Abstract.
Early diagnoses of Alzheimer’s disease (AD) would confer many benefits. Several biomarkers have been proposed to achieve such a task, where features extracted from magnetic resonance imaging (MRI) have played an important role. However, studies have focused exclusively on morphological characteristics. This study aims to determine whether features relating to the signal and texture of the image could predict mild cognitive impairment (MCI) to AD progression. Clinical, biological, and positron emission tomography information and MRI images of 62 subjects from the AD neuroimaging initiative were used in this study, extracting 4150 features from each MRI. Within this multimodal database, a feature selection algorithm was used to obtain an accurate and small logistic regression model, generated by a methodology that yielded a mean blind test accuracy of 0.79. This model included six features, five of them obtained from the MRI images, and one obtained from genotyping. A risk analysis divided the subjects into low-risk and high-risk groups according to a prognostic index. The groups were statistically different (). These results demonstrated that MRI features related to both signal and texture add MCI to AD predictive power, and supported the ongoing notion that multimodal biomarkers outperform single-modality ones.
Keywords: Alzheimer’s disease, mild cognitive impairment, multimodal biomarker, magnetic resonance imaging biomarker, magnetization-prepared rapid acquisition with gradient echo
1. Introduction
It has been estimated that 44.35 million people are living with dementia worldwide.1 Alzheimer’s disease (AD) is the most common cause for this condition and is estimated to currently affect 5.2 million people in the United States.2 Its hallmark abnormalities are deposits of the protein fragment amyloid beta () and twisted strands of the protein tau, histopathological evidence that can only be obtained from a post-mortem biopsy. Because of this, physicians in clinical settings cannot diagnose subjects with definite, but probable AD, as defined by the NINCDS-ADRDA Work Group criteria.3 This has led to the current search for robust and accurate AD biomarkers.
An early diagnosis of cognitive impairment confers many benefits: it allows prompt evaluation and treatment of reversible or treatable conditions (e.g., depression or vitamin B12 deficiency), allows potential management of symptoms with medication, and enables potential inclusion in clinical trials.2 Therefore, many efforts are being made to develop a biomarker capable of diagnosing AD as early as possible. To do so, many studies have focused on distinguishing between subjects with a mild cognitive impairment (MCI) which will progress to AD, and those who will not. MCI is a condition in which a person has problems with language, memory, or another cognitive ability and has been established as a risk factor for AD, representing in some cases a transitional stage between normal aging and AD.4
Several biomarkers from different information modalities (e.g., biological samples, imaging features, and clinical data) have been proposed for the early diagnosis of AD, and multimodal ones have been found to achieve the best accuracies.5–8 Among these, features extracted from magnetic resonance imaging (MRI) have played an important role, with studies focusing on the morphometry of brain structures and patterns of brain atrophy.9 However, changes in neural tissue properties, measured as signal- and texture-related features, could be able to detect earlier and more subtle changes.10
The objective of this work was to determine whether using a multivariate feature selection strategy within a multimodal database, including MRI signal- and texture-related features among many others, MCI to AD progression could be accurately predicted, and if such imaging features would play an important role in the aforementioned prediction.
2. Methods
Data used in the preparation of this article were obtained from the Alzheimer’s disease neuroimaging initiative (ADNI) database.11 The ADNI was launched in 2003 by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, the Food and Drug Administration, and private pharmaceutical companies and nonprofit organizations as a $60 million, 5-year public-private partnership. The primary goal of ADNI has been to test whether serial MRI, positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessments can be combined to measure the progression of MCI and early AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials. The principal investigator of this initiative is the Weiner VA Medical Center and the University of California – San Francisco. ADNI is the result of the efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the United States and Canada. The initial goal of ADNI was to recruit 800 subjects, but ADNI has been followed by ADNI-GO and ADNI-2. To date, these three protocols have recruited over 1500 adults, ages 55 to 90 to participate in the research. These recruits consist of cognitively normal older individuals, people with early or late MCI, and people with early AD. The follow-up duration of each group is specified in the protocols for ADNI-1, ADNI-2, and ADNI-GO. Subjects originally recruited for ADNI-1 and ADNI-GO had the option to be followed in ADNI-2. For up-to-date information, see Ref. 12.
