 million cubic meters of gas, representing the major base for production, refining and shipping center in the Bohai Bay Rim area.
Alkanes and aromatic hydrocarbons (AHs) are the primary pollutants of concern associated with crude oil, and account for approximately 80% of the total petroleum hydrocarbons (TPHs) in crude oils3. Alkanes with carbon chain lengths ranging from C8 ~ C40 can cause hardening and limit wetting of soils, and can result in toxicity to plants and/or soil invertebrates, as well as pose risks to humans and wildlife through direct contact with these soils or soil organisms4. Another constituent of concern associated with crude oils are polycyclic aromatic hydrocarbons (PAHs), which have been shown to be of significant toxicological concern at environmental concentrations of more than 1000 μg kg−1, dm in soils5. PAHs can cause a range of toxic effects, including mutagenicity and carcinogenicity6,7. PAHs adhere to colloids of organic matter in soils, which can result in a decrease in bioavailability to PAHs-degrading microorganisms and an increase in recalcitrance of these compounds8.
Microbes that degrade petroleum hydrocarbons are widely distributed in the environment and constitute dynamic changes in structures of communities according to their divergent abilities biodegradate oils. Many indigenous oil-degrading microorganisms such as Pseudomonas GPo1 (C5–C12, n-alkanes)9, Acinetobacter sp. strain DSM17874 (C10–C40, n-alkanes)10, Rhodococcus sp. strain Q15 (C12–C32, n-alkanes)11, Pseudomonas putida G7 (naphthalene)12, Burkholderia sp. strain JS15 (BTEX)13 and Pseudomonas aeruginosa JI104 (toluene)14 and corresponding metabolic substrates have been characterized in detail. In addition, technologies based on abilities of certain microorganisms to enzymatically degrade PHs have been favored for remediation of soils because they are minimally invasive, require little disturbance of soils, are cost-effective and result in minimal secondary contamination15. Genes encoding for a variety of metabolizing enzyme systems that can degrade PHs play critical roles in aerobic mineralization activity, and may represent promising alternative approaches for the characterization of bioremediation capacities of microbes.
Alkane monooxygenase gene AlkB and naphthalene dioxygenase gene Nah are the predominant genes corresponding to first-step hydroxylases involved in metabolism of alkanes and AHs, respectively16,17. Previous studies have reported that indigenous microorganisms expressing alkane catabolic genes were able to degrade oil in contaminated soils or sediments18,19, even under extreme environmental conditions in the Arctic or Antarctic20,21. Alternatively, naphthalene dioxygenase can add both atoms of molecular oxygen to the aromatic ring as a first step in aerobic degradation, and subsequently transfer the corresponding dihydric alcohol to catechol17,22. Genes involved in degrading PHs have been successfully used as biomarkers for characterization of the magnitude of contamination of soils by PHs18,23. Abundance of AlkB and PAH-ring-hydroxylating dioxygenases genes have been shown to be positively correlated to most probable number (MPN) counts of microorganisms that can degrade PHs, and to degradation of hexadecane and naphthalene24. Understanding relationships between abundance/expression of genes and biodegradation of PHs is important for predicting potential effects of contamination ecosystems with oil and the metabolic remediation capability of aerobic soil microorganisms.
This study was designed to characterize major classes of constituents in crude oil including total petroleum hydrocarbons (TPHs), saturated hydrocarbons (SHs), aromatic hydrocarbons (AHs) and concentrations of other components of TPHs in soils in three zones of the Dagang oilfield. To evaluate the presence and metabolic potential of oil degrading microorganisms, Q-PCR was used to quantify abundances of AlkB and Nah. Microbial communities were also characterized by use of PCR and Denaturing Gradient Gel Electrophoresis (DGGE). Finally, multivariate statistical techniques were applied to characterize relationships between numbers of copies of oil degrading genes, soil parameters as well as concentrations of PHs.
Results
Petroleum contamination and other physico-chemical properties
Soils collected from the Dagang Oilfield were variously contaminated with PHs (Table 1). The range in concentrations of TPHs, as measured gravimetrically in the three zones were: oil-producing zone, (2.0 ± 0.5) × 104 ~ (3.7 ± 0.7) × 104 mg kg−1 dm.; residential zone, (1.3 ± 0.3) × 104 ~ (3.4 ± 0.5) × 104 mg kg−1 dm.; oil-refinery and transportation zone, (2.0 ± 0.5) × 104 ~ (2.3 ± 0.4) × 104 mg kg−1 dm. Concentrations of TPH at S1 and S2 less than10 meters away from the oil well, as well as S8 near oil storage tanks in the residential zone exceeded 3.0 × 104 mg kg−1 dm, which were significantly greater than those at all other sites (p < 0.05). Concentrations of TPHs in soils near the oil tank and other petroleum transportation routes were greater than those in soils along the canal and in the residential area. Due to the run-off affect, soils collected next to the Banqiao canal at S6 contained the lesser concentration of TPHs with (1.3 ± 0.3) × 104 mg kg−1 dm.
Table 1. Characteristics of petroleum hydrocarbons (PHs) in soils of the Dagang oilfield.
