Abstract.
We present a platform for designing and executing studies that compare pathologists interpreting histopathology of whole slide images (WSIs) on a computer display to pathologists interpreting glass slides on an optical microscope. eeDAP is an evaluation environment for digital and analog pathology. The key element in eeDAP is the registration of the WSI to the glass slide. Registration is accomplished through computer control of the microscope stage and a camera mounted on the microscope that acquires real-time images of the microscope field of view (FOV). Registration allows for the evaluation of the same regions of interest (ROIs) in both domains. This can reduce or eliminate disagreements that arise from pathologists interpreting different areas and focuses on the comparison of image quality. We reduced the pathologist interpretation area from an entire glass slide (10 to ) to small ROIs (). We also made possible the evaluation of individual cells. We summarize eeDAP’s software and hardware and provide calculations and corresponding images of the microscope FOV and the ROIs extracted from the WSIs. The eeDAP software can be downloaded from the Google code website (project: eeDAP) as a MATLAB source or as a precompiled stand-alone license-free application.
Keywords: digital pathology, whole slide imaging, reader studies, technology evaluation, validation, microscopy
1. Introduction
Digital pathology (DP) incorporates the acquisition, management, and interpretation of pathology information generated from a digitized glass slide. DP is enabled by technological advances in whole slide imaging (WSI) systems, also known as virtual microscopy systems, which can digitize whole slides at microscopic resolution in a short period of time. The potential opportunities for DP are well documented and include telepathology, digital consultation and slide sharing, pathology education, indexing and retrieval of cases, and the use of automated image analysis.1–3 The imaging chain of a WSI system consists of multiple components including the light source, optics, motorized stage, and a sensor for image acquisition. WSI systems also have embedded software for identifying tissue on the slide, auto-focusing, selecting and combining different fields of view (FOVs) in a composite image, and image processing (color management, image compression, etc.). Details regarding the components of WSI systems can be found in Gu and Ogilvie4 There are currently numerous commercially available WSI systems as reviewed by Rojo et al. in terms of technical characteristics.5
A number of studies (many cited in Refs. 6 and 7) have focused on the validation of WSI systems for primary diagnosis, with findings generally showing high concordance between glass slide and digital slide diagnoses. A common drawback of current validation studies of WSI systems is that they sometimes combine diagnoses from multiple pathology tasks performed on multiple tissue types. Pooling cases can lead to the undersampling of clinical tasks as discussed in the study by Gavrielides et al.8 It can also dilute differences in reader performance that might be specific to certain tasks. Another issue from current validation studies is that agreement was typically determined by an adjudication panel comparing pathology reports from the WSI and microscope reads head-to-head. Guidelines are sometimes developed for defining major and minor discrepancies, but there is a considerable amount of interpretation and judgment required of the adjudication panel as the pathology reports are collected as real-world, sign-out reports (free text). Additionally, the focus of most validation studies is on primary diagnosis, with minor emphasis on related histopathology features that might be affected by image quality. The quantitative assessment of a pathologist’s ability to evaluate histopathology features in WSI compared to the microscope would be useful in identifying possible limitations of DP for specific tasks. Related work includes the study of Velez et al.9 where discordances in the diagnosis of melanocytic skin lesions were attributed to difficulty in identifying minute details such as inflammatory cells, apoptosis, organisms, and nuclear atypia. Finally, studies focusing on primary diagnosis do not typically account for differences in search patterns or FOV reviewed by observers. The selection of different areas to be assessed by different observers has been identified as a source of interobserver variability.10
In this paper, we present an evaluation environment for digital and analog pathology that we refer to as eeDAP. eeDAP is a software and hardware platform for designing and executing digital and analog pathology studies where the digital image is registered to the real-time view on the microscope. This registration allows for the same regions of interest (ROIs) to be evaluated in digital mode or in microscope mode. Consequently, it is possible to reduce or eliminate a large source of variability in comparing these modalities in the hands of the pathologist: the FOV (the tissue) being evaluated. In fact, the current registration precision of eeDAP allows for the evaluation of the same individual cell in both domains. As such, a study can be designed where pathologists are asked to evaluate a preselected list of individual cells or groups of cells in the digital mode and with the microscope. Consequently, paired observations from coregistered FOV are collected allowing for a tight comparison between WSI and optical microscopy.
A reader study with eeDAP is intended to evaluate the scanned image, not the clinical workflow of a pathologist or lab. Instead of recording a typical pathology report, eeDAP enables the collection of explicit evaluation responses (formatted data) from the pathologist corresponding to very narrow tasks. This approach removes the ambiguity related to the range of language and the scope that different pathologists use in their reports. At the same time, this approach requires the study designer to narrow the criteria for cases (slides, ROIs, cells) to be included in the study set.
