Abstract
Background: Vitamin B-6 interconversion enzymes are important for supplying pyridoxal 5′-phosphate (PLP), the co-enzyme form, to tissues. Variants in the genes for these enzymes [tissue nonspecific alkaline phosphatase (ALPL), pyridoxamine 5′-phosphate oxidase, pyridoxal kinase, and pyridoxal phosphatase] could affect enzyme function and vitamin B-6 status.
Objectives: We tested whether single-nucleotide polymorphisms (SNPs) in these genes influence vitamin B-6 status markers [plasma PLP, pyridoxal (PL), and 4-pyridoxic acid (PA)], and explored potential functional effects of the SNPs.
Methods: Study subjects were young, healthy adults from Ireland (n = 2345). We measured plasma PLP, PL, and PA with liquid chromatography–tandem mass spectrometry and genotyped 66 tag SNPs in the 4 genes. We tested for associations with single SNPs in candidate genes and also performed genome-wide association study (GWAS) and gene-based analyses.
Results: Seventeen SNPs in ALPL were associated with altered plasma PLP in candidate gene analyses (P < 1.89 × 10−4). In the GWAS, 5 additional ALPL SNPs were associated with altered plasma PLP (P < 5.0 × 10−8). Gene-based analyses that used the functional linear model β-spline (P = 4.04 × 10−15) and Fourier spline (P = 5.87 × 10−15) methods also showed associations between ALPL and altered plasma PLP. No SNPs in other genes were associated with plasma PLP. The association of the minor CC genotype of 1 ALPL SNP, rs1256341, with reduced ALPL expression in the HapMap Northern European ancestry population is consistent with the positive association between the CC genotype and plasma PLP in our study (P = 0.008). No SNP was associated with altered plasma PL or PA.
Conclusions: In healthy adults, common variants in ALPL influence plasma PLP concentration, the most frequently used biomarker for vitamin B-6 status. Whether these associations are indicative of functional changes in vitamin B-6 status requires more investigation.
Keywords: vitamin B-6, pyridoxal phosphate, pyridoxal, pyridoxic acid, alkaline phosphatase, dietary supplements
Introduction
Vitamin B-6 functions as an enzyme cofactor in numerous biochemical reactions, including the synthesis and degradation of neurotransmitters, homocysteine transsulfuration, heme synthesis, and the metabolism of amino acids, lipids, and glycogen (1). It is present in humans as 6 vitamers: 3 related pyridine compounds (pyridoxal, pyridoxine, and pyridoxamine) and their phosphate esters (2). Pyridoxal 5′-phosphate (PLP)12, the co-enzyme form of vitamin B-6, is required for >150 enzyme-catalyzed reactions (3).
Humans use enzymes encoded by 3 genes to perform the interconversion of vitamin B-6 forms: pyridoxal (pyridoxine, vitamin B-6) kinase (PDXK), pyridoxamine 5′-phosphate oxidase (PNPO), and tissue nonspecific alkaline phosphatase (ALPL) (Supplemental Figure 1) (4). After intestinal absorption of B-6 vitamers and their transport to the liver as nonphosphorylated forms, pyridoxal (PL), pyridoxine, and pyridoxamine are phosphorylated by PDXK [Online Mendelian Inheritance in Man (OMIM) 179020] to PLP, pyridoxine 5′-phosphate, and pyridoxamine 5′-phosphate. PNPO (OMIM 603287) then oxidizes pyridoxine 5′-phosphate and pyridoxamine 5′-phosphate to PLP. Hepatic PLP is exported into the circulation (bound to albumin) for delivery to all tissues. Before it can enter cells, PLP is dephosphorylated by ALPL (OMIM 171760). In addition, PLP can be hydrolyzed inside cells by pyridoxal (pyridoxine, vitamin B-6) phosphatase (PDXP; OMIM 609246), an enzyme with widespread expression in human tissues (5).
Proper functioning of the enzymes that interconvert B-6 vitamers is critical for an adequate supply of PLP in tissues. Genetic variants that alter the functions of these enzymes could affect vitamin B-6 status, as measured by circulating concentrations of PLP, PL, and 4-pyridoxic acid (PA; vitamin B-6 catabolite), the main forms of vitamin B-6 in plasma (6). Both rare and common variants in the genes for these enzymes are known to be associated with altered vitamin B-6 concentration. Rare loss-of-function variants in the PNPO gene lead to low PLP concentrations in the brain that cause neonatal epileptic encephalopathy (7), and inactivating variants in the ALPL gene cause a hypophosphatasia that results in impaired skeletal mineralization and also elevated plasma PLP concentrations (8, 9). In addition, 4 common variants in ALPL are associated with plasma PLP concentration in genome-wide association studies (GWASs) (10–12). We hypothesized that common genetic variants in the vitamin B-6 interconversion enzymes could affect vitamin B-6 status in the general population. Therefore, we investigated whether single-nucleotide polymorphisms (SNPs) in the ALPL, PNPO, PDXK, and PDXP genes are associated with plasma PLP, PL, and PA concentrations in a group of healthy, young adults. We also performed GWASs and gene-based analyses to determine whether other analytic approaches would produce results similar to those obtained from candidate gene analyses. For SNPs showing statistically significant associations, we performed in silico analyses to explore their potential regulatory functions. Further, we examined intake of vitamin B-6 from fortified foods/supplements and tested for associations between this intake and plasma B-6 vitamer concentrations.
Methods
Study subjects.
A sample of university students who attended Trinity College, Dublin (TCD), Ireland, between 2003 and 2004 formed the study population. Eligible subjects were between 18 and 28 y of age at the time of study enrollment, did not report a serious medical condition, and were ethnically Irish. All study participants provided written informed consent. The Research Ethics Committee of the Dublin Federated Hospitals, which is affiliated with TCD, and the Institutional Review Board of the National Human Genome Research Institute gave ethical approval for the study. Further details of this study were published previously (13–15).
Data collection.
Subjects completed a questionnaire to provide data on age at study enrollment, sex, height, weight, medical conditions that could affect the absorption of nutrients from food, unusual diets (e.g., vegetarian), smoking, alcohol use, and consumption of supplements and fortified foods. We calculated BMI (kg/m2) from verified reports of height and weight. We also assessed oral contraceptive use in women because these medications are associated with decreased blood PLP concentrations (16). Women were classified as oral contraceptive users if they indicated filling a prescription for an oral contraceptive in the past 12 mo and also did not indicate that they had stopped using the contraceptive.