| Samples | TPH (mg kg−1 dm) | TSH (mg kg−1 dm) | TAH (mg kg−1 dm) | Alkanes (C8 ~ C40)(μg kg−1 dm) | 16 PAHs (μg kg−1 dm) | naphthalene (μg kg−1 dm) | phenanthrene(μg kg−1 dm) | pyrene(μg kg−1 dm) | Alkanes (C8 ~ C19) (μg kg−1 dm) | Alkanes (C20 ~ C32) (μg kg−1 dm) | Alkanes (C33 ~ C40) (μg kg−1 dm) | pH | salinity(g kg−1 dm) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Oil-producing zone | S1 | (3.7 ± 0.7) × 104 | (1.1 ± 0.2) × 104 | (5.1 ± 0.8) × 103 | (1.9 ± 0.1) × 104 | (1.3 ± 0.05) × 103 | (1.2 ± 0.2) × 102 | 7.7 × 101 | 2.5 × 102 | 7.7 × 103 | 9.8 × 103 | 1.9 × 103 | 8.55 | 16.56 |
| S2 | (3.3 ± 0.7) × 104 | (6.5 ± 0.3) × 103 | (4.9 ± 0.2) × 103 | (1.1 ± 0.1) × 104 | (4.0 ± 0.3) × 102 | (7.9 ± 1.2) × 101 | 7.4 × 101 | 8.2 × 101 | 4.1 × 103 | 7.0 × 103 | 4.0 × 102 | 8.54 | 7.97 | |
| S3 | (2.2 ± 0.6) × 104 | (3.5 ± 0.4) × 103 | (3.4 ± 0.5) × 103 | (6.6 ± 0.2) × 103 | (4.3 ± 0.2) × 102 | (5.9 ± 0.9) × 101 | 8.9 × 101 | 6.9 × 101 | 3.2 × 103 | 3.2 × 103 | 1.0 × 102 | 8.53 | 7.10 | |
| S4 | (2.0 ± 0.5) × 104 | (2.6 ± 0.8) × 103 | (2.6 ± 0.4) × 103 | (1.2 ± 0.1) × 104 | (4.0 ± 0.7) × 102 | (1.2 ± 0.0) × 102 | 8.3 × 101 | 7.0 × 101 | 8.1 × 103 | 3.7 × 103 | 2.0 × 102 | 8.39 | 8.76 | |
| S5 | (2.1 ± 0.2) × 104 | (7.8 ± 0.8) × 103 | (1.7 ± 0.3) × 103 | (1.3 ± 0.1) × 104 | (1.7 ± 0.1) × 102 | (2.7 ± 0.1) × 101 | 3.9 × 101 | 5.1 × 101 | 6.3 × 103 | 6.9 × 103 | 2.2 × 102 | 8.63 | 11.78 | |
| Residential zone | S6 | (1.3 ± 0.3) × 104 | (9.7 ± 0.9) × 102 | (2.2 ± 0.6) × 103 | (2.8 ± 0.2) × 102 | (4.6 ± 0.4) × 102 | (5.8 ± 0.6) × 101 | 7.1 × 101 | 1.0 × 102 | 1.4 × 102 | 1.1 × 102 | 3.4 × 101 | 8.51 | 53.83 |
| S7 | (2.1 ± 0.5) × 104 | (5.4 ± 0.8) × 103 | (1.8 ± 0.4) × 103 | (5.9 ± 0.1) × 103 | (2.7 ± 0.3) × 102 | (4.7 ± 0.9) × 101 | 7.4 × 101 | 3.6 × 101 | 3.6 × 103 | 2.3 × 103 | 6.3 × 101 | 8.43 | 26.72 | |
| S8 | (3.4 ± 0.5) × 104 | (5.2 ± 0.4) × 103 | (1.1 ± 0.5) × 103 | (7.2 ± 0.7) × 103 | (1.9 ± 0.2) × 102 | (3.7 ± 0.2) × 101 | 4.6 × 101 | 2.9 × 101 | 4.4 × 103 | 2.7 × 103 | 7.4 × 101 | 8.56 | 33.23 | |
| S9 | (1.8 ± 0.2) × 104 | (6.0 ± 1.4) × 103 | (1.8 ± 0.3) × 103 | (2.9 ± 0.3) × 103 | (3.7 ± 0.1) × 102 | (1.3 ± 0.1) × 102 | 6.6 × 101 | 6.3 × 101 | 2.0 × 103 | 9.1 × 102 | 5.0 × 101 | 8.44 | 3.32 | |
| Oil-refinery and transportation zone | S10 | (2.3 ± 0.4) × 104 | (9.0 ± 1.3) × 103 | (2.2 ± 0.5) × 103 | (1.8 ± 0.2) × 104 | (3.2 ± 0.3) × 102 | (8.3 ± 0.7) × 101 | 7.6 × 101 | 6.3 × 101 | 9.5 × 103 | 8.6 × 103 | 1.9 × 102 | 8.38 | 4.82 |
| S11 | (2.2 ± 0.1) × 104 | (6.0 ± 0.8) × 103 | (5.4 ± 0.2) × 103 | (1.5 ± 0.4) × 104 | (2.8 ± 0.3) × 103 | (1.3 ± 0.1) × 102 | 2.6 × 102 | 4.4 × 102 | 6.0 × 103 | 8.4 × 103 | 2.8 × 102 | 8.41 | 14.37 | |
| S12 | (2.3 ± 0.2) × 104 | (7.6 ± 0.8) × 103 | (1.8 ± 0.2) × 103 | (1.5 ± 0.04) × 104 | (1.8 ± 0.2) × 102 | (3.7 ± 0.5) × 101 | 3.9 × 101 | 5.3 × 101 | 7.6 × 103 | 7.6 × 103 | 1.3 × 102 | 8.61 | 13.48 | |
| S13 | (2.2 ± 0.3) × 104 | (8.5 ± 0.8) × 103 | (3.4 ± 0.6) × 103 | (1.6 ± 0.1) × 104 | (7.1 ± 0.5) × 102 | (8.8 ± 1.1) × 101 | 1.1 × 102 | 1.1 × 102 | 7.7 × 103 | 8.0 × 103 | 2.2 × 102 | 8.4 | 1.42 | |
| S14 | (2.0 ± 0.5) × 104 | (6.6 ± 0.9) × 103 | (1.9 ± 0.9) × 103 | (1.3 ± 0.1) × 104 | (2.8 ± 0.7) × 102 | (4.0 ± 0.9) × 101 | 5.4 × 101 | 6.2 × 101 | 7.1 × 103 | 6.1 × 103 | 8.8 × 101 | 8.62 | 27.32 | |
Mean ± standard deviation for triplicate experiments.
Concentrations of TPHs in soils collected from oil-refinery and transportation zones, where there were fixed transport routes and oil-refinery workshops, were more homogeneous compared to the other zones, with concentrations ranging from (2.0 ± 0.5) × 104 to (2.3 ± 0.2) × 104 mg kg−1 dm. The variance of TPHs among the three sampling zones was examined by LSD-t method, and found that there was no significant difference between the oil producing zone and other zones.
SHs and AHs were the primary constituents of PHs (Table S1). Proportions of total hydrocarbons (THCs) contributed by SHs in the oil-producing, residential and oil-refinery and transportation zones were 23.3%, 20.5%, 34.1%, respectively, and those derived from AHs were 13.3%, 8.12% and 13.2%, respectively. The greatest concentration of SHs (1.1 ± 0.2) × 104 mg kg−1 dm was observed at S1, and the least concentration of (2.6 ± 0.8) × 103 mg kg−1 dm was observed at S4. S11 had the greatest concentrations of TAHs (5.4 ± 0.2) × 103 mg kg−1 dm, and the least concentration of (1.1 ± 0.5) × 103 mg kg−1 dm was observed at S8.
The 35 alkanes (C8 ~ C40) consisted predominantly of carbon chain lengths of C8 ~ 32, with C8 ~ C19 and C20 ~ C32 alkanes, which accounted for 47.0% and 50.4%, respectively. Concentrations of alkanes (C8 ~ C40) in the residential zone, ranging from 2.8 ± 0.2 × 102 to 7.2 ± 0.7 × 103 μg kg−1 dm, showed a significantly lesser range compared those in the oil producing zone, which ranged from 1.9 ± 0.1 × 104 to 6.6 ± 0.2 × 103 μg kg−1 dm, and oil-refinery and transportation zones, where they ranged from (1.8 ± 0.2) × 104 to (1.3 ± 0.1) × 104 μg kg−1 dm) (p < 0.01). However, there was no significant difference in concentrations of ∑16 PAHs between the oil-producing zone, where they ranged from (1.7 ± 0.1) × 102 to (1.3 ± 0.05) × 103 μg kg−1 dm, residential zone, where they ranged from (1.9 ± 0.2) × 102 to (4.6 ± 0.4) × 102 μg kg−1 dm) and oil-refinery and transportation zone, ranging from (1.8 ± 0.2) × 102 to (2.8 ± 0.3) × 103 μg kg−1 dm. PAHs were composed mainly of naphthalene, phenanthrene and pyrene. These three PAHs constituted approximately 55.7% of ∑16 PAHs in analyzed soils.