Reader studies utilizing eeDAP can focus on the evaluation of specific histopathology features. Since certain features challenge image quality properties such as color fidelity, focus quality, and depth of field, such reader studies can provide valuable information for the assessment of WSI and its role in clinical practice. The presented framework allows for the formulation of different types of tasks, many of which are currently available and customizable in eeDAP: free-text, integer input for counting tasks, a slider in a predefined range for a confidence scoring task (ROC task, receiver operating characteristic task), check boxes of specific categories for a classification task, and marking the image for a search task. Figure 1 shows the examples of the GUI presentation for two scoring tasks that we have explored: on the left, the reader would be asked to provide a score between 1 and 100 reflecting their confidence that the cell within the reticle is a plasma cell [in hematoxylin and eosin (H&E) stained, formalin-fixed, paraffin-embedded colon tissue], whereas on the right, the reader would provide a score reflecting their confidence that the cell within the reticle is a mitotic figure (in H&E stained, formalin-fixed, paraffin-embedded sarcoma).
Fig. 1.
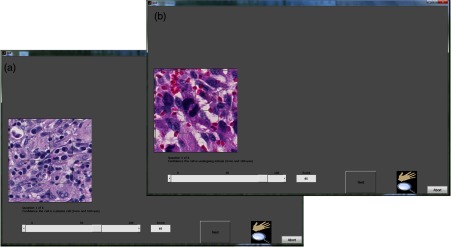
Here are the two windows, each showing the eeDAP presentation of a slider task: the image on the left is of colon tissue, the image on the right is of sarcoma.
In this paper, we outline the key software and hardware elements of eeDAP. First, we discuss the eeDAP software as a publicly available resource and describe software specifications and requirements. We next talk about the tone reproduction curves that characterize eeDAP and the native viewers: the curves showing the lightness in the output image given the transmittance of the input slide. In Sec. 2.3, we summarize the local and global registration methods that are key to pairing ROIs across the digital and microscope domains. In Sec. 2.4, we provide the key hardware specifications that eeDAP requires and then demonstrate the differences in FOVs and image sizes between the two domains: the digital image and the glass slide. These calculations and corresponding representative images help to provide a sense of scale across the digital and analog domains. Finally, we talk about reticles and their important role in narrowing the evaluation area to a small ROI or an individual cell.
2. Methods
In this section, we summarize the key elements of the eeDAP software and hardware. The eeDAP software is made up of three graphical user interfaces (GUIs) written in MATLAB (Mathworks, Natick, Massachusetts).
The first interface establishes the evaluation mode (Digital or MicroRT) and reads in the study input file. The input file contains the file names of the WSIs, hardware specifications, and the list of tasks with corresponding ROI locations that will be interpreted by the pathologist. Each ROI is defined by a location, width, and height in pixel coordinates of the WSI, and all are automatically extracted on the fly from the WSIs named. There are installation requirements that make the ROI extraction possible from the proprietary WSI file formats. We also discuss a color gamut comparison between eeDAP and a native WSI viewer (a viewer designed by a WSI scanner manufacturer).
The second interface is executed only for studies run in the MicroRT mode. This interface globally registers each WSI to its corresponding glass slide. For each global registration of each WSI, a study administrator must interactively perform three local registrations. The local and global registration methods are described in Sec. 2.3.
The third interface runs the study in accordance to the list of tasks given in the input file. If the study is run in the Digital mode, the pathologist views the ROIs on the computer display in the GUI and enters the evaluations therein. If the study is run in MicroRT mode, the pathologist views the ROIs through the microscope (calibrated for Köhler illumination) and is responsible for any focusing in the -plane. Although the pathologist is engaged with the microscope in the MicroRT mode, the study administrator is viewing the ROIs on the computer display in the GUI and enters the evaluations there as dictated by the pathologist. The study administrator also monitors a live camera image of what the pathologist sees through the microscope. This allows the study administrator to confirm and maintain a high level of registration precision in MicroRT mode.
The eeDAP hardware consists of an upright brightfield microscope, a digital camera, a computer-controlled stage with a joystick, a computer monitor, and a computer (see Fig. 2). The microscope requires a port for mounting the camera that allows for simultaneous viewing with the eyepiece. eeDAP currently supports a Ludl controller and compatible stage, and an IEEE 1394 FireWire camera communicating according to a DCAM interface (RGB, 8 bits-per-channel, minimum width 640, minimum height 480). Setup instructions and example specifications can be found in the user manual.
Fig. 2.
The evaluation environment for digital and analog pathology (eeDAP) hardware: microscope, camera, computer-controlled stage with joystick, and a computer with monitor (not shown).
Below we summarize how these components are used in registration and how the WSI and real microscope image appear to the pathologist. We also identify an important part of the microscope, the reticle. The reticle is housed in the microscope eyepiece. One reticle that we use identifies ROIs in the microscope FOV and another points at individual cells.
2.1. eeDAP Availability and Technical Requirements
The software component of eeDAP is publicly available as MATLAB source code or as a precompiled stand-alone license-free MATLAB application.11 Running eeDAP source code requires the MATLAB image acquisition toolbox and the installation of third party software to extract ROIs from WSIs. WSIs are often extremely large (several GB) and are stored as large layered TIFF files embedded in proprietary WSI file formats. eeDAP uses ImageScope, a product of Aperio (a Leica Biosystems Division) to read images scanned with WSI scanners from Aperio (.svs) and other formats, including .ndpi (Hamamatsu). ImageScope contains an ActiveX control named TIFFcomp that allows for the extraction and scaling of ROIs. A consequence of using TIFFcomp is that the MATLAB version must be 32 bits.