Available information from biological samples, PET analyses and clinical data were obtained. Neuropsychological assessments were not taken into account, given that the clinical diagnosis of MCI and AD was based on some of these. Magnetization-prepared rapid acquisition with gradient echo (MP-RAGE) images and their corresponding segmentation masks were downloaded. The segmentation masks were provided by Heckemann et al13 using multiatlas propagation enhanced registration, an automatic whole-brain multiregion segmentation method. In order to continue studying the same population as in previous works,14 only data and images uploaded up to June 2012 were considered. The 98 MCI subjects studied in such works were considered for this study, where those with a reported progression to AD within the 24 months following their baseline visit (BL) were considered as cases and those who maintained a cognitive stability during the same period of time were considered as controls. The 27 subjects who left the study early were excluded, provided they had not already shown progression.
Each MP-RAGE image was divided into 83 regions of interest (ROI) using their corresponding segmentation masks. Four cases and five controls were discarded due to misalignment issues between the image and the segmentation mask. This resulted in a database with 62 subjects whose demography is shown in Table 1, where the average mini-mental state examination (MMSE) score at the BL and the proportion of subjects with an allele in the apolipoprotein E gene () are also detailed. Each ROI was measured for a set of 13 morphometrical features (e.g., volume, superficial area, and compactness), 28 features related to the signal distribution (e.g., entropy, mean, and skewedness), and 9 texture-related features (e.g., superficial volume, area, and compactness of the intensity projection map). These 4150 features, in addition to 208 biological features, 78 clinical features and 47 PET-related features, were -standardized ( and ) and used to construct a multimodal database from which the feature selection algorithm would select a predictive set. The biological features were obtained from apolipoprotein E (APOE) genotyping, homocysteine and isoprostanes concentrations, urine and blood laboratory data, cerebrospinal fluid (CSF) laboratory data, rules-based medicine plasma data, University of Pennsylvania’s CSF and plasma biomarker data, and TOMM40 polymorphism data;15 the clinical features were obtained from a symptoms checklist, the family dementia history, neurological and physical exams, demographic information, and vital signs data; the PET-related features were obtained from the Banner Alzheimer’s Institute NMRC summaries analysis, the University of Utah PET analysis, and the New York University’s FDG-PET hippocampus analysis.16
Table 1.
Characteristics of key variables. Third and fourth rows show the -value of the Mann-Whitney test, indicating the probability of being independent to the class, and the area under receiver operating characteristic curve (AUC), respectively, for gender, age, , and MMSE.
| Population (women) | Mean age () | Mean MMSE score () | ||
|---|---|---|---|---|
| Controls | 37 (10) | 74.6 (6.4) | 41% | 27.8 (1.4) |
| Cases | 25 (6) | 74.6 (7.7) | 64% | 26.6 (1.7) |
| Mann-Whitney test -value | 0.8 | 1 | .07 | .01 |
| AUC | 0.52 | 0.5 | 0.61 | 0.69 |
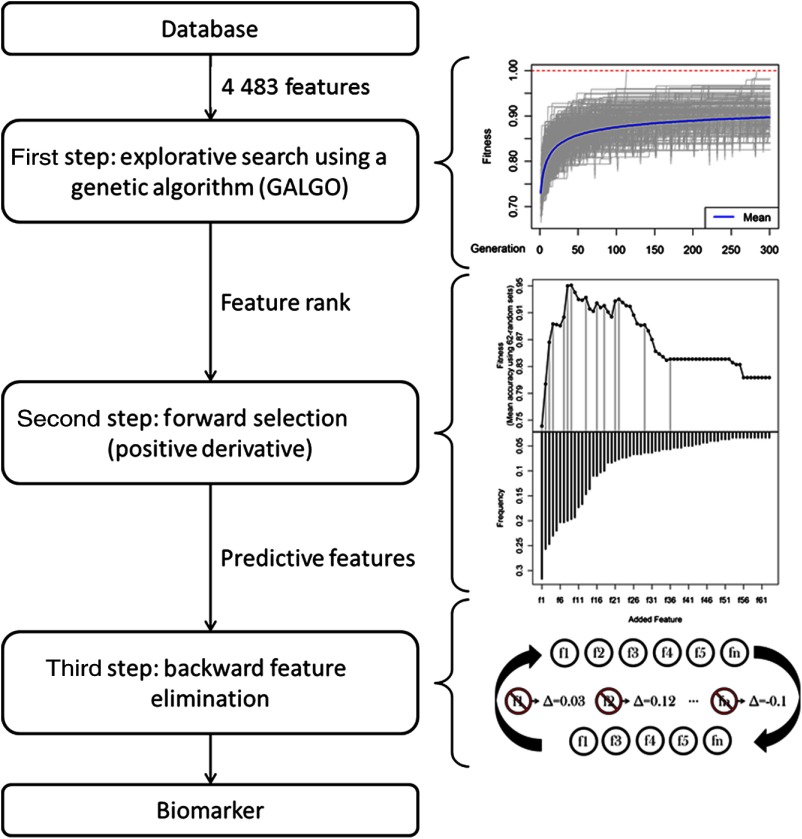
In order to select an accurate and compact set of features capable of classifying controls and cases, a three-step multivariate feature selection strategy was performed, as shown in Fig. 1. The strategy includes a genetic algorithm used to rank features, a customized forward selection (FS) methodology that selected predictive features, and a backward feature elimination (BFE) algorithm that removed redundant information.