Participants were asked to recall their fortified food and supplement intake in the past week and over an average month. Data from intakes in the past week were used in this study. We obtained active nutrient information for each fortified food and supplement to determine the amount of each nutrient consumed. Subjects were provided with a list of fortified foods and definitions of serving sizes, and nutrient intake from fortified foods was determined on the basis of the fortified food consumed, frequency of consumption, and reported serving size. Similarly, subjects were asked to report their supplement intake within the past week. Active nutrient information for supplements was converted to microgram of nutrient per portion (tablet or liquid) or recorded in International Unit format. Data in International Unit format were converted to microgram of nutrient per portion, according to standard conversion rates. Data on the quantity and frequency of intake of both fortified foods and supplements in the past week were used to calculate the average amount of individual nutrients from these dietary sources that each subject consumed per day.
Laboratory assays.
Each subject provided 30 mL of blood (nonfasting) for measurement of metabolites and DNA extraction. The concentrations of PLP, PL, PA, and creatinine in plasma and serum cotinine concentrations were measured by liquid chromatography–tandem mass spectrometry, as described previously (17, 18). These analyses were performed by the BEVITAL AS laboratory by using an Agilent series 1100 HPLC instrument outfitted with a thermostatted autosampler and degasser and a triple-quadrupole tandem mass spectrometer from Applied Biosystems/MSD SCIEX with electrospray ionization source.
The Tagger function in Haploview 4.2 (19) was used to select tag SNPs (minor allele frequency > 0.05, r2 < 0.8) for each candidate gene plus their 10-kb flanks on the basis of the HapMap sample of Utah residents with Northern and Western European ancestry (CEU) (HapMap Data Release 27) (20). SNP genotyping was performed by detecting allele-specific extension products via matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (Sequenom). We attempted to genotype 76 tag SNPs in ALPL, PNPO, PDXK, and PDXP, but 10 SNPs failed the assay. Of the 66 SNPs successfully genotyped (39 in ALPL, 11 in PNPO, 15 in PDXK, and 1 in PDXP), the genotype call rate was ≥95% for 56 SNPs and between 92% and 95% for 10 SNPs. After replating 3% of DNA samples, genotype concordance was ≥95% for all 66 SNPs. All discordant genotypes were excluded from analysis. Testing for Hardy-Weinberg equilibrium showed that 2 SNPs had P < 0.01: ALPL rs2071424 (P = 0.0023) and PDXK rs3746954 (P = 0.0018).
Statistical analysis.
We used the Wilcoxon’s rank-sum test to compare continuous variables, and Fisher’s exact test to compare categorical variables between subgroups defined by vitamin B-6 intake from fortified foods/supplements and by SNP genotype. To examine the correlation between plasma PLP, PL, and PA concentrations, we used linear regression and the Spearman rank correlation test after performing an inverse normal rank transformation to produce normally distributed data.
For the candidate gene approach, we used an additive genetic model in linear regression to test the association between plasma B-6 vitamer concentrations and each of the 66 tag SNPs, adjusting for amount of vitamin B-6 intake from fortified foods/supplements (mg/d), sex, BMI, serum cotinine (nmol/L), and alcohol intake (g/d). In a subgroup analysis restricted to women, use of oral contraceptives (yes, no) was added as a covariate. We performed an inverse normal rank transformation of the variables for plasma PLP, PL, and PA concentrations before proceeding with linear regression analyses because the distributions of these variables were skewed to the right. The Bonferroni method was used to adjust for multiple comparisons (66 × 4 = 264 tests; P < 1.89 × 10−4 was considered statistically significant).
For the GWAS analysis of 2232 subjects recruited at TCD (13–15), details of genotyping, data cleaning, and tests for population substructure are described in the Supplemental Methods and in Desch et al. (15). We used genotypes for 757,337 autosomal SNPs that had a minor allele frequency >0.01. SNPs with extreme deviation from Hardy-Weinberg equilibrium (P < 1.0 × 10−4) were flagged for future reference and remained in the analysis (n = 3512). We performed single-SNP tests of association with log10 transformed plasma B-6 vitamer concentrations on 2158 of these individuals by using an additive genetic model in linear regression analyses. We included age at study enrollment, sex, and vitamin B-6 intake from fortified/foods supplements as covariates in the regression analyses. We considered P < 5.0 × 10−8 to have met the threshold for genome-wide statistical significance.
In addition, to perform a gene-based analysis, we applied F-distributed tests of functional linear models (FLMs) to data for all subjects. The details of the F-distributed tests of FLMs are available in Fan et al. (21). Briefly, we considered a person’s observed genotypes to be the outcome of a stochastic process, and, treating a person’s genetic data as a stochastic function, we modeled the genetic effects of the genetic variants as a function termed the genetic variant function. To estimate the genetic variant function, we used the observed genotype data and functional data analysis techniques in the form of β-spline or Fourier spline basis functions. We then used the estimated genetic variant function in FLM regression analyses to test for association between the genetic variants and the quantitative trait (inverse normal rank transformation of plasma B-6 vitamer concentration). We measured statistical significance of the association tests by applying F-distributed test statistics to the FLMs. We considered P < 3.1 × 10−6 to be statistically significant in accordance with a previous report (22) that used this P value threshold in a meta-analysis of gene-based association tests.
Statistical analyses were performed with SAS 9.3 (SAS Institute), PLINK (23), and R 2.12.0 (24).
Results
After excluding 3 subjects with bowel resection, Crohn disease, or ulcerative colitis, 16 subjects with missing data for plasma B-6 vitamer concentrations, and 78 siblings (so that only 1 sibling from a family was included in the analysis data set), there were 2411 study subjects. We considered that for persons with extremely high intakes of vitamin B-6 from fortified foods/supplements, plasma B-6 vitamer concentrations are more likely to reflect high vitamin B-6 intake rather than variation in the activities of enzymes involved in vitamin B-6 metabolism. Therefore, we chose a value 10 times the Estimated Average Requirement (1.1 mg/d for adults aged 19–50 y) (25) as a cutoff for high intake and excluded 66 persons whose reported intake of vitamin B-6 from fortified foods/supplements was >11 mg/d. After these exclusions, 2345 subjects remained for analysis.