The precompiled stand-alone application requires that the MATLAB compiler runtime (MCR) library be installed. It is important that the version of the MCR correspond exactly to that used for the stand-alone application (refer to the user manual).
2.2. Tone Reproduction Curves
Manufacturers of WSI scanners typically provide software for viewing their proprietary file formats. These viewers may include specialized color management functions. In fact, we observed color differences when we viewed .ndpi images with the native Hamamatsu viewer (NDP.view) side-by-side with the Aperio viewer (ImageScope) and MATLAB (with the Aperio ImageScope Active X component TIFFcomp). In an attempt to understand the native viewer and correct for these differences (so that we can show the images as they would be seen in the native viewer), we considered the image adjustments that may have caused them. From these, we observed that the images appeared the same in the three viewers when we adjusted the gamma setting. To confirm our observations, we measured the tone reproduction curves of NDP.view ( and ) and ImageScope (no adjustments made; equivalent to the MATLAB).
Following the work of Cheng et al.,12 we measured the transmittance of the 42 color patches of a color phantom (film on a glass slide, see Fig. 3). Using an FPGA board, we then retrieved the sRGB values of a Hamamatsu scanned image of the color phantom from the NDP.view with gamma set to 1.8 (default), gamma set to 1.0 (turning off the gamma adjustment), and ImageScope (default, no gamma correction). We then converted the sRGB values to the CIELAB color space and plotted the normalized lightness channel against the normalized transmittance. The results (Fig. 3) supported our visual observations:
-
•
There is good agreement between the tone reproduction curves of NDP.view with and ImageScope.
-
•
The tone reproduction curve of NDP.view with appears to be linearly related to transmittance.
-
•
The tone reproduction curve of NDP.view with appears to be 1/1.8 gamma transformation of transmittance.
-
•
The default images displayed by NDP.view and ImageScope (and MATLAB by equivalence) differ only in the gamma setting.
Fig. 3.
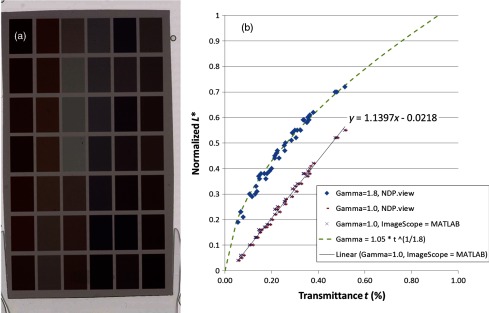
(a) Hamamatsu scanned image of a color phantom (film on a glass slide). (b) The transmittance of the 42 color patches plotted against the normalized lightness in the CIELAB color space (derived from the average sRGB values in a patch).
2.3. Registration
eeDAP uses registration to link the stage (glass slide) coordinates to the WSI coordinates. eeDAP has two levels of registration: global and local. The global registration is equivalent to finding the transformation between the stage and WSI coordinates. The global registration requires three anchors, three pairs of stage-WSI registered coordinates. Each anchor is generated by a local registration: a stage coordinate and a WSI coordinate that correspond to the same specimen location.
eeDAP conducts two levels of global registration: low and high resolutions. Low resolution corresponds to microscope magnifications such as , , and ; the entire WSI image is scaled to fit in the GUI. High resolution registration corresponds to the microscope magnifications such as and ; the low-resolution registration results are used to limit the amount of the WSI shown in the GUI, sequentially zooming in on the location of the low-resolution anchors.
eeDAP uses local registration for two purposes. The first purpose is to support global registration as discussed. The second purpose is to maintain a high level of registration precision throughout data collection. During our pilot studies, we observed that the precision of the global registration was deteriorating as the stage moved throughout the study. Therefore, we implemented a button that could be pressed during data collection that could register the current microscope view to the current task-specific ROI. The current level of precision appears to allow for the reliable evaluation of individual cells. Technical details of local and global registrations are provided below.
2.3.1. Local registration
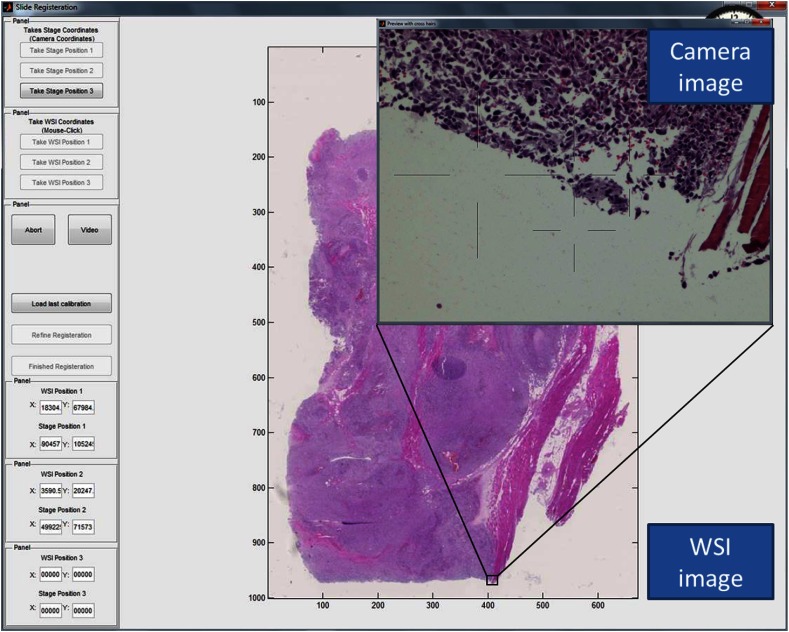
A local registration is accomplished by taking a snapshot of the microscope FOV with the microscope-mounted camera and by finding a search region containing the corresponding location in the WSI (see Fig. 4). The search region is identified by the study administrator and avoids searching the entire (very large) image. A local registration yields a coordinate in the WSI and a coordinate on the microscope stage that identify the same location.