Values of pH of soils of the Dagang oilfield ranged from 8.38 to 8.63, and were homogeneous among zones. Salt contents of soils from different zones was more variable with concentrations ranging from 1.42 to 53.83 g kg−1 dm. There were significant (p < 0.05) differences in salinity between those of the residential zone and other zones, with a large variability of salinity in the residential zone where concentrations ranged from 3.32 to 58.83 g kg−1 dm. Compared with the salt content of other major Chinese oilfields: Shengli (9.8 g kg−1 dm), Changqing (4.3 g kg−1 dm), Daqing (7.6 g kg−1 dm), Yumen (4.4 g kg−1 dm), Jianghan (5.3 g kg−1 dm)25, the Dagang oilfield had markedly greater salinities with an average of 16.47 g kg−1 dm in this study.
Distribution of degradation genes
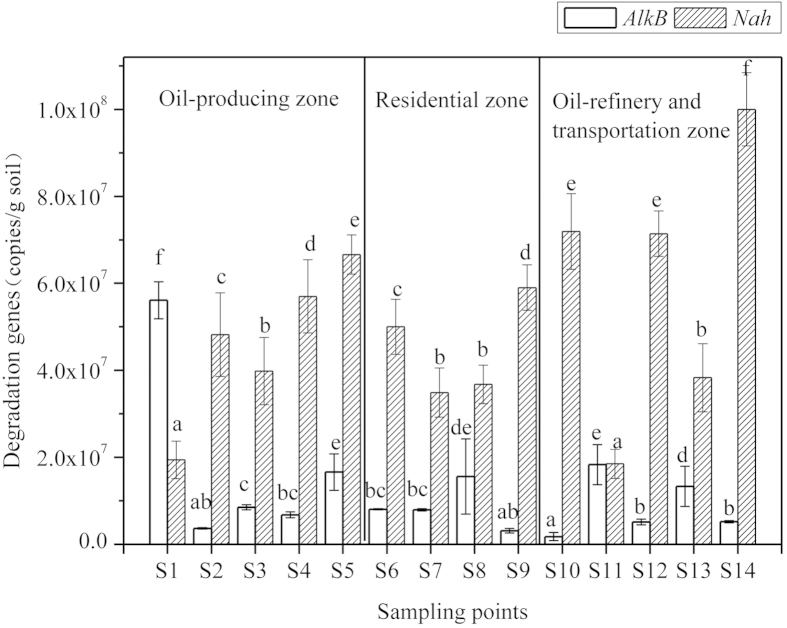
Numbers of copies of genes AlkB and Nah, coding for PH-degrading enzymes, that were detected by qPCR in soils of the Dagang Oilfield differed significantly among sampling sites (Fig. 1), and the relative abundance of AlkB and Nah genes were normalized to total 16S rDNA (Table S2). The standard curve for quantification of numbers of copies of PH-degrading genes (AlkB and Nah) and the 16S rDNA is presented in the supplementary materials (Figure S1). Abundances of AlkB exhibited large variations in different oil contaminated areas, ranging from (1.8 ± 0.9) × 106 to (5.6 ± 0.4) × 107 copies/g dm soil. Numbers of copies of AlkB in soils collected from S1, S5, S8, S11 and S13 were significantly (p < 0.05) greater than those at other sampling sites. The residential zone, which had a lesser concentration of alkanes, exhibited a lesser relative abundance of AlkB. Mean relative abundances of AlkB were as follows: oil-producing zones (3.0 ± 0.4) × 10−3 > oil-refinery and transportation zones (2.3 ± 0.3) × 10−3 > residential zones (4.0 ± 0.3) × 10−4.
Figure 1. Distribution of AlkB and Nah degradation genes in all the Dagang Oilfield soil samples.
Error bars represent standard deviations of efficiencies for PCR. Letters on the columns indicate a significant difference among sites at p < 0.05 according to Duncan’s multiple range tests of One-Way ANOVA.
Numbers of copies of Nah ranged from (1.9 ± 0.5) × 107 to (1.0 ± 0.08) × 108 copies/g dm soil, and abundance of Nah at S4, S5, S10, S12, S14 significantly (p < 0.05) greater than other sites. Distribution of Nah was relatively even across soils of oil-producing, residential, and oil-refinery and transportation zones ((4.6 ± 0.7) × 107, (4.5 ± 0.5) × 107, (6.2 ± 0.6) × 107 copies/g dm soil, respectively). However, the relative abundance of Nah fluctuated among S9 with the least value of (2.9 ± 0.4) × 10−4 and S14 with the greatest value of (8.1 ± 0.5) × 10−2.
DGGE analysis
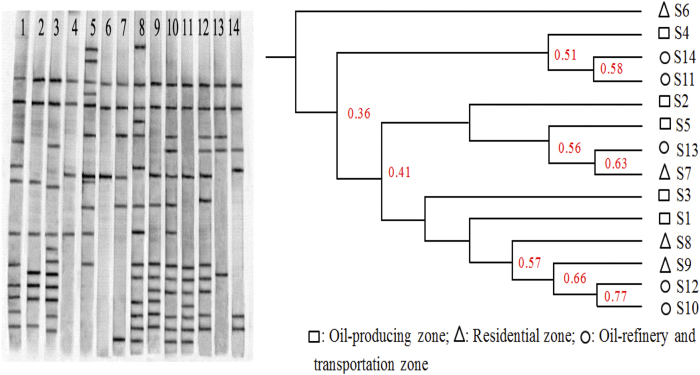
Diversity of microbial communities in soils, based on DGGE, was estimated by the number of amplified 16S rDNA bands (Fig. 2a), in which each band was assumed to represent a single operational taxonomic unit (OTU)26. Samples S1, S2, S3, S8, S9 and S10 displayed a greater number of bands, whereas samples S4, S6, S7 and S13 produced fewer distinct bands. Partitioning of features of microbial community structure can be seen in the dendrogram that was created by illustrating the similarity of microbial communities in soils (Fig. 2b). Microbial communities could be grouped into three major phylogenetic clusters. S10 and S12, which both were located in the oil refinery and transportation zones, revealed similar community structure with the highest similarity (85.9%), whereas S6 and S10 displayed the divergent communities with only 17.2% similarity. Moreover, the soils in the same zones with different concentrations of PHs and salinity were grouped into different clusters.
Figure 2. DGGE analysis result of different soil samples in Dagang oil field.
(a) Community profiles based on DGGE analysis of the V3 region of the 16S rDNA amplified by PCR of DNA extracts from soil samples of S1 to S14 (lanes 1–14). (b) Dendrogram analysis (UPGMA) of DGGE banding patterns from different soil samples.
Values of the Shannon-Wiener Index of 14 soils from the Dagang Oilfield were between 1.5 and 3, and Uniformity Indexes were almost equivalent in each soil (Table S3). In general, greater Shannon-Wiener Indexes of petroleum contaminated soils at locations S1, S2, S5, S8 and S10 were more similar to soils S3 and S9, which had lesser salinities and differ greatly from soils S6, S7 and S14, which had greater salinities.