Fig. 1.
Diagram of the three-step multivariate feature selection strategy. First step: evolution of the fitness of the 1000 models throughout 300 generations. Second step: features are ranked and added into nested models, only the features whose addition improve the fitness are kept (gray vertical line). Last step: model is reduced in size.
During the first step, an explorative search was performed using GALGO, the genetic algorithm in Ref. 17. In total, 1000 logistic regression models, each one with five features, were generated. They evolved from an initial set of models with random features throughout 300 generations, during which replication, recombination and mutation processes took place, and as with evolution, the fittest models survived and reproduced. Fitness was defined as the accuracy of the logistic regression model using a sevenfold validation strategy, randomly selecting in every fold six-sevenths of the subjects to train the classifier and the rest to test its accuracy.
Then, features were ranked according to their frequencies, defined as the ratio of models in which they were included to the number of models generated. An adjustment was made to account for correlations. If any pair of features had a significant Kendall correlation () with a coefficient larger than 0.8, the least frequent feature was removed, and the frequency of the remaining feature was recalculated as the ratio of models in which any of those two features were included to the number of models generated.
The second step involved using the aforementioned rank to generate a representative model by applying a customized FS strategy. The classical FS strategy generates nested models, adding the next best ranked feature one at a time, and selects the model that results in the maximum fitness. To avoid the inclusion of futile features, the classical FS strategy was modified. A model was built using the features whose addition represented any improvement in fitness with respect to their corresponding parent model. In other words, only features with a positive derivative value of the fitness curve were selected. Finally, in the last step, the resulting model was reduced in size using a BFE strategy, intended to remove features with redundant information. During each cycle of this shrinking process, the fitness of the FS model was evaluated after removing one feature at a time. The feature whose elimination from its parent model resulted in the largest fitness increase or in no fitness reduction was permanently removed. This procedure was carried on until no feature removal resulted in an equal or better fitness than its parent model. Fitness for the FS and BFE strategies was defined as the average accuracy of the test partitions from 62 random sets, each with a training and test proportion of two-thirds and one-third the size of the population, respectively. The final model was assessed in the same way to obtain the final performance of the model.
Since the subjects used as a test in each split of the second and third steps of this methodology included subjects that had already been used to train when obtaining the rank, they could not actually be considered independent test subjects, and thus the performance could be optimistically biased.18,19 To evaluate the performance of a model using this same methodology in a previously not seen population, the entire feature selection process was performed 50 times, each time using only 90% of the population. Once a final model had been obtained, the remaining 10% of the population was used to test such a model.
Moreover, we tested whether the final model was powerful enough to predict the risk of progressing from MCI to AD over a period of time longer than the 2-year window used to define controls and cases. In order to do this, subjects were ranked according to their prognostic index (PI). Subjects with a PI larger than the median were considered as high-risk cases and subjects with a PI smaller than the median were considered as low-risk cases. The PI was defined as the fitted mean value of the logistic regression model with the features from the proposed biomarker in the entire population of the experiment. Events were defined as any MCI to AD progression and the time to event was defined as the number of days between the BL and the visit in which they were first diagnosed with AD, or their last recorded visit (had the subjects not shown cognitive decline). Since there was no available information on whether the latter subjects would eventually progress to AD, they were considered as right-censored subjects. Having done this, a Kaplan-Meier curve was plotted,20 and a concordance index (CI) obtained.21 The CI was obtained by analyzing all possible pairs of subjects (except those where the subject with the shorter time to event was right censored) and evaluating whether the subject with the longer time to event also had the smaller PI, defining it as the proportion of agreements to the number of evaluations.