Nearly 86% of subjects reported intake of vitamin B-6 in the past week from either fortified foods or supplements (Table 1). The median intake amount was 0.56 mg/d. Approximately 80% of subjects obtained vitamin B-6 from fortified foods, whereas only ∼20% used supplements that contained vitamin B-6 (Table 1). Consumers and nonconsumers of vitamin B-6 from fortified foods/supplements were similar for the percentage of men, age, BMI, alcohol intake, serum cotinine, and plasma creatinine (Supplemental Table 1). Plasma PLP, PL, and PA concentrations increased for all subjects with the amount of intake of vitamin B-6 from fortified foods/supplements (Supplemental Figure 2). Fifty-eight subjects (2.5%) had a plasma PLP concentration <30 nmol/L, a suggested minimum value that represents adequate vitamin B-6 status in adults (26), and only 3 subjects (0.1%) had a concentration <20 nmol/L, the value used for adequate status by the Food and Nutrition Board in setting the recommended daily intake for vitamin B-6 (25).
TABLE 1.
Vitamin B-6 intake from fortified foods and supplements among healthy Irish adults1
| Vitamin B-6 intake |
||
| Source of vitamin B-6 | Subjects reporting intake | Amount of intake from vitamin B-6 source, mg/d |
| Any fortified foods or supplements | 2008 (85.6) | 0.56 [0.26–1.33] |
| Fortified foods | 1915 (81.7) | 0.41 [0.10–0.69] |
| Supplements | 496 (21.2) | 2.00 [1.43–3.87] |
Values are n (%) or median [IQR] for those subjects who reported vitamin B-6 intake. n = 2345.
In a regression model that tested for associations with tag SNPs in candidate genes and that was adjusted for the amount of vitamin B-6 intake from fortified foods/supplements, sex, BMI, serum cotinine, and alcohol intake, 17 SNPs in ALPL (of 39 tag SNPs investigated) were associated with plasma PLP concentration after correction for multiple comparisons (Table 2). The β coefficients showed that the minor alleles of 10 SNPs were associated with reduced plasma PLP concentration; the minor alleles of the other 7 SNPs were positively associated with plasma PLP concentration. The minor allele frequency was >0.10 for 15 of the 17 SNPs and was between 0.06 and 0.10 for 2 SNPs (Table 2). In a subgroup analysis restricted to women and that included use of oral contraceptives as a covariate, 13 of the 17 SNPs were significantly associated with plasma PLP. The 17 PLP-associated SNPs span the ALPL gene, which in turn is divided in the first intron into 2 blocks of D’ (Supplemental Figure 3). SNPs in the other 3 genes were not associated with plasma PLP (Supplemental Table 2). None of the SNPs in ALPL, PNPO, PDXK, or PDXP was associated with plasma PL or PA concentrations (Supplemental Table 3).
TABLE 2.
ALPL SNPs associated with plasma pyridoxal 5′-phosphate concentration in healthy Irish adults, based on candidate gene analyses1
| All subjects 3 (n = 2345) |
Women 4 (n = 1368) |
|||||||||
| SNP | Base pair position2 on chromosome 1 | Gene region | Major, minor alleles | Minor allele frequency | β | SE | P | β | SE | P |
| rs4654748 | 21786068 | 5′ near gene (NBPF3 intron) | C, T | 0.48 | 0.15 | 0.03 | 4.61 × 10−8 | 0.15 | 0.04 | 7.71 × 10−5 |
| rs6664097 | 21834693 | 5′ near gene | C, A | 0.22 | −0.15 | 0.03 | 6.47 × 10−6 | −0.18 | 0.05 | 7.68 × 10−5 |
| rs67039765 | 21836934 | Intron 1 | C, T | 0.10 | −0.21 | 0.05 | 2.23 × 10−6 | −0.22 | 0.06 | 4.15 × 10−4 |
| rs8691805 | 21839730 | Intron 1 | G, A | 0.32 | 0.13 | 0.03 | 1.54 × 10−5 | 0.14 | 0.04 | 5.59 × 10−4 |
| rs75365455 | 21844822 | Intron 1 | C, T | 0.38 | 0.11 | 0.03 | 1.77 × 10−4 | 0.13 | 0.04 | 1.47 × 10−3 |
| rs12145027 | 21847548 | Intron 1 | T, C | 0.24 | 0.18 | 0.03 | 4.67 × 10−8 | 0.19 | 0.04 | 1.60 × 10−5 |
| rs9628653 | 21848962 | Intron 1 | T, A | 0.23 | 0.15 | 0.03 | 8.84 × 10−6 | 0.18 | 0.05 | 1.34 × 10−4 |
| rs17803155 | 21863956 | Intron 1 | T, C | 0.29 | −0.14 | 0.03 | 1.63 × 10−6 | −0.13 | 0.04 | 1.62 × 10−3 |
| rs1256341 | 21866829 | Intron 1 | T, C | 0.27 | 0.16 | 0.03 | 1.09 × 10−6 | 0.20 | 0.04 | 4.56 × 10−6 |
| rs1256342 | 21876492 | Intron 1 | C, T | 0.15 | −0.17 | 0.04 | 1.89 × 10−5 | −0.21 | 0.05 | 9.46 × 10−5 |
| rs1256335 | 21890386 | Intron 5 | A, G | 0.22 | −0.24 | 0.03 | 7.39 × 10−13 | −0.27 | 0.05 | 2.00 × 10−9 |
| rs1256331 | 21895008 | Intron 7 | C, T | 0.23 | 0.16 | 0.03 | 3.47 × 10−7 | 0.16 | 0.04 | 1.24 × 10−4 |
| rs1780318 | 21895625 | Intron 7 | T, A | 0.06 | −0.29 | 0.06 | 2.22 × 10−7 | −0.29 | 0.08 | 1.31 × 10−4 |
| rs2275370 | 21900420 | Intron 9 | G, A | 0.21 | −0.21 | 0.03 | 1.17 × 10−9 | −0.23 | 0.05 | 3.43 × 10−7 |
| rs1780329 | 21902950 | Intron 10 | C, A | 0.19 | −0.16 | 0.03 | 2.89 × 10−6 | −0.18 | 0.05 | 9.18 × 10−5 |
| rs1772719 | 21904374 | 3′ UTR | A, C | 0.23 | −0.22 | 0.03 | 4.70 × 10−2 | −0.26 | 0.04 | 2.06 × 10−9 |
| rs1772720 | 21905007 | 3′ near gene | G, A | 0.07 | −0.29 | 0.05 | 2.66 × 10−8 | −0.31 | 0.07 | 1.04 × 10−5 |
ALPL, tissue nonspecific alkaline phosphatase gene; NBPF3, neuroblastoma breakpoint family, member 3 gene; SNP, single-nucleotide polymorphism; UTR, untranslated region.
Based on National Center for Biotechnology Information–Genome Reference Consortium Human genome build 37/human genome 19 assembly.