Relationship between degradation genes and other parameters
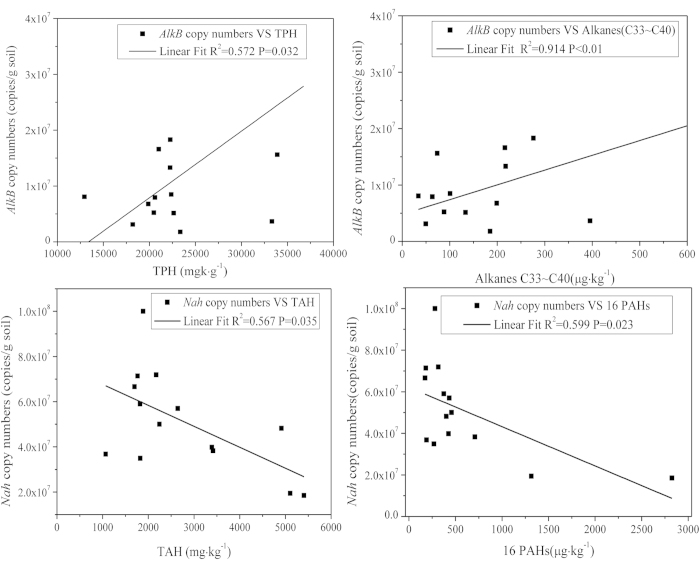
Numbers of copies of AlkB were positively correlated with various petroleum components, including TPHs (R2 = 0.573, p = 0.032) and long-chain alkanes (C33 ~ C40) (R2 = 0.914, p < 0.01) (Fig. 3). Soils collected from the oil-producing zone, which contained greater concentrations of alkanes exhibited the greatest numbers of copies of AlkB. Numbers of copies of Nah were negatively correlated with TAH (R2 = −0.567, p = 0.035) and ∑16 PAHs (R2 = −0.599, p = 0.023) (Fig. 3).
Figure 3. Correlations between numbers of copies of genes involved in degradation with the responding petroleum components.
Factor analysis
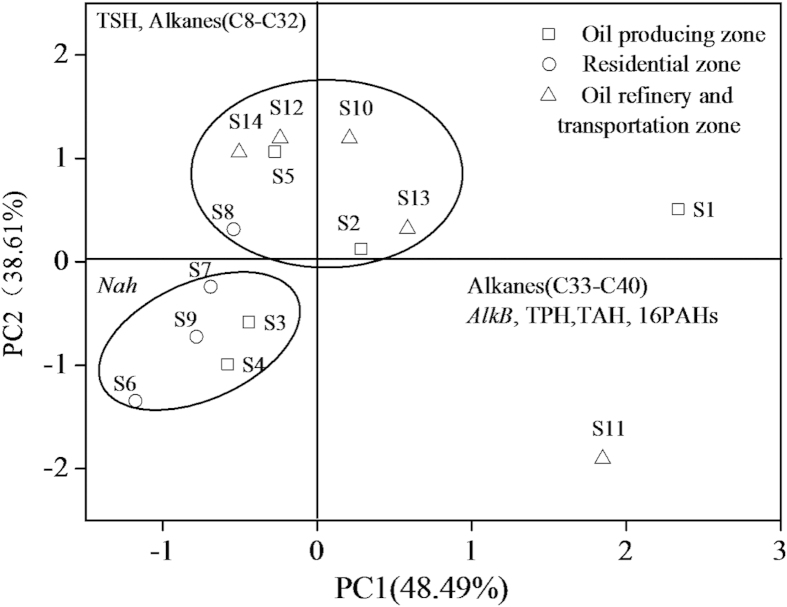
When the eleven measures of contamination of soils by PHs and two classes of genes involved in degradation of PHs were analyzed by factor analysis, the first two principal components explained 73.7% of the total variance (Fig. 4). Sites S1 and S11 were ordinated differently from all other locations, which indicated different sources of PHs. S1 was the most contaminated site, and had the greatest abundance of AlkB. S6, S7, S9 in soils of the residential zone as well as S3, S4 grouped together in the lower left quadrants of the graph, an area that was characterized by greater concentrations of lesser molecular weight PHs and a general accumulation of microorganisms containing the Nah gene. Locations S2, S5, S8, S10, S12, S13 and S14 were ordinated together in the upper portion of the diagram, which indicated that these samples were predominated by AHs and alkanes (C8 ~ C32). Copies of AlkB had larger factor scores compared with abundance of Nah in these areas. Even though some sites, such as S2, S3, S4 and S5 were relatively close together, they were ordinated differently except samples from the residential zone, which were clustered together. Factor analysis showed that different sites were not plotted according to the located zones.
Figure 4. Factor analysis of degradation genes and the petroleum components.
PC1 and PC2: first and second principal components in the analysis
Discussion
The correlation between abundance of oil-degrading genes and oil pollution in different zones represented a useful method for describing factors involved in bioremediation soils contaminated with oil. Numbers of copies of the AlkB and Nah genes in soils differed among the three zones. Soils in the residential zone with lesser concentrations of alkanes and relative abundance of AlkB might be explained by lesser oil emission and leakage than other sampling sites. Dagang Oilfield soils exhibited abundances of AlkB similar to those in soils from other oil exploring areas, and greater than those in sediment contaminated with crude oil that were investigated previously. The number of copies of the AlkB gene varied from 2.56 × 106 to 9.37 × 107 copies per gram dry soil in Daqing Oilfield soils, and 7.48 × 105 to 6.63 × 107 copies per gram dry soil in Karamay Oilfield soils27. The number of copies of the AlkB gene ranged from 1.1 × 105 to 2.9 × 105 copies g−1 in Timor Sea sediment contaminated with oil28.
Compared with numbers of copies of AlkB, numbers of copies of Nah were approximately 10-fold greater in soils of all three zones. Multimeric, naphthalene dioxygenase was the major enzyme involved in aerobic metabolism of naphthalene that can also mineralize phenanthrene, BTEX and other PAHs through ring-opening, and terminal oxidation29. The Nah degradation gene is generally found in soils and sediments contaminated with PHs. However, the terminal monooxygenase encoded by the AlkB gene has greater specificity in degradation of constituents of PHs than the enzyme encoded by the Nah gene, and degradation targets are primarily short and medium-chain alkanes (C6 ~ C15)16,23,30,31,32. Although the alkane monooxygenase enzyme can also mineralize longer-chain alkanes (C30 ~ C40), appropriate sets of primers were designed for specific PHs in particular soils28,33. In conclusion, naphthalene dioxygenases with a wider range of target substrates compared to alkane monooxygenase are likely to be more common in oil contaminated soils, and thus, are hypothesized to have resulted in greater numbers of copies of Nah reported here.
Differences in numbers of DGGE bands suggest that variations in bacterial communities might be affected synthetically by concentrations of petroleum and salinity. Communities of microbes in S1, S2, S8, S9 and S10 with greater species diversity were selectively enriched by higher concentration of petroleum. Soils from S6, S7 and S14, which had lesser species diversities were affected by greater salinity, whereas, S3 and S9 which had lesser salinities, exhibited richer biodiversity, even though both of these sites had lesser concentrations of PHs. A previous study identified only a few halophilic microbes, including Halomonas and Dietzia, by use of polymorphism community fingerprinting PCR in saline soils with 20% NaCl near Comodoro Rivadavia in Patagonia that were contaminated with diesel fuel30. Other studies have found inhibition of growth of microbes and diversities of genes involved in degradation of PHs and a decrease in oil degradation rates at sites with a greater salt content (>5% NaCl)31,34. Thus, greater salinity and pH are likely limiting factors for bioremediation PHs in soils of the Dagang. The diversity indexes observed were consistent with the conclusion that the presence of greater concentrations of TPHs enriched the microbial community, while salinity tended to inhibit the microbial communities of soils.