3. Results
The proposed biomarker, found from a multimodal database using a multivariate feature selection strategy, included six features: one related to genotyping, three related to the MP-RAGE image signal distribution, and two related to the MP-RAGE image texture. It achieved an average accuracy and area under the receiver operating characteristic curve (AUC) of 0.96 and 0.97, respectively. Tables 2 and 3 show a complete list of the features found in the proposed model and the detailed performance, respectively. Table 3 also includes the train and blind-test performances of the 50 models obtained to evaluate the bias of the methodology, resulting in an average blind-test accuracy and AUC of 0.79.
Table 2.
Features found in the proposed biomarker. The abbreviation that describes each feature was defined as ID.
| Feature | Modality | ROI | ID |
|---|---|---|---|
| Precision range | MRIa | Right middle frontal gyrus | PR RMFG |
| Probability of value being greater than | MRIa | Left amygdala | PG3 LA |
| MRIb | Right cerebellum | RM110 RC | |
| MRIb | Left medial orbital gyrus | RM200 LMOG | |
| TOMM40 maximum allele length | Genotyping | – | TOMM40 |
| Value at 0.1% | MRIa | Third ventricle | V001 TV |
Features related to the signal distribution of the image.
Features related to the texture of the image.
Table 3.
Performance of the proposed biomarker and the 50 blind-tested models. The concordance index (CI) and the logrank-test -value were obtained from the risk analysis, the other measures via classification.
| Mean accuracy (95% C.I.) | Mean sensitivity (95% C.I.) | Mean specificity (95% C.I.) | Mean AUC (95% C.I.) | CI | Logrank-test -value | |
|---|---|---|---|---|---|---|
| Proposed biomarker | 0.96 (0.84–1) | 0.96 (0.75–1) | 0.96 (0.82–1) | 0.97 (0.87–1) | 0.74 | |
| 50 models (train) | 0.99 (0.94–1) | 0.99 (0.96–1) | 0.99 (0.9–1) | 1 (1–1) | – | – |
| 50 models (test) | 0.79 (0.6–1) | 0.85 (0.5–1) | 0.7 (0–1) | 0.79 (0.4–1) | – | – |
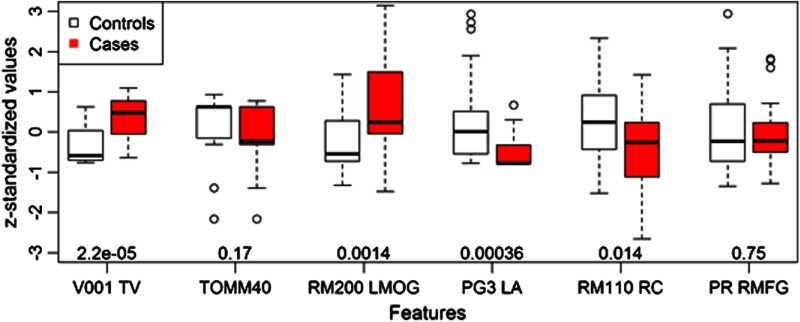
The features from this model were neither exclusively in the top of the rank obtained during the three-step feature selection strategy, nor were they of significant importance for classifying controls and cases on their own. Table 4 shows the top-ranked features and a few other relevant features that will be discussed later. Figure 2 shows the distribution of each feature for both controls and cases, including their -value for the Mann-Whitney test; that is the probability of such feature being independent to the class of the subjects.
Table 4.
Features ranked by their frequency. Last column (MW) shows -values of the Mann-Whitney test.