Fig. 4.
Screen shot of the registration interface including the real-time microscope field of view (FOV) as seen with the mounted camera (“Camera image”).
The camera image contains some amount of specimen on the glass slide and is labeled by the coordinate of the current stage position. See, for example, the “Preview with cross hairs” window labeled “Camera image” depicted in Fig. 4. The camera image has three channels (RGB) and must be at least . The physical size of a (square) camera pixel is given by the manufacturer specifications. This size divided by any magnification by the microscope () determines the camera’s spatial sampling period in units of the specimen.
We extract a patch of the WSI image (RGB) that is larger than and contains the same content as captured by the camera. See, for example, the image labeled “WSI image” depicted in Fig. 4. The WSI’s spatial sampling period (often referred to as the WSI resolution) is given by the manufacturer specifications in units of the specimen and is often recorded in the WSI image.
An ROI extracted from a WSI image can be rescaled (interpolated) to have the same sampling period as the camera image using the ratio of the sampling periods. In other words, the number of pixels before and after rescaling is determined by
Given the camera image and the WSI image at the same scale, we perform normalized cross-correlation to find the shift that best registers the two images. In other words, we find the that maximize the following sum:
where the sum is over the pixels in the camera image, indexes the pixels in the image, is the shift in pixels, and and are the average and standard deviation of the elements of and considered in the sum.
2.3.2. Global registration
Global registration is done for each WSI in the input file and corresponding glass slide on the microscope stage. Each global registration is built on three local registrations. The three local registrations yield three pairs of coordinates that define the transformation (the change of basis) between the coordinate system of the WSI (image pixels) and the coordinate system of the stage (stage pixels).
Let the three pairs of coordinates be given by , for . Given these pairs, we define the two coordinate systems with the following matrices:
Then given a new location in the WSI coordinate system , we can determine the corresponding location in the stage coordinate system with the following transformation:
In words, we first shift the new point according to the origin in the WSI coordinate system . Next, we map the point from the WSI coordinate system to the standard one with and then map it to the stage coordinate system with . Finally, we shift the point according to the origin in the stage coordinate system . Consequently, the location of each ROI for each task given in the input file can be accessed in the WSI coordinate system or the stage coordinate system.
The study administrator determines each local registration by navigating the microscope with the joystick to an appropriate anchor, taking the camera image, and then approximately identifying the corresponding anchor in the WSI. An appropriate anchor is one that can be recognized in the WSI image and is surrounded by one or more salient features. Salient features increase the likelihood of a successful registration; repetitive features and homogeneous regions do not. Additionally, global registration is better when the set of three anchors are widely separated; encompassing the entirety of the tissue is best. The most challenging aspect in finding the appropriate anchors is navigating the microscope stage with the joystick, focusing on the specimen, and determining the corresponding location in the WSI image.
In Fig. 4, we see the “Camera image” and the “WSI image.” The study administrator has clicked on the WSI image to indicate where in the WSI to search for the camera image. A patch of the WSI image is extracted from the WSI at the full scanning resolution, the patch is scaled to the resolution of the camera, and a local registration produces the shift that identifies the corresponding WSI location to pair with the current stage location.
2.4. Comparing FOV and Image Sizes
In the following, we provide the key hardware specifications that eeDAP requires and demonstrates the calculation of different FOVs and image sizes. These calculations provide the relationships regarding scale across the digital and analog domains.
2.4.1. Microscope FOV
An important parameter of an optical microscope is the field number (FN); it is the diameter of the view field in millimeters at the intermediate image plane, which is located in the eyepiece. The FN is a function of the entire light path of the microscope starting with the glass slide, through the objective, and ending at the intermediate image plane in the eyepiece; the FN is often inscribed on the eyepiece. To get the FOV in units of the specimen being viewed, we divide the FN by the objective magnification. We currently have an Olympus BX43 microscope () and a Zeiss Axioplan2 Imaging microscope (). At magnification due to the objective, the FOV covered in the specimen plane is given by
-
•Olympus FOV at
-
•,
-
•,
-
•
-
•Zeiss FOV at
-
•,
-
•.