Linear regression model adjusted for amount of vitamin B-6 intake from fortified foods/supplements, sex, BMI, serum cotinine, and alcohol intake; outcome variable was inverse normal rank transformation of plasma pyridoxal 5′-phosphate concentration.
Linear regression model adjusted for amount of vitamin B-6 intake from fortified foods/supplements, BMI, serum cotinine, alcohol intake, and use of oral contraceptives; outcome variable was inverse normal rank transformation of plasma pyridoxal 5′-phosphate concentration.
For analyses restricted to women, P values failed to reach statistical significance (P < 1.89 × 10−4).
Two alternative methods produced results consistent with those from the candidate gene analysis: a GWAS and a gene-based method. With the use of the GWAS approach, 9 ALPL SNPs were associated with log-transformed plasma PLP concentration at P values that reached genome-wide statistical significance (Table 3). Four of these SNPs (rs1772719, rs1255335, rs1772710, and rs2275370) were also associated with plasma PLP in our candidate gene analyses (Table 2). Additional GWAS results and quantile–quantile plots are in Supplemental Table 4 and Supplemental Figures 4–7. Of the 10 tag SNPs that failed the genotyping assay on the matrix-assisted laser desorption/ionization–time-of-flight mass spectrometer, 6 were directly genotyped in the GWAS, and the remaining 4 were covered in the GWAS by proxy SNPs with varying amounts of linkage disequilibrium (LD) (r2 between 0.52 and 0.93) in persons of European ancestry in the 1000 Genomes Project (27). One of the 10 SNPs (rs4021228 that was directly genotyped in the GWAS) was associated with plasma PLP at genome-wide statistical significance (Table 3). The other 9 SNPs were not among our strongest signals from the GWAS (Table 3; Supplemental Table 5). Combining results from the candidate gene and GWAS approaches, we observed associations between 22 ALPL SNPs and plasma PLP. In the GWAS, SNPs in other genes were not associated with plasma PLP. We observed positive correlations between plasma PLP, PL, and PA concentrations (Supplemental Figure 8). However, no SNP reached genome-wide statistical significance in GWAS analyses for plasma PL or PA (Supplemental Figures 9 and 10).
TABLE 3.
Genome-wide association study results of ALPL SNPs associated with plasma pyridoxal 5′-phosphate in healthy Irish adults1
| SNP | Base pair position2 on chromosome 1 | Gene region | Major, minor alleles | Minor allele frequency | β3 | SE | P |
| rs1772719 | 21904374 | 3′ untranslated region | A, C | 0.23 | −0.06 | 0.01 | 2.48 × 10−16 |
| rs1256335 | 21890386 | Intron 5 | A, G | 0.22 | −0.06 | 0.01 | 4.38 × 10−14 |
| rs1780321 | 21824108 | 5′ near gene | A, G | 0.33 | −0.05 | 0.01 | 2.80 × 10−12 |
| rs1780324 | 21821757 | 5′ near gene | C, T | 0.49 | −0.04 | 0.01 | 1.17 × 10−11 |
| rs1697421 | 21823292 | 5′ near gene | G, A | 0.49 | 0.04 | 0.01 | 3.40 × 10−11 |
| rs1772720 | 21905007 | 3′ near gene | G, A | 0.08 | −0.07 | 0.01 | 8.15 × 10−10 |
| rs2275370 | 21900420 | Intron 9 | G, A | 0.21 | −0.05 | 0.01 | 1.74 × 10−9 |
| rs4021228 | 21863691 | Intron 1 | G, T | 0.47 | 0.04 | 0.01 | 4.25 × 10−9 |
| rs1780316 | 21889635 | Exon 5 | C, T | 0.07 | −0.07 | 0.01 | 8.11 × 10−9 |
n = 2158 subjects. ALPL, tissue nonspecific alkaline phosphatase gene; SNP, single-nucleotide polymorphism.
Based on National Center for Biotechnology Information–Genome Reference Consortium Human genome build 37/human genome 19 assembly.
Linear regression model adjusted for age, sex, and vitamin B-6 intake from fortified/foods supplements; outcome variable was log10 transformation of plasma pyridoxal 5′-phosphate concentration.
For the gene-based approach, all variants in a gene are grouped together to test for association; therefore, we restricted the analyses to the 3 candidate genes (ALPL, PDXK, and PNPO) that had genotype data available for multiple SNPs. In these analyses (involving use of genotype data for 2345 subjects), the ALPL gene’s association with plasma PLP, based on the FLM β-spline (P = 4.04 × 10−15) and Fourier spline (P = 5.87 × 10−15) methods, had a higher level of statistical significance than the single-SNP associations from the candidate gene and GWAS approaches (Supplemental Table 6). The 2 other genes tested in gene-based analyses, PDXK and PNPO, were not associated with plasma PLP. None of the 3 genes was associated with plasma PL or PA in gene-based analyses (Supplemental Table 6).
Tryptophan catabolism through the kynurenine pathway depends on PLP as an enzyme cofactor. With the use of an additive genetic model in linear regression analyses, the 22 PLP-associated SNPs in ALPL were assessed for association with plasma concentrations of the kynurenine pathway metabolites, 3-hydroxykynurenine and the 3-hydroxykynurenine/xanthurenic acid ratio, reported to correlate inversely with plasma PLP and to be possible vitamin B-6 status markers (28, 29). No significant effects were observed (data not shown).