Numbers of copies of genes, quantified by Q-PCR, that are involved in degradation of PH, can be used as the biomarker to reflect the bioremediation potential of microbes in oil contaminated soils. The statistically significant, positive correlation between the number of copies of the AlkB gene and concentrations of alkanes clearly illustrated that oil-degrading microorganisms containing AlkB gene were a good indicator of long-term exposure to alkanes in contaminated soils. This conclusion is consistent with that of Pérez-de-Mora et al. who found a positive correlation between alkane contents and the abundance of AlkB in soils at four forest sites co-contaminated with mineral oil hydrocarbons and metals35. Genes involved in degrading longer-chain n-alkane were also found in soils contaminated with PHs33,36.
Abundances of Nah genes was between 3 × 101 and 9 × 104 copies/g dm soil at sites in the vicinity of Trollberget, Etna and Sköldvik, southern Finland and were positively correlated with the rate of aerobic mineralization of 14C-naphthalene37. S1 and S11, which had the greatest abundances of 16 PAHs, also had the least number of copies of Nah. This result indicated that greater concentrations of PAHs resulted in lesser abundance of microorganisms capable of degrading PHs. Soils with lesser concentrations of naphthalene (10 μg g−1) exhibited greater expression of naphthalene dioxygenase during vermicompost remediation for 30 days, while greater naphthalene concentrations of 100 μg g−1 led to lesser expression of naphthalene dioxygenase38. It is hypothesized that the greater concentrations of PAHs observed in soils at sites S1 and S11 were toxic to certain microbes that express the Nah gene, and thus, resulted in a lesser abundance of this gene.
In this study, correlations between oil degradation genes (AlkB and Nah) and different oil pollution in oil producing, residential, and oil-refinery and transportation zones was studied. Concentration of oil and salinity both affected expression of genes involved in degradation of oil as well as biodiversity of indigenous microorganisms. Dynamic quantification of oil degradation genes can be used as biomarker to estimate the bioremediation capacity of indigenous microbes in oil contaminated soils. In addition, metabolic enzymes encoded by responding oil degradation genes played crucial roles in the oil degradation process. Therefore, further investigation of the enzyme activity is of great significance.
Materials and Methods
Hydrocarbon-contaminated soils and sampling
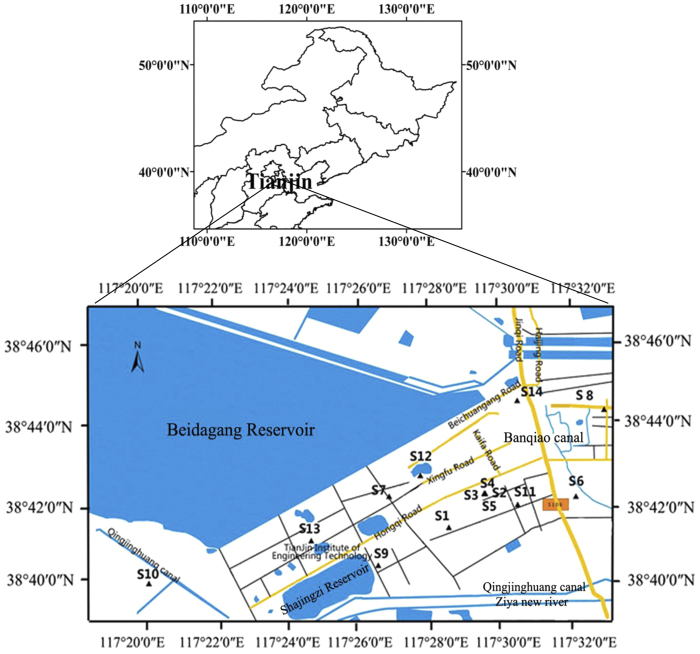
Fourteen soils contaminated with PHs were sampled from three different zones in the Dagang Oilfield area, which is situated in the southeast of Tianjin city, Northern China (lat.38°39′47.43″ ~ 38°44′42.37″N, long.117°20′1.47″ ~ 117°32′41.30″E): oil-producing, S1–S5 (Distance from the well: S1, S2 < 10 m; 20 < S3–S5 < 50 m); residential, S6–S9; and oil-refinery and transportation zones, S10–S14 (Table 2; Fig. 5). Soils were collected to a depth of ~10–20 cm. Four samples of soil were collected at each site using cross sampling methods by use of sterile spatulas. These four sub-samples were combined and then thoroughly homogenized to obtain a uniform composite for each site with total weight of 4 kg. Samples were transported to the laboratory on ice and stored at −20 °C for microbial analysis (DNA extraction and genetic characterization). Samples were air-dried for the determination of PHs and other physical and chemical properties such as pH and salt content.
Table 2. Latitude and longitude of sampling sites in Dagang Oilfield.
| Zones | Sampling number | Sampling sites | Latitude | Longitude |
|---|---|---|---|---|
| Oil-producing zone | S1 | Chuangxin Road | 38°41′17.92″ | 117°28′22.24″ |
| S2 | Lesser zone | 38°42′12.80″ | 117°29′21.14″ | |
| S3 | Middle zone | 38°42′12.69″ | 117°29′21.52″ | |
| S4 | Higher zone | 38°42′12.39″ | 117°29′22.39″ | |
| S5 | Test field zone | 38°42′12.53″ | 117°29′23.10″ | |
| Residential zone | S6 | Chuangxin Road-canal | 38°42′8.06″ | 117°31′54.31″ |
| S7 | Xingfu Road-pool | 38°42′7.77″ | 117°26′42.61″ | |
| S8 | Chuangye Road-oil tank | 38°44′28.13″ | 117°32′41.30″ | |
| S9 | Chuangxin Road | 38°40′16.79″ | 117°26′24.37″ | |
| Oil-refinery and transportation zone | S10 | Maxi Road | 38°39′47.43″ | 117°20′1.47″ |
| S11 | Chuangxin Road-living zones | 38°41′54.77″ | 117°30′17.01″ | |
| S12 | Xingfu Road1 | 38°42′41.63″ | 117°27′34.44″ | |
| S13 | Xingfu Road2 | 38°40′56.46″ | 117°24′32.47″ | |
| S14 | Yard 2 bridge | 38°44′42.37″ | 117°30′16.16″ |
Figure 5. Sampling area and location of sampling points.
The figure map was generated by using software ArcGIS 10 (Environmental Systems Research Institute, Inc. Redlands, US). The sampling sites were located by using Global Positioning System (GPS).
Measurement of salinity and pH
Salinity was determined by use of a gravimetric method. In brief, a suspension of soil (1:5 soil: deionized water, w:w) was heated in water bath at 100 °C to dryness, and 10% H2O2 was added as oxidant subsequently for three times. The residual was then weighted and salinity was calculated. The pH of the soil suspension was measured by a pH meter (Sartorius, Germany).