| Rank | Modality | Feature | ROI | Freq | MW |
|---|---|---|---|---|---|
| 1a | MRI | Value at 0.1% | Third ventricle | 32% | |
| 2 | PET | Normalized CMRgl | Right insula | 26% | |
| 3a | Genotyping | Max TOMM40 length | – | 25% | 0.17 |
| 4 | MRI | Right amygdala | 23% | ||
| 5 | MRI | Left inferiolateral remainder of parietal lobe | 22% | 0.35 | |
| 6 | PET | Normalized CMRgl | Left middle temporal gyrus | 20% | |
| 7 | Genotyping | Mean TOMM40 length | – | 20% | 0.37 |
| 8a | MRI | Left medial orbital gyrus | 20% | ||
| 9a | MRI | Left amygdala | 20% | ||
| 10 | MRI | Skewness | Left parahippocampal and ambient gyri | 19% | 5.90e−5 |
| 11 | MRI | Right subgenual frontal cortex | 17% | ||
| 12 | PET | Normalized CMRgl | Left superior and medial frontal giry | 17% | |
| 13 | MRI | Right parahippocampal and ambient giry | 15% | ||
| 14 | MRI | Left parahippocampal and ambient giry | 14% | ||
| 15 | MRI | Right amygdala | 11% | ||
| 16a | MRI | Right cerebellum | 11% | ||
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | |
| 21a | MRI | Precision range | Right middle frontal gyrus | 8% | 0.75 |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | |
| 30 | MRI | Compactness | Right hippocampus | 6% | |
| 31 | MRI | Left thalamus | 6% | ||
| 32 | Genotyping | – | 6% | ||
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | |
| 53 | Biospecimen | Platelets | – | 3% | |
| 54 | Biospecimen | Epidermal growth factor | – | 3% | |
| 55 | Biospecimen | Neutrophil gelatinase-associated lipocal | – | 3% | 0.19 |
| 56 | Clinical | Participant has siblings | – | 3% | 0.53 |
Features found in the proposed model.
Fig. 2.
Value distribution of features found in the proposed model across controls and cases. The -value of the Mann-Whitney test for every feature is shown at the bottom of the plot.
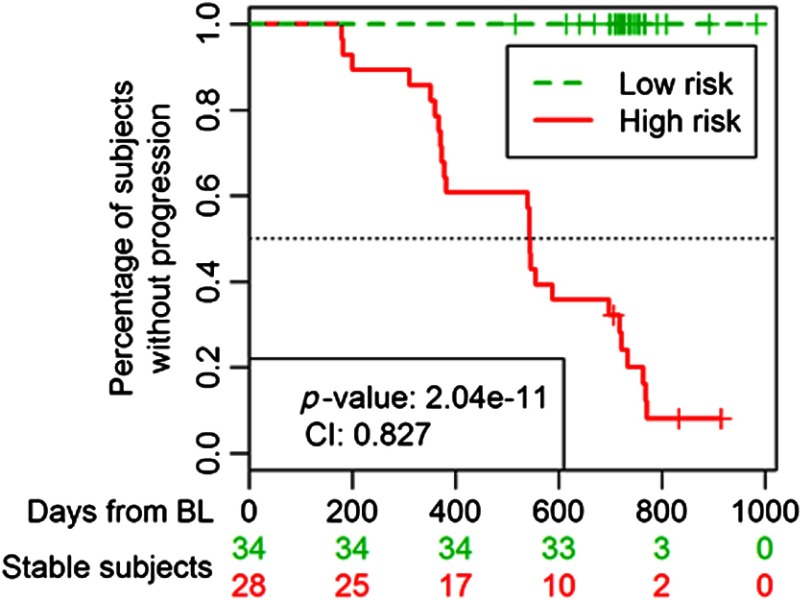
The high- and low-risk populations were found to have significantly different times to event, with a logrank-test -value of , rejecting the null hypothesis that the two groups have identical survival functions, and a concordance index of 0.827. The resulting Kaplan-Meier curve is shown in Fig. 3, which also shows the number of nonprogressing (stable) subjects at each tick of the -axis for both groups.
Fig. 3.
Kaplan-Meier curve showing the different times to event between low- and high-risk groups. The median of the PI was used as the threshold to define group membership. The PI was defined as the linear predictors of the logistic regression model with the features of the proposed biomarker applied to the whole population in this study.
4. Discussion
This work showed that brain MRI MP-RAGE signal- and texture-related features aid in the prediction of MCI to AD progression. To achieve this conclusion, a set of image analyses and feature selection tools were used to explore the complex multidimensional space of a multimodal database composed of biological, clinical, PET-, and MRI-related features. The proposed model included a nonimaging feature and MRI features related to signal and texture. It is important to notice that even though there are two features that have no individual statistical power, all features are shown to be significant when evaluated inside the logistic regression model. That means that all features in the model have coefficients in the logistic regression model with a probability lower than 5% of being zero.