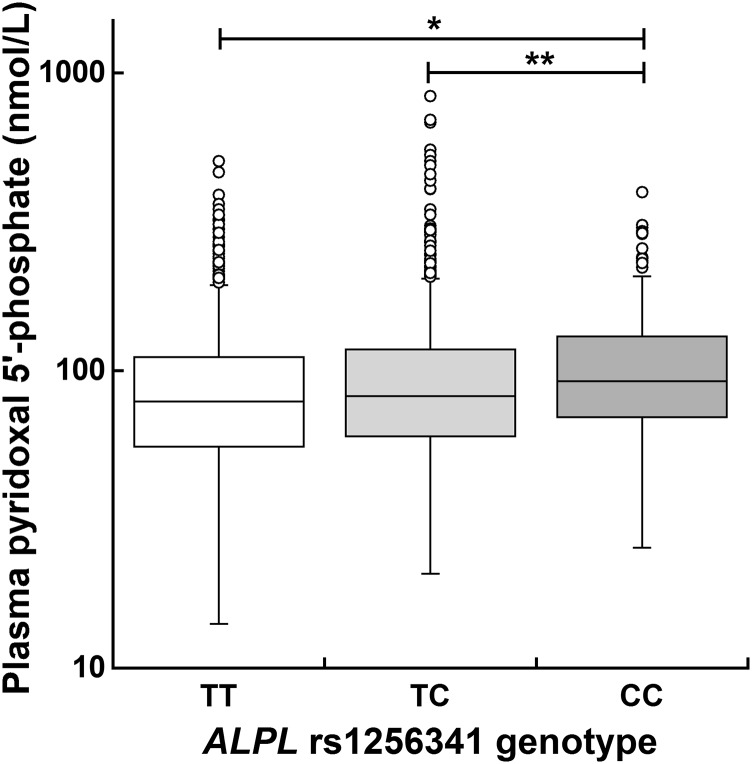
We used the Genevar database (30), which contains data on gene expression in lymphoblastoid cell lines from HapMap individuals (31), to explore whether the ALPL SNPs associated with plasma PLP in our Irish study population were also associated with ALPL expression in other persons of European ancestry (HapMap CEU population). Data were available for 17 of the 22 SNPs (Supplemental Figure 11). The only statistically significant association was for rs1256341 (P = 0.016) (Supplemental Figure 11G). Homozygosity for the minor C allele was associated with decreased ALPL expression. The subset (6.5%) of our study population who had the minor CC genotype also had a median plasma PLP concentration (92.2 nmol/L) that was significantly greater than that of subjects with either the TT (78.9 nmol/L; P = 0.00012) or TC (82.0 nmol/L; P = 0.0084) genotype (Figure 1). This observation is congruent with the CC genotype’s association with decreased ALPL expression because this decreased expression is likely to result in reduced PLP dephosphorylation by the ALPL-encoded enzyme.
FIGURE 1.
Plasma pyridoxal 5′-phosphate concentration according to the ALPL rs1256341 genotype in healthy Irish adults. One hundred thirty-two (5.63%) of the 2345 study subjects had missing genotype data for ALPL rs1256341. Of the 2213 subjects with rs1256341genotype data, 1178 (53.2%) had the TT genotype, 892 (40.3%) had the TC genotype, and 143 (6.5%) had the CC genotype. The median plasma pyridoxal 5′-phosphate concentration was 78.9 nmol/L for the TT genotype, 82.0 nmol/L for the TC genotype, and 92.2 nmol/L for the CC genotype. *P = 0.00012, **P = 0.0084; Wilcoxon’s rank-sum test. ALPL, tissue nonspecific alkaline phosphatase gene.
To investigate further the potential regulatory functions of the 22 SNPs, we used the rSNPBase database (32) which reports the location of SNPs relative to putative regulatory regions of the genome annotated by the Encyclopedia of DNA Elements project (33) that used DNA from various human cell lines. One or more of 18 of the SNPs were located in regions of open chromatin (accessible to regulatory factors), histone modification (often found near promoters, enhancers, and other regulatory elements), and RNA polymerase II interactions with chromatin (RNA polymerase II binding site that could participate in transcriptional regulation) and in regions that contain binding sites for transcription factors and an RNA-binding protein encoded by the poly(A) binding protein, cytoplasmic 1 gene (Supplemental Tables 7–12). With the use of the University of California at Santa Cruz genome browser (34), we determined that none of the SNPs was located in CpG islands (often found near transcription start sites and promoter regions) or in the ALPL target site for hsa-mir-204–5p, a microRNA that was shown to influence ALPL expression (35).
Most (>95%) circulating PLP is bound to serum albumin; this binding protects PLP from dephosphorylation by phosphatases such as the enzyme encoded by ALPL (36). Because unbound, circulating PLP is likely to be dephosphorylated to PL, the binding of PLP to albumin is a factor in determining circulating PLP concentration. We sought to determine whether genetic variants associated with serum albumin concentration are also associated with changes in plasma PLP; therefore, we performed additional analyses by using 6 SNPs that influenced serum albumin concentrations in a prior GWAS that involved a study population with European ancestry (37). Genotype data were available for 4 of the SNPs (rs1260326, rs16948098, rs11078597, rs4806073) in our GWAS data set. For the other 2 SNPs (rs13381710, rs739347), we used genotype data for nearby SNPs (rs12954590, rs10403394) selected to act as proxies (r2 > 0.92). On the basis of an additive genetic model in linear regression analyses, none of the SNPs was associated with plasma PLP concentration in our study population (Table 4).
TABLE 4.
Test for association between serum albumin GWAS SNPs and plasma PLP concentration in healthy Irish adults1
| Results from serum albumin GWAS (n > 38,000) |
Results from this study for plasma PLP (n = 2219) |
|||||||
| SNP | Chr | Base pair position2 | Candidate genes | Major, minor alleles | β | P | β3 | P |
| rs1260326 | 2 | 27730940 | GCKR-FNDC4 | C, T | 0.01 | 2.9 × 10−14 | 0.05 | 0.11 |
| rs16948098 | 15 | 44219607 | FRMD5-WDR76 | G, A | 0.02 | 1.9 × 10−8 | −0.07 | 0.33 |
| rs11078597 | 17 | 1618363 | SERPINF2-WDR81 | T, C | 0.02 | 6.8 × 10−13 | −0.01 | 0.79 |
| rs4806073 | 19 | 35555190 | HPN-SCN1B | C, T | 0.03 | 3.3 × 10−15 | −0.08 | 0.94 |
| rs129545904 | 18 | 60155750 | TNFRSF11A-ZCCHC2 | C, T | 0.01* | 3.9 × 10−9* | 0.01 | 0.84 |
| rs104033945 | 19 | 49999345 | RPS11-FCGRT | G, A | 0.02** | 3.2 × 10−8** | −0.07 | 0.23 |
Serum albumin GWAS SNPs were reported in Franceschini et al. (37). Chr, chromosome number; FCGRT, Fc fragment of IgG, receptor, transporter, α gene; FNDC4, fibronectin type III domain containing 4 gene; FRMD5, FERM domain containing 5 gene; GCKR, glucokinase (hexokinase 4) regulator gene; GWAS, genome-wide association study; HPN, hepsin gene; PLP, pyridoxal 5′-phosphate; RPS11, ribosomal protein S11 gene; SCN1B, sodium channel, voltage-gated, type I, β subunit gene; SERPINF2, serpin peptidase inhibitor, clade F (α-2 antiplasmin, pigment epithelium derived factor), member 2 gene; SNP, single-nucleotide polymorphism; TNFRSF11A, tumor necrosis factor receptor superfamily, member 11a, nuclear transcription factor κB activator gene; WDR76, WD repeat domain 76 gene; WDR81, WD repeat domain 81 gene; ZCCHC2, zinc finger, CCHC domain containing 2 gene.