Gravimetric quantification of total petroleum hydrocarbon (TPH)
Five-gram aliquots of soils were Soxhlet-extracted for 18 h with 125 ml dichloromethane at 54 °C39,40. Extracts were concentrated to dryness by use of a rotary evaporator, and concentrations of TPHs were determined gravimetrically41. The increment in mass of round-bottom flasks after evaporation of the extracts was defined as the final TPHs concentration. All extracts were analyzed in triplicate.
Gravimetric identification and quantification of components of PHs
Methods for purification and separation of constituents of PHs were modified from those previously published42,43. In brief, concentrated extracts were eluted from a glass column (dimensions: 20 mm × 400 mm) containing pre-rinsed activated silica gel and neutral aluminum (12 g:6 g, soaked with hexane) using the following sequence of solvents: 20 mL of hexane, 70 mL of hexane and dichloromethane (1:1), 50 mL of methanol to separate SHs, AHs and polar components. After purification and separation, the three solvent fractions were reduced to dryness and concentrations of PHs were determined by gravimetric measurement of the extracted residues as described above.
Quantification of PAHs and saturated hydrocarbons by GC/MS
Concentrations of PAHs and SHs were determined by use of a 6850 Agilent HP gas chromatograph connected to a 5975 Agilent HP mass spectrometer (Agilent, CA, USA). Components of PHs were separated by a Thermo Trace GC Ultra system equipped with a Thermo DB-5MS capillary column coated with 5% diphenyl and 95% dimethyl polysiloxane stationary phase (30 m × 0.25 mm, i.d. 0.25 μm film thickness; Thermo Scientific, Runcorn, UK), operating with helium (99.99% purity) as the carrier gas at a constant flow of 1.0 ml min−1. 1 μl aliquots of each were injected at 280 °C in pulsed, splitless mode (1 min, then split ratio 1:50 to the end of analysis). The GC oven temperature was held at 60 °C for 3 min, and then temperature was increased by 15 °C/min from 60 °C to 180 °C, followed by 6 °C/min from 180 °C to 300 °C, and then temperature was held at 300 °C for 10 min. The mass spectrometer was operated with the ion source at 220 °C with an ionization energy of 70 eV. Five different concentrations of mixtures of 16 target PAHs and 33 target alkanes (C8 ~ C40) were used as external standards for determination of components of extracts. PAHs and SHs were quantified in single ion monitoring (SIM) mode, with the molecular ion of each PAH and SH component corresponding to the elution retention time of the external standard.
Quantification of petroleum degrading genes and 16S rDNA
Real-time q-PCR based on fluorescent dye SYBR green I was used to quantify two PH degradation genes (AlkB and Nah) and 16S rDNA. Degenerated primer sets, designed for AlkB amplication in 17 microbes and Nah amplication in 33 PAHs degradation bacterial species were used according to previously studies44,45. Primer sets for 16S rDNA were adapted from Suzuki46. The sequence, amplicon size and annealing temperature conditions of the PCR primers are given (Table S4).
DNA was extracted from soils by use of a ZR Soil Microbe DNA MiniPrep™ (Zymo Research, USA) following the protocol provided by the manufacturer. Conventional PCR was used to verify and recover the target genes. Each PCR reaction mixture contained 10× Easy Taq buffer, 0.2 mM total concentrations of dNTPs mixture, 5 mU Easy Taq DNA polymerase, 0.4 μmol of both forward and reverse primers, and 2 μL DNA per plate. Conditions for conventional PCR were set according to previously published methods47: initial denaturation at 94 °C for 5 min; 35 cycles of denaturing at 94 °C for 20 s, annealing temperature of target genes were set as Table S1, for 2 min; and final extension at 72 °C for 7 min. PCR products were examined by 1.5% agarose gel electrophoresis.
Target genes were recovered from agarose gel by using gel extraction kit (Axygen, USA), and were combined with pEASY-T1 vectors (Trans Gen Biotech, China) and transformed into Escherichia coli JM109 (Takara, Japan). Plasmids carrying target genes were extracted with Plasmid kit (Omega, USA) and quantified by use of a nucleic acid analyzer (Bibby Scientific Limited, United Kingdom) to construct standard curves. Because lengths of the vector and target gene inserts were known, gene copy numbers were calculated directly from extracted plasmid DNA concentration. For every set of primers, standard curves were obtained from ten-fold serial dilutions of template DNA prepared from plasmids containing target genes (Figure S1; Supplemental Materials).
Quantification of 16S rDNA, AlkB, and Nah degradation genes by qPCR were carried out on a BioRad CFX96 (Hercules, USA) with a C1000 thermal cycler iCycler by using Quantitect® SYBR green PCR kits (Trans Gen Biotech, China) following the manufacturer’s directions. Reaction mixtures contained 12.5 μL 2 × trans startTM top green qPCR super mix, 0.5 μL passive reference dye, and 1 μL of recombinant plasmids DNA and soil DNA as template for the construction of standard curve and quantification of target genes, respectively. Concentrations of primers were optimized to 0.2 μmol for both the forward and reverse primer. Cycling conditions for real-time qPCR were as follows: hold for 30 s at 94 °C followed by 40 cycles of denaturing at 94 °C for 5 s, annealing temperature of primers target genes were set as Table S4, for 15 s, extension and first plate read at 72 °C for 10 s; hold at 55 °C for 30 s; a second plate read at 55 °C hold for 5 s followed by a melt curve from 55 °C to 95 °C (increment = 0.5 °C/10 s).
Every template plasmid DNA was run in triplicate and each experiment was repeated at twice in order to generate a reproducible dataset and avoid false detections in environmental samples. Melt curve analyses were used to detect the formation of primer dimmers and other amplification of nonspecific sequences. Data were analyzed with CF Manager Software (version 2.1, Bio-Rad, US). The limit of detection for quantification of each gene was determined by comparing the linear relationship between the base-10 logarithm of diluted concentrations of the plasmid DNA and the fluorescence signal.
Analysis of microbial community by PCR-DGGE
PCR- DGGE technology was adopted to assess the microbial community of soil samples targeting the V3 region of bacterial 16S rDNA. The primer set targeting the V3 region of bacterial 16S rDNA consisted of GC-338f (5′-GCclamp- CACGGGGGGACTCCTACGGGAGGCAGCAG-3′) (GC clamp = CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGG) and 518r (5′-ATTACCGCGGCTGCTGG-3′). PCR amplicons were loaded with loading dye into 8% polyacrylamide gel (37.5:1 acrylamide: bisacrylamide) with a denaturing gradient from 15% to 45% (100% denaturant consisted of 7 M urea and 40% formamide, v/v). The DGGE analysis was carried out on a Universal Mutation Detection System D-code (BioRad, CA, USA) at140 V and 60 °C for 3 h. Gels were then stained with ethidium bromide, visualized with an UV transilluminator (ATTO Corporation, Japan). Dendrogram and intensities analysis of DGGE banding patterns was performed using Quantity One 4.6 (Bio-Rad Laboratories, CA, USA). Calculation of the pair-wise similarities was based on the Dice correlation coefficient. Dendrograms were created using the algorithm of un-weighted pair-group method with the arithmetic averages (UPGMA)48.