Interestingly, a few features that have been previously found to be associated to AD were not found in the final model, and some were not even found in the rank, meaning that not a single model from the 1000 ones originally generated found such information valuable. Hippocampal volume is one of such cases, a measure that has been associated with progression from MCI to AD with a relative risk of 0.69,22 but which was not used by the feature selection strategy. However, the compactness of the right hippocampus was found at the middle of the rank, as shown in Table 2, and although the volume is the usually discussed predictor, this feature is still a measure of shape. Another example is , which has been linked to late-onset familial and sporadic AD23 and even though it was found in the rank, it was not included in the final model. Regarding biospecimens, the ones found highest in the rank are shown in Table 2, but CSF levels of neither nor tau, measurements recognized as potential predictors of risk for developing AD in healthy and MCI subjects,24,25 were included. This shows that there exists more information with AD predictive power than that previously found using univariate analyses guided by biological hypotheses, such as the ones used to identify the aforementioned predictors, and that combinations of MRI features related to the signal distribution and texture were shown to have a larger predictive power than any combination of these predictors, as was made obvious by the lack of such features in the models.
The performance of the identified model is promising, but the test partitions used to evaluate it were not truly blind to the methodology. When tested in a new population, it will probably underperform. However, the experiment in which 50 models were obtained, each with a 10% of the population remaining completely blind to the feature selection methodology, showed that a test accuracy of almost 80% could be expected when evaluating a different population using the suggested methodology. Thus, it is viable for the proposed model to achieve such an accuracy when faced with a new population, or even achieve a slightly better one, considering that such a performance was calculated for models trained using a smaller population than the population with which the proposed model was trained. This gap in performance estimates, shown with more detail in Table 3, is an example of the optimistic results that may be encountered in exploratory studies versus results obtained under more stringent testing conditions.
The fact that almost all features are signal- or texture-related indicates that these novel features could be helpful for the prediction of MCI to AD progress. Interestingly enough, the regions of the brain involved in this model are not often considered as relevant for AD etiology in the current literature unlike the already mentioned hippocampus or the entorihnal cortex, a structure shown to gradually improve the prognostic efficiency compared with hippocampal volumetry.26 Thus, although these regions may not show significant atrophy in either early or late stages of AD, they may suffer changes in the properties of their neural tissue which can be observed and quantified with these signal- and texture-related features. This is especially relevant, since such changes are thought to occur earlier than morphological ones, allowing for an earlier diagnosis. Regarding the biological feature found in the model, TOMM40, it has already been reported to be associated to the prediction of the age of late-onset AD in carriers of the APOE gene.27 Additionally, the fact that the proposed model was constructed using features from different information modalities, namely MRI and genotyping, supports the idea that multimodal biomarkers outperform single-modality ones.
The risk analysis showed that the model could divide the subjects into high- and low-risk groups. But, even more importantly, though subjects were studied up to 983 days after their BL visit, only three were categorized as high risk, indicating that this model could accurately predict MCI to AD progression, even 3 years before progressing.
One limitation of this study was the population size, which did not allow for a blind test within the ADNI population for the proposed biomarker, although the average test performance of the 50 models obtained using only a subset of the population allowed for a valid expectation of how the proposed biomarker would perform when classifying new subjects. Even though ADNI has recruited more than 1000 subjects, the specific requirements of this study greatly limited the number of participants. An experiment in which the amount of information required from each subject is reduced, thus allowing the population size to be largely increased, is underway. It will challenge this model and allow the identification of a more robust biomarker using a similar strategy. Despite this, the results and the strategy used in this study are promising, and so is the fact that people were accurately predicted to progress to AD up to 2 years prior to their clinical diagnosis.
Additionally, Table 1 shows that gender, age, and were not significantly different between controls and cases, but that MMSE scores were. This difference implies that cases were slightly more cognitively impaired than controls at BL, a somewhat expected result since diagnoses were partially based on this score. Regardless, this difference only results in an AUC of 0.69.
Also, there were a few parameters in the methodology that, if varied, could have resulted in a different biomarker. Such is the case for the number of models generated in the first step of the methodology and the size of each one of these models. However, the number of models was sufficiently large to allow for the rank to reach stability. On the other hand, it could not be assured that a change in the size of the models would not generate different results. However, even if that were to happen, the conclusions from this study would still be valid. The proposed biomarker is just one solution from a universe of possible solutions to this problem, but the fact that novel MRI features related to the signal distribution and texture aided in predicting the MCI to AD progression would still apply.