Based on National Center for Biotechnology Information–Genome Reference Consortium Human genome build 37/human genome 19 assembly.
Linear regression used inverse normal rank transformation of plasma pyridoxal 5′-phosphate concentration as the outcome variable; model adjusted for age and sex.
Nearby rs13381710 (base pair position 60153329 on chromosome 18). *GWAS result for rs13381710.
Nearby rs739347 (base pair position 50001385 on chromosome 19). **GWAS result for rs739347.
Discussion
In our study population of healthy, young adults, we observed statistically significant associations between 22 ALPL SNPs and plasma PLP concentration. These include 4 SNPs (rs4654748, rs1256335, rs1697421, and rs1780316) that were associated with plasma PLP in previous GWASs of one-carbon metabolites (10–12). The results of the 3 different analytic methods (candidate gene, GWAS, and gene-based approaches) applied to our data all strongly suggest that ALPL is the most important gene that influences plasma PLP concentration. We had high statistical power to rule out the effects of common variants in other genes on plasma PLP concentration and of SNPs in any gene on plasma PL and PA concentrations. We also observed that SNPs previously shown to influence serum albumin concentration were not associated with plasma PLP concentration in our study population.
The ALPL locus encodes tissue nonspecific alkaline phosphatase (TNAP), an enzyme (attached to the outer cell membrane) that can dephosphorylate PLP extracellularly (38). Multiple lines of evidence suggest that TNAP activity is a major determinant of plasma PLP concentration. First, investigations into hypophosphatasia, caused by inactivating mutations in ALPL, found that these mutations also caused elevated plasma PLP in affected patients (8, 9). Second, patients with liver disease often have a low plasma PLP concentration that is associated with elevated TNAP activity in the circulation (39). Third, in population-based studies, 4 ALPL SNPs are associated with plasma PLP at a genome-wide level of statistical significance (10–12), as mentioned, and we have obtained similar findings in our GWAS by using a cohort recruited at TCD.
The liver is suggested as a possible site of PLP dephosphorylation by TNAP (40), based on observations that TNAP activity is readily inducible in liver (41) and is further increased in patients with liver disease (in association with low plasma PLP concentrations) (39). The physiologic role of TNAP in liver is unknown, but an animal study has shown that TNAP from liver cell membranes is able to hydrolyze PLP (42). Most PLP in the circulation is normally bound to albumin (36), whereas only free PLP is a substrate for alkaline phosphatase (43). Human serum contains a substantial amount of TNAP protein (44), but it is uncertain to what extent TNAP activity in the circulation directly regulates plasma PLP concentration.
Reports of associations between genetic variants and circulating TNAP concentration in population-based studies suggest a role for genetic factors in regulating the amount of TNAP protein produced. One of the 22 PLP-associated SNPs (rs1780324) is among 3 SNPs (the other 2 are rs2242420 and rs1976403) in or near the ALPL gene that were associated with circulating alkaline phosphatase concentrations in prior GWASs (45–47). On the basis of data for persons of European ancestry in the 1000 Genomes Project (27), another 8 PLP-associated SNPs share LD (D’ > 0.90, r2 < 0.40) with rs2242420, and an additional 2 share LD (D’ > 0.85, r2 > 0.50) with rs1976403 (data not shown). However, no reports to date indicate whether rs1780324, rs2242420, or rs1976403 have a functional effect on alkaline phosphatase. The 22 PLP-associated SNPs and SNPs associated with circulating PLP or alkaline phosphatase concentrations in previous GWASs (10–12, 45–47) are present in both LD blocks that span the ALPL gene. Therefore, we were unable to define a specific region of the gene that could harbor functional variants that affect PLP dephosphorylation.
Only 1 of the 22 ALPL SNPs is located in a coding region: the synonymous rs1780316 (S110S) variant in exon 5 of ALPL. Therefore, none of the SNPs leads to amino acid changes, and it is unknown whether any has direct functional consequences. Our observation that the locations of 18 of the 22 SNPs coincided with at least 1 putative regulatory region identified by the Encyclopedia of DNA Elements project suggests that the SNPs could potentially affect the regulation of ALPL transcription. A genomic locus that regulates transcription is likely to be associated with changes in the amount of RNA copied from DNA (expression quantitative trait locus). We identified a potential expression quantitative trait locus among the 22 SNPs by using the Genevar database (30). The ALPL rs1256341 homozygous minor CC genotype is associated with decreased ALPL mRNA expression in lymphoblastoid cell lines from a European ancestry population. This is not proof of a functional effect of rs1256341, but the direction of the association between ALPL expression and the CC genotype is consistent with the positive association between the rs1256341 minor C allele and plasma PLP concentration in our study population. That is, reduced PLP dephosphorylation and a concomitant increase in plasma PLP concentration are likely to result from decreased ALPL expression associated with the minor CC genotype.
Our search for previous reports of functional effects associated with any ALPL SNP found that a functional effect was reported for 1 ALPL common variant, rs3200254 T > C, not included in our study. Of the 22 PLP-associated SNPs in our study, rs1780329 is the one most closely linked to rs3200254 [D’ = 0.97, r2 = 0.64; 1000 Genomes Project (27); data not shown]. The rs3200254 C allele frequency is 0.12 in the HapMap CEU population. This SNP is a missense variant (T263H) situated near the metal-binding domains of TNAP (48). Enzyme activity assays that compared the effects of the T and C alleles showed that the C allele decreased the Michaelis constant of the enzyme reaction (48). The reduced Michaelis constant in the presence of the rs3200254 minor C allele and the association between the rs1780329 minor A allele and decreased plasma PLP in our study suggest that these minor alleles are associated with increased TNAP activity. The reported phenotypes observed with the minor alleles of both SNPs may therefore be driven by a single functional allele and may be the source of the plasma PLP association signal we detect.
Strengths of this study were the ethnically homogenous, large study population of healthy adults who did not have diseases that could impair vitamin B-6 absorption in the gut and the collection of detailed data on the amount of intake of vitamin B-6 from fortified foods/supplements. A weakness was that our vitamin B-6 intake data did not include the vitamin content from unfortified food; therefore, we could not account for vitamin B-6 intake from all dietary sources in regression analyses.