-
•
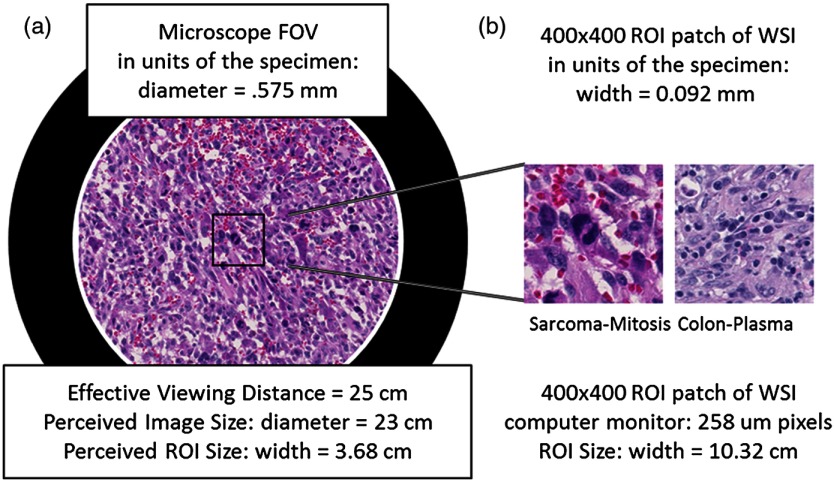
The FN can also be used to determine the perceived size of the microscope image at an effective viewing distance of 25 cm. The 25 cm effective viewing distance is a design convention13 that is not well documented or well known. The perceived size is then simply the FN times the eyepiece magnification. Since the eyepieces on both microscopes above have magnification, the perceived diameters of the intermediate images are 22 cm (Olympus) and 23 cm (Zeiss) at the effective viewing distance of 25. This corresponds to a visual angle (subtended angle of object at the eye) equal to . In Fig. 5, we show what the microscope FOV looks like for the sarcoma slide scaled to fit the page.
Fig. 5.
The two images in this figure depict the relative sizes of the microscope image as seen through the eyepiece at (a) and a regions of interest patch from a whole slide image as seen on a computer monitor with pixels at a viewing distance of 25 cm (b).
2.4.2. Size of scanner images
We have access to two WSI scanners: a Hamamatsu Nanozoomer 2.0HT and an Aperio CS. They both operate at and magnification equivalent settings with similar sampling periods:
-
•
at and at (Hamamatsu);
-
•
at and 0.2500 μm at (Aperio).
The Hamamatsu scanned images we have been using for pilot studies have pixels (10 GB) and pixels (2 GB). By multiplying the number of pixels by the sampling period, we get the size of the images in units of the specimen on the glass slide. These images correspond to the image areas of and . We have been extracting ROI patches that show patches of the specimen () for our most recent pilot study, which is 3.2% of the microscope FOV.
The size of a patch seen by a pathologist depends on the computer monitor pixel pitch (distance between pixels). For a computer monitor with a pixel pitch, the display size of a patch is (). If we assume a viewing distance of 25 cm from the computer monitors (to match the effective viewing distance in the microscope), we can compare the image size of the ROI on the computer monitor to the microscope perceived image size. Figure 5 shows the relative sizes of the two views side by side, demonstrating the apparent magnification of the specimen area in the displayed patches.
2.4.3. Size of camera images
We currently have a Point Grey Flea2 color camera (FL2G-50S5C-C) that has a default output format of with pixels. This format corresponds to binning of a camera with a native pixel size of . At magnification ( objective times camera adapter), the spatial sampling period in units of the specimen plane corresponds to (6.9/20) and the camera FOV is (), which is about 36% of the microscope FOV.
2.5. Reticles
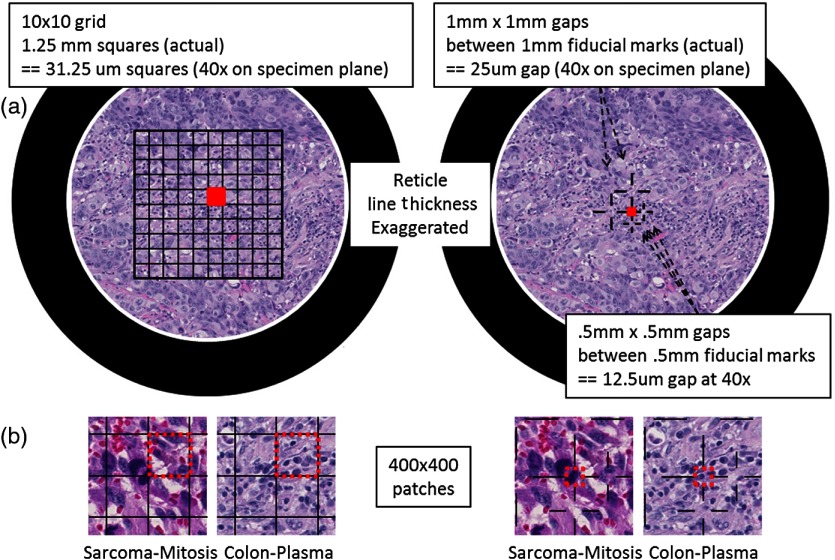
Reticles are pieces of glass that are inserted at the intermediate image plane in the eyepiece. They contain fine lines and grids that appear superimposed on the specimen. Reticles help to measure features or help to locate objects. The current version of eeDAP uses them to narrow tasks to very small regions and individual cells, allowing for an expansion of capabilities. In Fig. 6, we depict reticles as seen through the microscope (line thickness exaggerated) and as they appear in eeDAP ( patches). These reticles are described below and were studied in two feasibility reader studies for their functionality.