Statistical analysis
All mathematical and statistical computations were conducted using SPSS 16.0 (IBM, New York, USA). Comparison of concentrations of PHs among sites was accomplished by use of One-Way ANOVA. Differences among zones were analyzed by use of the nonparametric, Kruskal-Wallis test. Normality was confirmed by the Kolmogorov-Smirnov test and homogeneity of variance was confirmed by use of Levine’s test. In order to further investigate distributions of degradation genes and the correlation with the petroleum pollution status of Dagang Oilfield soils, 13 factors were measured to characterize soils. Inter-relationships were analyzed by factor analysis (FA) on the varimax-rotated factors, and factor loadings were calculated by use of eigenvalues greater than 1.049.
Additional Information
How to cite this article: Liu, Q. et al. Distribution of petroleum degrading genes and factor analysis of petroleum contaminated soil from the Dagang Oilfield, China. Sci. Rep. 5, 11068; doi: 10.1038/srep11068 (2015).
Supplementary Material
Acknowledgments
This work was supported by (1) National Natural Science Foundation of China (31270544, 41473070), (2) 863 Major Program (2013AA06A205), (3) Research Fund for the Doctoral Program of Higher Education. The research was supported by a Discovery Grant from the Natural Science and Engineering Research Council of Canada (Project # 326415-07) and a grant from the Western Economic Diversification Canada (Project # 6578, 6807 and 12711). The authors wish to acknowledge the support of an instrumentation grant from the Canada Foundation for Infrastructure. Prof. Giesy was supported by the program of 2014 “High Level Foreign Experts” (#GDT20143200016) funded by the State Administration of Foreign Experts Affairs, the P.R. China to Nanjing University and the Einstein Professor Program of the Chinese Academy of Sciences. Profs. Giesy and Hecker were also supported by the Canada Research Chair program.
Footnotes
Author Contributions Q.L. and J.T. wrote the main manuscript text. Z.B. prepared the Figure 5. J.P.G. and M.H. prepared the Discussion part of the manuscript. All authors reviewed the manuscript.
References
- Ji K. et al. Genotoxicity and endocrine-disruption potentials of sediment near an oil spill site: two years after the Hebei spirit oil spill. Environ Sci Technol 45, 7481–7488 (2011). [DOI] [PubMed] [Google Scholar]
- Council, N. R. Oil in the sea: Inputs, Fates, and Effects. National Academy Press, Washington DC. (2002). [Google Scholar]
- Libbey L. M. A paradox database for GC/MS data on components of essential oils and other volatiles. J Essen Oil Res 3, 193–194 (1991). [Google Scholar]
- Barrutia O. et al. Plant tolerance to diesel minimizes its impact on soil microbial characteristics during rhizoremediation of diesel-contaminated soils. Sci Total Environ 409, 4087–4093 (2011). [DOI] [PubMed] [Google Scholar]
- Maliszewska-Kordybach B. Polycyclic aromatic hydrocarbons in agricultural soils in Poland: Preliminary proposals for criteria to evaluate the level of soil contamination. Appl Geochem 11, 121–127 (1996). [Google Scholar]
- Abba E. J., Unnikrishnan S., Kumar R., Yeole B. & Chowdhury Z. Fine aerosol and PAH carcinogenicity estimation in outdoor environment of Mumbai City, India. Int J Environ Heal R 22, 134–149 (2012). [DOI] [PubMed] [Google Scholar]
- Carreras H. A., Calderón-Segura M. E., Gómez-Arroyo S., Murillo-Tovar M. A. & Amador-Muñoz O. Composition and mutagenicity of PAHs associated with urban airborne particles in Córdoba, Argentina. Environ Pollut 178, 403–410 (2013). [DOI] [PubMed] [Google Scholar]
- Balachandran C., Duraipandiyan V., Balakrishna K. & Ignacimuthu S. Petroleum and polycyclic aromatic hydrocarbons (PAHs) degradation and naphthalene metabolism in Streptomyces sp. (ERI-CPDA-1) isolated from oil contaminated soil. Bioresource Technol 112, 83–90 (2012). [DOI] [PubMed] [Google Scholar]
- Beilen J. B. v. et al.Analysis of Pseudomonas putida alkane degradation gene clusters and flanking insertion sequences : evolution and regulation of the alk genes. Microbiol 147, 1621–1630 (2001). [DOI] [PubMed] [Google Scholar]
- Throne-Holst M. et al. Utilization of n-alkanes by a newly isolated strain of Acinetobacter venetianus: the role of two AlkB-type alkane hydroxylases. Appl Microbiol Biot 72, 353–360 (2006). [DOI] [PubMed] [Google Scholar]
- Whyte L. G. et al. Physiological adaptations involved in alkane assimilation at a low temperature by Rhodococcussp. Strain Q15. Appl Environ Microb 56, 2961–2968 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrodi D. V. et al. Identification of the key genes of naphthalene catabolism in soil DNA. Microbiol 72, 597–604 (2003). [PubMed] [Google Scholar]
- Kahng H. Y., Malinverni J. C., Majko M. M. & Kukor J. J. Genetic and functional analysis of the tbc operons for catabolism of alkyl- and chloroaromatic compounds in Burkholderia sp. strain JS150. Appl Environ Microb 67, 4805–4816 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin B. R. et al. Enumeration of aromatic oxygenase genes to evaluate biodegradation during multi-phase extraction at a gasoline-contaminated site. J Hazard Mater 163, 524–530 (2009). [DOI] [PubMed] [Google Scholar]
- Balba M. T., Al-Awadhi N. & Al-Daher R. Bioremediation of oil-contaminated soil: microbiological methods for feasibility assessment and field evaluation. J Microbiol Meth 32, 155–164 (1998). [Google Scholar]
- Beilen J. B. & Funhoff E. G. Alkane hydroxylases involved in microbial alkane degradation. Appl Microbiol Biot 74, 13–21 (2007). [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. Cloning and expression of naphthalene dioxygenase genes from Comamonas sp. MQ for indigoids production. Process Biochem 48, 581–587 (2013). [Google Scholar]
- Paisse S., Duran R., Coulon F. & Goñi-Urriza M. Are alkane hydroxylase genes (alkB) relevant to assess petroleum bioremediation processes in chronically polluted coastal sediments? Appl Microbiol Biot 92, 835–844 (2011). [DOI] [PubMed] [Google Scholar]
- Kloos K., Munch J. C. & Schloter M. A new method for the detection of alkane-monooxygenase homologous genes (alkB) in soils based on PCR-hybridization. J Microbiol Meth 66, 486–496 (2006). [DOI] [PubMed] [Google Scholar]
- Panicker G., Mojib N., Aislabie J. & Bej A. K. Detection, expression and quantitation of the biodegradative genes in Antarctic microorganisms using PCR. Antonie van Leeuwenhoek 97, 275–287 (2009). [DOI] [PubMed] [Google Scholar]
- Powell S. M., Ferguson S. H., Bowman J. P. & Snape I. Using Real-Time PCR to assess changes in the hydrocarbon-degrading microbial community in antarctic soil during bioremediation. Microbial Ecol 52, 523–532 (2006). [DOI] [PubMed] [Google Scholar]