In summary, our finding that common variants that span the ALPL gene were associated with plasma PLP concentration adds to the body of evidence to suggest that TNAP, encoded by ALPL, has a role in regulating plasma PLP concentration. In comparison with previous GWAS reports, we have extended knowledge about ALPL SNPs and circulating PLP by using gene expression data to examine the possible functional effects of ALPL SNPs. The observation that the minor CC genotype of ALPL rs1256341 was associated with both increased plasma PLP and reduced ALPL expression suggests that the regulation of ALPL expression by common ALPL variants is a determinant of plasma PLP concentration. Most of the SNPs associated with plasma PLP had a minor allele frequency >0.10; therefore, the possible functional consequences of common ALPL variants should be investigated because these variants could influence plasma PLP concentration in a large percentage of the general population.
Supplementary Material
Acknowledgments
AMM, BS, LCB, and JLM designed the research; FP, AMM, ERG, ØM, and PMU conducted the research; TCC, FP, RF, YW, CDC, YK, AFW, and JEB-W analyzed the data; TCC wrote the paper; FP, AMM, BS, ERG, ØM, PMU, CDC, JEB-W, LCB, and JLM provided critical comments; TCC had primary responsibility for the final content. All authors contributed intellectually to the paper. All authors read and approved the final manuscript.
Footnotes
Abbreviations used: ALPL, tissue nonspecific alkaline phosphatase gene; CEU, sample of Utah residents with Northern and Western European ancestry in the HapMap project; FLM, functional linear model; GWAS, genome-wide association study; LD, linkage disequilibrium; OMIM, Online Mendelian Inheritance in Man; PA, 4-pyridoxic acid; PDXK, pyridoxal (pyridoxine, vitamin B-6) kinase gene; PDXP, pyridoxal (pyridoxine, vitamin B-6) phosphatase gene; PL, pyridoxal; PLP, pyridoxal 5′-phosphate; PNPO, pyridoxamine 5′-phosphate oxidase gene; SNP, single-nucleotide polymorphism; TCD, Trinity College, Dublin; TNAP, tissue nonspecific alkaline phosphatase protein.
References
- 1.John RA. Pyridoxal phosphate-dependent enzymes. Biochim Biophys Acta 1995;1248:81–96. [DOI] [PubMed] [Google Scholar]
- 2.di Salvo ML, Safo MK, Contestabile R. Biomedical aspects of pyridoxal 5′-phosphate availability. Front Biosci (Elite Ed) 2012;4:897–913. [DOI] [PubMed] [Google Scholar]
- 3.Percudani R, Peracchi A. A genomic overview of pyridoxal-phosphate-dependent enzymes. EMBO Rep 2003;4:850–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merrill AH Jr, Henderson JM. Vitamin B6 metabolism by human liver. Ann N Y Acad Sci 1990;585:110–7. [DOI] [PubMed] [Google Scholar]
- 5.Jang YM, Kim DW, Kang TC, Won MH, Baek NI, Moon BJ, Choi SY, Kwon OS. Human pyridoxal phosphatase. Molecular cloning, functional expression, and tissue distribution. J Biol Chem 2003;278:50040–6. [DOI] [PubMed] [Google Scholar]
- 6.Lumeng L, Lui A, Li TK. Plasma content of B6 vitamers and its relationship to hepatic vitamin B-6 metabolism. J Clin Invest 1980;66:688–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mills PB, Surtees RA, Champion MP, Beesley CE, Dalton N, Scambler PJ, Heales SJ, Briddon A, Scheimberg I, Hoffmann GF, et al. . Neonatal epileptic encephalopathy caused by mutations in the PNPO gene encoding pyridox(am)ine 5′-phosphate oxidase. Hum Mol Genet 2005;14:1077–86. [DOI] [PubMed] [Google Scholar]
- 8.Whyte MP, Mahuren JD, Vrabel LA, Coburn SP. Markedly increased circulating pyridoxal-5′-phosphate levels in hypophosphatasia. Alkaline phosphatase acts in vitamin B-6 metabolism. J Clin Invest 1985;76:752–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiss MJ, Cole DE, Ray K, Whyte MP, Lafferty MA, Mulivor RA, Harris H. A missense mutation in the human liver/bone/kidney alkaline phosphatase gene causing a lethal form of hypophosphatasia. Proc Natl Acad Sci USA 1988;85:7666–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka T, Scheet P, Giusti B, Bandinelli S, Piras MG, Usala G, Lai S, Mulas A, Corsi AM, Vestrini A, et al. . Genome-wide association study of vitamin B6, vitamin B12, folate, and homocysteine blood concentrations. Am J Hum Genet 2009;84:477–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hazra A, Kraft P, Lazarus R, Chen C, Chanock SJ, Jacques P, Selhub J, Hunter DJ. Genome-wide significant predictors of metabolites in the one-carbon metabolism pathway. Hum Mol Genet 2009;18:4677–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keene KL, Chen WM, Chen F, Williams SR, Elkhatib SD, Hsu FC, Mychaleckyj JC, Doheny KF, Pugh EW, Ling H, et al. . Genetic associations with plasma B12, B6, and folate levels in an ischemic stroke population from the Vitamin Intervention for Stroke Preventions (VISP) trial. Front Public Health 2014;2:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mills JL, Carter TC, Scott JM, Troendle JF, Gibney ER, Shane B, Kirke PN, Ueland PM, Brody LC, Molloy AM. Do high blood folate concentrations exacerbate metabolic abnormalities in people with low vitamin B-12 status? Am J Clin Nutr 2011;94:495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stone N, Pangilinan F, Molloy AM, Shane B, Scott JM, Ueland PM, Mills JL, Kirke PN, Sethupathy P, Brody LC. Bioinformatic and genetic association analysis of microRNA target sites in one-carbon metabolism genes. PLoS ONE 2011;6:e21851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desch KC, Ozel AB, Siemieniak D, Kalish Y, Shavit JA, Thornburg CD, Sharathkumar AA, McHugh CP, Laurie CC, Crenshaw A, et al. . Linkage analysis identifies a locus for plasma von Willebrand factor undetected by genome-wide association. Proc Natl Acad Sci USA 2013;110:588–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shane B, Contractor SF. Assessment of vitamin B 6 status. Studies on pregnant women and oral contraceptive users. Am J Clin Nutr 1975;28:739–47. [DOI] [PubMed] [Google Scholar]
- 17.Midttun Ø, Hustad S, Ueland PM. Quantitative profiling of biomarkers related to B-vitamin status, tryptophan metabolism and inflammation in human plasma by liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom 2009;23:1371–9. [DOI] [PubMed] [Google Scholar]
- 18.Midttun Ø, Kvalheim G, Ueland PM. High-throughput, low-volume, multianalyte quantification of plasma metabolites related to one-carbon metabolism using HPLC-MS/MS. Anal Bioanal Chem 2013;405:2009–17. [DOI] [PubMed] [Google Scholar]
- 19.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–5. [DOI] [PubMed] [Google Scholar]
- 20.International HapMap Consortium. The international HapMap project. Nature 2003;426:789–96. [DOI] [PubMed] [Google Scholar]
- 21.Fan R, Wang Y, Mills JL, Wilson AF, Bailey-Wilson JE, Xiong M. Functional linear models for association analysis of quantitative traits. Genet Epidemiol 2013;37:726–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu DJ, Peloso GM, Zhan X, Holmen OL, Zawistowski M, Feng S, Nikpay M, Auer PL, Goel A, Zhang H, et al. . Meta-analysis of gene-level tests for rare variant association. Nat Genet 2014;46:200–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. . PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2014 [cited 2015 Feb 23]. Available from: http://www.R-project.org.