Fig. 6.
Reticles (line thicknesses exaggerated) as seen through simulated microscope FOV (a) and patches as they appear in eeDAP (b). The red squares in the simulated microscope FOVs and the red-dash boundary squares in the patches indicate the “evaluation” areas. In the sarcoma patch on the left, a majority of the obvious mitotic figure falls in the grid square to the left of the evaluation square. In the sarcoma patch on the right, the central cross hairs point to the obvious mitotic figure. In the colon patch on the left, there are several plasma-cell candidates in the “evaluation” square. In the colon patch on the right, the cross hairs point to a single plasma-cell candidate to be evaluated.
In the first feasibility study, we used a reticle containing a grid with squares that are 1.25 mm on a side (Klarmann Rulings: KR-429). At , these squares are on a side in the specimen plane. When running in Digital mode, eeDAP digitally creates a reticle mask to create the same effect as the real reticle in the microscope. The instructions for this study were to score the reticle square that was immediately above and to the right of the center cross (red squares in Fig. 6). Identifying the center cross in the grid in MicroRT mode is challenging; it is accomplished by rotating the eyepiece as the center cross remains fixed. Additionally, the instructions to score a square were to score the cell that was most likely the target (mitotic figure or plasma cell as shown in Fig. 1), considering cells with at least half their nuclei in the square.
In the second (similar) feasibility study, we used a custom reticle that has fiducials that point to gaps (Klarmann Rulings: KR-32536). Two gaps are and three gaps are . At , these gaps are and on a side. The instructions for this study were much more direct: score the cell at the center of the center fiducials (red squares in Fig. 6).
3. Results and Discussion
We have been using pilot studies to identify weaknesses and future improvements needed for eeDAP and the general study design. The main weakness that we identified was that the registration precision throughout data collection was not good enough: pathologists were not evaluating identical ROIs. We have addressed this in the current generation of eeDAP by incorporating the ability to do a local registration for every ROI during data collection. We have also created a custom reticle that allows us to point at individual cells. This reduces ambiguity and disagreements due to evaluations based on multiple different cells within an ROI.
We also observed that the .ndpi WSI images appeared darker when viewing with eeDAP (and ImageScope) compared to viewing with the native viewer, NDP.view. Through observation and subsequent measurement, we determined that the difference was a simple gamma adjustment, and we implemented a color look-up table to make this and any other color adjustment possible with eeDAP.
Our pilot studies emphasized the need for reader training. We found that pathologists needed to develop a level of comfort in scoring individual candidate cells, as this is not a part of a pathologist’s typical clinical work flow. This is especially true when we asked for a 101-point confidence rating instead of a yes–no decision. Consequently, we are focusing our efforts to creating training on the cell types and scorings. Training on cell types may include Power Point slides that contain verbal descriptions of typical features and sample images. Training may also include eeDAP training modules: the training modules may elicit scores of the typical features as well as the overall score and then provide feedback in the form of the scores of experts.
As we move beyond pilot studies to pivotal studies, we need to investigate and establish several methods and protocols to reduce the variability between the pilot study and the pivotal study, to reduce variability during a pivotal study, and to allow for a study to be replicated as closely as possible. Methods and protocols are needed on the following issues:
-
•Computer monitor QA/QC and calibration, including color
-
•It is understood in radiology that poor-quality displays can result in misdiagnosis, eye-strain, and fatigue.14 As such, it is common in the practice and evaluation of radiology to control, characterize, and document the display conditions. This culture has led to the creation of standards that treat displays.15 This issue is not yet fully enabled and appreciated within the culture of DP practice or evaluation. Study reports do very little to describe the display characteristics and calibration, with recent work being the exception.8 However some groups, including the International Color Consortium, are filling the void and addressing the challenging issue of display and color calibration.16–18
-
•
-
•Slide preparation.
-
•It is well known that there is significant variability in tissue appearances based on processing, sectioning, and staining differences and this variability leads to variability in diagnosis.19 Protocols for slide preparation are a part of standard lab practice and are changing with increased automation, driving standards in this space.20
-
•
-
•Tissue inclusion/exclusion criteria, including methods to objectively identify candidate cells for the evaluation task.
-
•Identifying inclusion/exclusion criteria for study patients (or in the current context, their tissue) is needed to convey the spectrum of the tissue being used, and thus the trial’s generalizability and relevance.21,22 Given the tissue, when the task is to evaluate individual cells, it is important to not bias the selection process. For our work, we intend to first identify the entire spectrum of presentations, not just presentations that are easy in one modality or another (as might result from pathologist identified candidates). Once the entire spectrum of presentations is identified, there may be reasons to subsample within to stress the imaging system evaluation and comparison. Future work may include the incorporation of algorithms for the automated identification of candidate cells to be classified or histopathological features to be evaluated. Such algorithms may be less biased and more objective in creating the study sets.