- Witzig R., Junca H., Hecht H. J. & Pieper D. H. Assessment of toluene/biphenyl dioxygenase gene diversity in benzene-polluted soils: links between benzene biodegradation and genes similar to those encoding isopropyl benzene dioxygenases. Appl Environ Microb 72, 3504–3514 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Wang L. & Shao Z. Diversity and abundance of oil-degrading bacteria and alkane hydroxylase (alkB) genes in the subtropical seawater of Xiamen Island. Microbial Ecol 60, 429–439 (2010). [DOI] [PubMed] [Google Scholar]
- Yergeau E. et al. Microarray and Real-Time PCR analyses of the responses of high-Arctic soil bacteria to hydrocarbon pollution and bioremediation treatments. Appl Environ Microb 75, 6258–6267 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y. et al. Functional gene diversity of soil microbial communities from five oil-contaminated fields in China. ISME J 5, 403–413 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer C., Dobbs F. C. & Karl D. M. Estimation of diversity and community structure through restriction fragment length polymorphism distribution analysis of bacterial 16S rRNA genes from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microb 60, 871–879 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Wang J., Liao J., Xie S. & Huang Y. Abundance and diversity of soil petroleum hydrocarbon-degrading microbial communities in oil exploring areas. Appl Microbiol Biot 99, 1935–1946 (2014). [DOI] [PubMed] [Google Scholar]
- Wasmund K., Burns K. A., Kurtboke D. I. & Bourne D. G. Novel Alkane Hydroxylase Gene (alkB) Diversity in Sediments Associated with Hydrocarbon Seeps in the Timor Sea, Australia. Appl Environ Microb 75, 7391–7398 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn Y., Sanseverino J. & Sayler G. S. Analyses of polycyclic aromatic hydrocarbon-degrading bacteria isolated from contaminated soils. Biodegr 10, 149–157 (1999). [DOI] [PubMed] [Google Scholar]
- Kleinsteuber S., Riis V., Fetzer I., Harms H. & Muller S. Populationdynamics within a microbial consortium during growth on diesel fuel in saline environments. Appl Environ Microb 72, 3531–3542 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich A. C. et al. Effect of salt on aerobic biodegradation of petroleum hydrocarbons in contaminated groundwater. Biodegr 20, 27–38 (2008). [DOI] [PubMed] [Google Scholar]
- Cappelletti M. et al. Analyses of both the alkB gene transcriptional start site and alkB promoter-inducing properties of Rhodococcus sp. Strain BCP1 grown on n-alkanes. Appl Environ Microb 77, 1619–1627 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throne-Holst M., Wentzel A., Ellingsen T. E., Kotlar H. K. & Zotchev S. B. Identification of novel genes involved in long-chain n-alkane degradation by Acinetobacter sp. strain DSM 17874. Appl Environ Microb 73, 3327–3332 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minai-Tehrani D., Minoui S. & Herfatmanesh A. Effect of salinity on biodegradation of polycyclic aromatic hydrocarbons (PAHs) of heavy crude oil in soil. B Environ Contam Tox 82, 179–184 (2008). [DOI] [PubMed] [Google Scholar]
- Pérez-de-Mora A., Schulz S. & Schloter M. MPN- and Real-Time-based PCR methods for the quantification of alkane monooxygenase homologous genes (alkB) in environmental samples. Methods Mol Biol 599, 59–68 (2010). [DOI] [PubMed] [Google Scholar]
- Margesin R. & Schinner F. Biodegradation and bioremediation of hydrocarbons in extreme environments. Appl Microbiol Biot 56, 650–663 (2001). [DOI] [PubMed] [Google Scholar]
- Tuomi P. M., Salminen J. M. & Jørgensen K. S. The abundance of nahAc genes correlates with the 14C-naphthalene mineralization potential in petroleum hydrocarbon-contaminated oxic soil layers. FEMS Microbiol Ecol 51, 99–107 (2004). [DOI] [PubMed] [Google Scholar]
- Di Gennaro P. et al. Dynamic changes in bacterial community structure and in naphthalene dioxygenase expression in vermicompost-amended PAH-contaminated soils. J Hazard Mater 172, 1464–1469 (2009). [DOI] [PubMed] [Google Scholar]
- Wang P. et al. Evaluation of Soxhlet extraction, accelerated solvent extraction and microwave-assisted extraction for the determination of polychlorinated biphenyls and polybrominated diphenyl ethers in soil and fish samples. Anal Chim Acta 663, 43–48 (2010). [DOI] [PubMed] [Google Scholar]
- Luque de Castro M. D. & Priego-Capote F. Soxhlet extraction: Past and present panacea. J Chromatogr A 1217, 2383–2389 (2010). [DOI] [PubMed] [Google Scholar]
- Rajakovic V., Aleksic G., Radetic M. & Rajakovic L. Efficiency of oil removal from real wastewater with different sorbent materials. J Hazard Mater 143, 494–499 (2007). [DOI] [PubMed] [Google Scholar]
- Villalobos M., Avila-Forcada A. P. & Gutierrez-Ruiz M. E. An improved gravimetric method to determine total petroleum hydrocarbons in contaminated soils. Water Air Soil Pol 194, 151–161 (2008). [Google Scholar]
- Peng S., Zhou Q., Cai Z. & Zhang Z. Phytoremediation of petroleum contaminated soils by Mirabilis Jalapa L. in a greenhouse plot experiment. J Hazard Mater 168, 1490–1496 (2009). [DOI] [PubMed] [Google Scholar]
- Powell S. M., Bowman J. P., Ferguson S. H. & Snape I. The importance of soil characteristics to the structure of alkane-degrading bacterial communities on sub-Antarctic Macquarie Island. Soil Biol Biochem 42, 2012–2021 (2010). [Google Scholar]
- Park J.-W. & Crowley D. E. Dynamic changes in nahAc gene copy numbers during degradation of naphthalene in PAH-contaminated soils. Appl Microbiol Biot 72, 1322–1329 (2006). [DOI] [PubMed] [Google Scholar]
- Suzuki M. T., Taylor L. T. & Delong E. F. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 59-nuclease assays. Appl Environ Microb 66, 4605–4614 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamura N., Fukui M., Ward D. M. & Inskeep W. P. Assessing soil microbial populations responding to crude-oil amendment at different temperatures using phylogenetic, functional gene (alkB) and physiological analyses. Environ Sci Technol 42, 7580–7586 (2008). [DOI] [PubMed] [Google Scholar]
- Labb´e D., Margesin R., Schinner F., Whyte L. G. & Greer C. W. Comparative phylogenetic analysis of microbial communities in pristine and hydrocarbon-contaminated Alpine soils. FEMS Microbiol Ecol 59, 466–475 (2007). [DOI] [PubMed] [Google Scholar]
- Huang F., Wang X., Lou L., Zhou Z. & Wu J. Spatial variation and source apportionment of water pollution in Qiantang River (China) using statistical techniques. Water Res 44, 1562–1572 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.