In conclusion, brain MRI MP-RAGE images were analyzed and several features were extracted from them, describing the morphology, signal distribution, and texture of the brain. An MCI to AD progression biomarker based in some of these imaging features and in one biological feature has been presented. The model was selected from a multimodal database containing almost 4500 features using a multivariate feature selection strategy based on genetic algorithms. The performance of the proposed biomarker is very encouraging, and a risk analysis showed that using these features, subjects could be accurately classified as having a high risk or a low risk of MCI to AD progression, even 3 years before clinical diagnosis.
Acknowledgments
This work was partially supported by the Consejo Nacional de Ciencia y Tecnología (CONACYT), by Grant No. 16864 Ciencia Básica from CONACYT, and by Catédra de Bioinformática (CAT220) from Tecnológico de Monterrey. Data collection and sharing for this project was funded by ADNI (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Amorfix Life Sciences Ltd.; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd. and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC; Johnson & Johnson Pharmaceutical Research & Development LLC; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study Rev October 16, 2012 at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH Grants P30 AG010129 and K01 AG030514.
Biographies
Antonio Martinez-Torteya received his BSc degree in biomedical engineering from Tecnológico de Monterrey in 2008 and his MSc degree in electronic systems from the same institution in 2010. Currently, he is pursuing his PhD degree in computer science at the same institution. His academic interests are quantitative analysis of medical images and techniques for the discovery of image-based biomarkers.
Juan Rodriguez-Rojas received his BSc degree in biomedical engineer from Tecnológico de Monterrey in 2007. Currently, he is pursuing his PhD degree in computer science at the same institution. His academic interests are quantitative analysis of medical images and techniques for the discovery of image-based biomarkers.
José M. Celaya-Padilla is a PhD student at Tecnológico de Monterrey. He received his BSc degree in computer engineering and his MSc degree in engineering from the Universidad Autónoma de Zacatecas in 2008 and 2010, respectively. Currently, he is studying for his PhD degree at Tecnológico de Monterrey in the Bioinformatics Department. His current research interests include medical imaging, data mining, and image registration.
Jorge I. Galván-Tejada received his engineering degree in electronics and communications and his master’s degree in engineering from the Universidad Autónoma de Zacatecas. Currently, he is pursuing his PhD degree in information technology and communications at Tecnológico de Monterrey. His areas of interest include signal processing, intelligent systems, bioinformatics, and image processing.
Victor Treviño is currently a professor in the Computer Science Department at Tecnológico de Monterrey. He received his BSc degree in electronic systems from Tecnológico de Monterrey in 1994, his MSc degree in molecular biology and genetic engineering from Universidad Autónoma de Nuevo León in 1999, and his PhD degree from Birmingham University in 2007. His academic interests include bioinformatics, molecular medicine, and artificial intelligence.
Jose Tamez-Peña is currently a research scientist at the Escuela de Medicina del Tecnológico de Monterrey. He received his BSc degree in physics engineering from Tecnológico de Monterrey in 1989, and his PhD degree in electrical engineering from the University of Rochester in 1999. His current research interests include quantitative medical imaging, computer vision, and artificial intelligence.
References
- 1.Prince M., Guerchet M., Prina M., “Policy brief for heads of government: the global impact of dementia 2013–2050,” Alzheimers Disease International, London, United Kingdom: (2013). [Google Scholar]
- 2.Thies W., Bleiler L., “2011 Alzheimer’s disease facts and figures,” Alzheimer’s Dementia 7, 208–244 (2011). 10.1016/j.jalz.2011.02.004 [DOI] [PubMed] [Google Scholar]
- 3.McKhann G., et al. , “Clinical diagnosis of Alzheimer’s disease report of the NINCDS‐ADRDA Work Group* under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease,” Neurology 34, 939–939 (1984). 10.1212/WNL.34.7.939 [DOI] [PubMed] [Google Scholar]
- 4.Petersen R. C., et al. , “Aging, memory, and mild cognitive impairment,” Int. Psychogeriatr. 9, 65–69 (1997). 10.1017/S1041610297004717 [DOI] [PubMed] [Google Scholar]
- 5.Zhang D., et al. , “Multimodal classification of Alzheimer’s disease and mild cognitive impairment,” NeuroImage 55(3), 856–867 (2011). 10.1016/j.neuroimage.2011.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinrichs C., et al. , “Predictive markers for AD in a multi-modality framework: an analysis of MCI progression in the ADNI population,” Neuroimage 55(2), 574–589 (2010). 10.1016/j.neuroimage.2010.10.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walhovd K. B., et al. , “Combining MR imaging, positron-emission tomography, and CSF biomarkers in the diagnosis and prognosis of Alzheimer disease,” Am. J. Neuroradiol. 31, 347–354, (2010). 10.3174/ajnr.A1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaffer J. L., et al. , “Predicting cognitive decline in subjects at risk for Alzheimer disease by using combined cerebrospinal fluid, MR imaging, and PET biomarkers,” Radiology, 266, 583–591, (2013). 10.1148/radiol.12120010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braskie M. N., Thompson P. M., “A focus on structural brain imaging in the Alzheimer’s disease neuroimaging initiative,” Biol. Psychiatry 75(7), 527–533 (2014). 10.1016/j.biopsych.2013.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salat D. H., et al. , “Age-associated alterations in cortical gray and white matter signal intensity and gray to white matter contrast,” Neuroimage 48, 21–28 (2009). 10.1016/j.neuroimage.2009.06.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ADNI—Alzheimer’s Disease Neuroimaging Initiative, “Sharing Alzheimer’s Research with the World,” adni.loni.ucla.edu.