-
•
Finally, a coherent analysis method is needed that does not require a gold standard, since one is typically not available for the tasks being considered. To address this need, we are investigating agreement measures, such as concordance, that compare pathologist performance with WSI to conventional optical microscopy. The goal is to develop methods and tools for multireader, multicase analysis of agreement measures, similar to the methods and tools for the area under the ROC curve23 and the rate of agreement.24
4. Conclusions
In this paper, we presented the key software and hardware elements of eeDAP, a framework that allows for the registration and display of corresponding tissue regions between the glass slide and the WSI. The goal was to remove search as a source of observer variability that might dilute differences between modalities. The software part of eeDAP can be downloaded from the google code website (project: eeDAP) as a MATLAB source or as a precompiled stand-alone license-free application.11 This software can be paired with the required hardware (microscope, automated stage, and camera) and used to design and execute reader studies to compare DP to traditional optical microscopy.
Acknowledgments
This research project was partially funded through a Critical Path grant from the U.S. Food and Drug Administration (FDA/CDRH). The mention of commercial entities, or commercial products, their sources, or their use in connection with material reported herein is not to be construed as either an actual or implied endorsement of such entities or products by the Department of Health and Human Services or the U.S. Food and Drug Administration.
Biographies
Brandon D. Gallas provides mathematical, statistical, and modeling expertise to the evaluation of medical imaging devices at the FDA. His main areas of contribution are in the design and statistical analysis of reader studies, image quality, computer-aided diagnosis, and imaging physics. Before working at the FDA, he was in Dr. Harrison Barrett’s radiology research group at the University of Arizona earning his PhD degree in applied mathematics from the Graduate Interdisciplinary Program.
Marios A. Gavrielides received his PhD degree in biomedical engineering from Duke University in 2001. From 2002 to 2004, he was a postdoctoral researcher at the Aristotle University of Thessaloniki, Greece. Since 2004, he has been a NIBIB/CDRH fellow and then a staff scientist with the Division of Imaging, Diagnostics, and Software Reliability, OSEL/CDRH at the Food and Drug Administration. His research interests include the development and assessment of quantitative imaging methodologies.
Catherine M. Conway provides expertise in digital pathology evaluation, validation, and study design. Her main areas of contribution are in histology study design and prep, digital pathology evaluation design, and computer-aided diagnosis. Before working at the NIH, she worked in industry in the area of digital pathology. She earned a PhD degree from Dublin City University in the subject area of development of image analysis software for the evaluation of IHC across digital slides.
Adam Ivansky is a PhD student at the University of Rochester. His main research interests include classical microscopy, nonlinear microscopy, and image processing. He is a member of a research group under Dr. Wayne H. Knox that focuses on development of noninvasive refractive laser eye surgery. His main contribution to the eeDAP project was coding of a portion of the software.
Tyler C. Keay received his BS degree in bioengineering from University of Maryland’s Fischell School of Engineering in 2011. After graduating, he was an ORISE fellow at the Food and Drug Administration, where he began his work in digital pathology under the guidance of Marios Gavrielides, Wei-Chung Cheng, and Brandon Gallas. Since 2013, he has been a clinical affairs scientist at Omnyx, LLC. His research interests include quantitative imaging analysis and color science.
Wei-Chung Cheng is a research scientist at the FDA. Since 2009, he has been developing assessment methods for medical imaging devices in various modalities including digital pathology, endoscopy, and radiology. He received his PhD degree in electrical engineering from the University of Southern California. Before joining the FDA, he was an assistant professor in the Department of Photonics and Display Institute, National Chiao-Tung University, Taiwan. His research interests include color science, applied human vision, and display technology.
Jason Hipp served as a Rabson fellow in the Laboratory of Pathology at the National Cancer Institute (NCI), National Institutes of Health (NIH). He earned his MD/PhD degree from Wake Forest University School of Medicine. He then completed his residency in anatomic pathology at the NCI/NIH, and subsequently completed a 2-year pathology informatics fellowship at the University of Michigan, School of Medicine.
Stephen M. Hewitt is a clinical investigator in the Laboratory of Pathology, National Cancer Institute. His research interests include tissue-based biomarkers and whole-slide imaging. He is a treasurer of the Histochemical Society, Program Committee Chair for the Association for Pathology Informatics and a consultant and collaborator to the Center for Devices and Radiological Health, Food and Drug Administration. He has coauthored over 200 articles and serves on the editorial board of five peer-reviewed journals.