- 25.Institute of Medicine. Dietary reference intakes for thiamin, riboflavin, niacin, vitamin B6, folate, vitamin B12, pantothenic acid, biotin, and choline. Washington (DC): National Academy Press; 1998. [PubMed] [Google Scholar]
- 26.Leklem JE. Vitamin B-6: a status report. J Nutr 1990;120:(Suppl 11):1503–7. [DOI] [PubMed] [Google Scholar]
- 27.1000 Genomes Project Consortium, Xue Y, Cartwright RA, Abecasis GR, Altshuler D, Keebler J, Kokko-Gonzales P, Nickerson DA. A map of human genome variation from population-scale sequencing. Nature 2010;467:1061–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Midttun O, Ulvik A, Ringdal Pedersen E, Ebbing M, Bleie O, Schartum-Hansen H, Nilsen RM, Nygard O, Ueland PM. Low plasma vitamin B-6 status affects metabolism through the kynurenine pathway in cardiovascular patients with systemic inflammation. J Nutr 2011;141:611–7. [DOI] [PubMed] [Google Scholar]
- 29.Ulvik A, Theofylaktopoulou D, Midttun O, Nygard O, Eussen SJ, Ueland PM. Substrate product ratios of enzymes in the kynurenine pathway measured in plasma as indicators of functional vitamin B-6 status. Am J Clin Nutr 2013;98:934–40. [DOI] [PubMed] [Google Scholar]
- 30.Yang TP, Beazley C, Montgomery SB, Dimas AS, Gutierrez-Arcelus M, Stranger BE, Deloukas P, Dermitzakis ET. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics 2010;26:2474–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stranger BE, Montgomery SB, Dimas AS, Parts L, Stegle O, Ingle CE, Sekowska M, Smith GD, Evans D, Gutierrez-Arcelus M, et al. . Patterns of cis regulatory variation in diverse human populations. PLoS Genet 2012;8:e1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo L, Du Y, Chang S, Zhang K, Wang J. rSNPBase: a database for curated regulatory SNPs. Nucleic Acids Res 2014;42:D1033–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res 2002;12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Q, Liu W, Sinha KM, Yasuda H, de Crombrugghe B. Identification and characterization of microRNAs controlled by the osteoblast-specific transcription factor Osterix. PLoS ONE 2013;8:e58104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lumeng L, Brashear RE, Li TK. Pyridoxal 5′-phosphate in plasma: source, protein-binding, and cellular transport. J Lab Clin Med 1974;84:334–43. [PubMed] [Google Scholar]
- 37.Franceschini N, van Rooij FJ, Prins BP, Feitosa MF, Karakas M, Eckfeldt JH, Folsom AR, Kopp J, Vaez A, Andrews JS, et al. . Discovery and fine mapping of serum protein loci through transethnic meta-analysis. Am J Hum Genet 2012;91:744–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fedde KN, Whyte MP. Alkaline phosphatase (tissue-nonspecific isoenzyme) is a phosphoethanolamine and pyridoxal-5′-phosphate ectophosphatase: normal and hypophosphatasia fibroblast study. Am J Hum Genet 1990;47:767–75. [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson BB, O’Brien H, Griffin GE, Mollin DL. Hydrolysis of pyridoxal-5′-phosphate in plasma in conditions with raised alkaline phosphate. Gut 1980;21:192–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merrill AH Jr, Henderson JM. Diseases associated with defects in vitamin B6 metabolism or utilization. Annu Rev Nutr 1987;7:137–56. [DOI] [PubMed] [Google Scholar]
- 41.Pekarthy JM, Short J, Lansing AI, Lieberman I. Function and control of liver alkaline phosphatase. J Biol Chem 1972;247:1767–74. [PubMed] [Google Scholar]
- 42.Lumeng L, Li TK. Characterization of the pyridoxal 5′-phosphate and pyridoxamine 5′-phosphate hydrolase activity in rat liver. Identity with alkaline phosphatase. J Biol Chem 1975;250:8126–31. [PubMed] [Google Scholar]
- 43.Lumeng L, Schenker S, Li TK, Brashear RE, Compton MC. Clearance and metabolism of plasma pyridoxal 5′-phosphate in the dog. J Lab Clin Med 1984;103:59–69. [PubMed] [Google Scholar]
- 44.Gordon T. Factors associated with serum alkaline phosphatase level. Arch Pathol Lab Med 1993;117:187–90. [PubMed] [Google Scholar]
- 45.Yuan X, Waterworth D, Perry JR, Lim N, Song K, Chambers JC, Zhang W, Vollenweider P, Stirnadel H, Johnson T, et al. . Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet 2008;83:520–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamatani Y, Matsuda K, Okada Y, Kubo M, Hosono N, Daigo Y, Nakamura Y, Kamatani N. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat Genet 2010;42:210–5. [DOI] [PubMed] [Google Scholar]
- 47.Chambers JC, Zhang W, Sehmi J, Li X, Wass MN, Van der Harst P, Holm H, Sanna S, Kavousi M, Baumeister SE, et al. . Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet 2011;43:1131–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goseki-Sone M, Sogabe N, Fukushi-Irie M, Mizoi L, Orimo H, Suzuki T, Nakamura H, Orimo H, Hosoi T. Functional analysis of the single nucleotide polymorphism (787T>C) in the tissue-nonspecific alkaline phosphatase gene associated with BMD. J Bone Miner Res 2005;20:773–82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.