References
- 1.Pantanowitz L., et al. , “Review of the current state of whole slide imaging in pathology,” J. Pathol. Inf. 2, 36 (2011). 10.4103/2153-3539.83746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinstein R. S., et al. , “Overview of telepathology, virtual microscopy, and whole slide imaging: prospects for the future,” Hum. Pathol. 40(8), 1057–1069 (2009). 10.1016/j.humpath.2009.04.006 [DOI] [PubMed] [Google Scholar]
- 3.Al-Janabi S., Huisman A., Van Diest P. J., “Digital pathology: current status and future perspectives,” Histopathology 61(1), 1–9 (2012). 10.1111/his.2012.61.issue-1 [DOI] [PubMed] [Google Scholar]
- 4.Gu J., Ogilvie R. W., Eds., Virtual Microscopy and Virtual Slides in Teaching, Diagnosis, and Research, CRC Press, Boca Raton, Florida: (2005). [Google Scholar]
- 5.Rojo M. G., et al. , “Critical comparison of 31 commercially available digital slide systems in pathology,” Int. J. Surg. Pathol. 14(4), 285–305 (2006). 10.1177/1066896906292274 [DOI] [PubMed] [Google Scholar]
- 6.Pantanowitz L., et al. , “Validating whole slide imaging for diagnostic purposes in pathology: guideline from the College of American Pathologists Pathology and Laboratory Quality Center,” Arch. Pathol. Lab. Med. 137(12), 1710–1722 (2013). 10.5858/arpa.2013-0093-CP [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jara-Lazaro A. R., et al. , “Digital pathology: exploring its applications in diagnostic surgical pathology practice,” Pathology 42(6), 512–518 (2010). 10.3109/00313025.2010.508787 [DOI] [PubMed] [Google Scholar]
- 8.Gavrielides M. A., et al. , “Observer performance in the use of digital and optical microscopy for the interpretation of tissue-based biomarkers,” Anal. Cell. Pathol., Epub ahead of print (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Velez N., Jukic D., Ho J., “Evaluation of 2 whole-slide imaging applications in dermatopathology,” Hum. Pathol. 39(9), 1341–1349 (2008). 10.1016/j.humpath.2008.01.006 [DOI] [PubMed] [Google Scholar]
- 10.van Diest P. J., et al. , “A scoring system for immunohistochemical staining: consensus report of the task force for basic research of the EORTC-GCCG. European Organization for Research and Treatment of Cancer-Gynaecological Cancer Cooperative Group,” J. Clin. Pathol. 50(10), 801–804 (1997). 10.1136/jcp.50.10.801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallas B. D., eeDAP v2.0: Evaluation Environment for Digital and Analog Pathology (code.google.com/p/eedap), Division of Imaging and Applied Mathematics, CDRH, FDA, Silver Spring, Maryland: (2013). [Google Scholar]
- 12.Cheng W.-C., et al. , “Assessing color reproducibility of whole-slide imaging scanners,” Proc. SPIE 8676, 86760O (2013). 10.1117/12.2007215 [DOI] [Google Scholar]
- 13.Kapitza H. G. Microscopy from the Very Beginning, 2nd ed., Carl Zeiss, Jena, Germany: (1997). [Google Scholar]
- 14.Hirschorn D., Krupinski E., Flynn M., “Displays,” in IT Reference Guide for the Practicing Radiologist (2013). [Google Scholar]
- 15.Norweck J. T., et al. , “ACR-AAPM-SIIM technical standard for electronic practice of medical imaging,” J. Digit Imaging 26(1), 38–52 (2013). 10.1007/s10278-012-9522-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.ICC, “ICC White Paper 7: The Role of ICC Profiles in a Colour Reproduction System, International Color Consortium,” Reston, VA (2004), http://www.color.org/ICC_white_paper_7_role_of_ICC_profiles.pdf (27 October 2014); ; Green P., Ed., Color Management: Understanding and Using ICC Profiles, Wiley, New York, NY: (2010). [Google Scholar]
- 17.Krupinski E. A., et al. , “Observer performance using virtual pathology slides: impact of LCD color reproduction accuracy,” J. Digit Imaging 25(6), 738–743 (2012). 10.1007/s10278-012-9479-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Badano A., et al. , “Consistency and standardization of color in medical imaging: a consensus report,” J. Digit Imaging, Epub ahead of print (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allison K. H., et al. , “Understanding diagnostic variability in breast pathology: lessons learned from an expert consensus review panel,” Histopathology 65(2), 240–251 (2014). 10.1111/his.2014.65.issue-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.I/LA28-A2: Quality Assurance for Design Control and Implementation of Immunohistochemistry Assays; Approved Guideline, 2nd ed., Clinical and Laboratory Standards Institute, Wayne, Pennsylvania: (2011). [Google Scholar]
- 21.Bossuyt P. M., et al. , “Toward complete and accurate reporting of studies of diagnostic accuracy: the STARD initiative,” Am. J. Clin. Pathol. 119(1), 18–22 (2003). 10.1309/8EXCCM6YR1THUBAF [DOI] [PubMed] [Google Scholar]
- 22.Schulz K. F., et al. , “Consort 2010 statement: updated guidelines for reporting parallel group randomised trials,” BMJ 340, c332 (2010). 10.1136/bmj.c332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallas B. D., et al. , “A framework for random-effects ROC analysis: biases with the bootstrap and other variance estimators,” Commun. Stat. A 38(15), 2586–2603 (2009). 10.1080/03610920802610084 [DOI] [Google Scholar]
- 24.Chen W., Wunderlich A., iMRMC_Binary v1.0_Beta1: A Matlab Software Package for Simulating, Analyzing, and Sizing a Multi-Reader Multi-Case Reader Study with Binary Assessments, Division of Imaging and Applied Mathematics, CDRH, FDA, Silver Spring, Maryland: (2014). [Google Scholar]