- 12.ADNI Home, “Alzheimer’s Disease Neuroimaging Initiative,” www.adni-info.org.
- 13.Heckemann R. A., et al. , “Automatic morphometry in Alzheimer’s disease and mild cognitive impairment,” Neuroimage 56, 2024–2037 (2011). 10.1016/j.neuroimage.2011.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martínez-Torteya A., Treviño-Alvarado V. M., Tamez-Peña J. G., “Improved multimodal biomarkers for Alzheimer’s disease and mild cognitive impairment diagnosis: data from ADNI,” Proc. SPIE 8670 86700S (2013). 10.1117/12.2008100 [DOI] [Google Scholar]
- 15.Shaw L. M., “PENN biomarker core of the Alzheimer’s disease neuroimaging initiative,” NeuroSignals 16, 19–23 (2007). 10.1159/000109755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jagust W. J., et al. , “The Alzheimer’s Disease Neuroimaging Initiative positron emission tomography core,” Alzheimers Dement. 6(3), 221–229 (2010). 10.1016/j.jalz.2010.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trevino V., Falciani F., “GALGO: an R package for multivariate variable selection using genetic algorithms,” Bioinformatics 22, 1154–1156 (2006). 10.1093/bioinformatics/btl074 [DOI] [PubMed] [Google Scholar]
- 18.Simon R., et al. , “Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification,” J. Natl. Cancer Inst. 95, 14–18 (2003). 10.1093/jnci/95.1.14 [DOI] [PubMed] [Google Scholar]
- 19.Kupinski M. A., Giger M. L., “Feature selection with limited datasets,” Med. Phys. 26, 2176–2182 (1999). 10.1118/1.598821 [DOI] [PubMed] [Google Scholar]
- 20.Kaplan E. L., Meier P., “Nonparametric estimation from incomplete observations,” J. Am. Stat. Assoc. 53, 457–481 (1958). 10.1080/01621459.1958.10501452 [DOI] [Google Scholar]
- 21.Harrell F. E., Lee K. L., Mark D. B., “Tutorial in biostatistics multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors,” Stat. Med. 15, 361–387 (1996). [DOI] [PubMed] [Google Scholar]
- 22.Jack C. R., et al. , “Prediction of AD with MRI-based hippocampal volume in mild cognitive impairment,” Neurology 52, 1397–1397 (1999). 10.1212/WNL.52.7.1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rocchi A., et al. , “Causative and susceptibility genes for Alzheimer’s disease: a review,” Brain Res. Bull. 61, 1–24 (2003). 10.1016/S0361-9230(03)00067-4 [DOI] [PubMed] [Google Scholar]
- 24.Hampel H., et al. , “Biological markers of amyloid -related mechanisms in Alzheimer’s disease,” Exp. Neurol. 223, 334–346 (2010). 10.1016/j.expneurol.2009.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hampel H., et al. , “Total and phosphorylated tau protein as biological markers of Alzheimer’s disease,” Exp. Gerontol. 45, 30–40 (2010). 10.1016/j.exger.2009.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hampel H., et al. , “Core candidate neurochemical and imaging biomarkers of Alzheimer’s disease,” Alzheimers Dement. 4, 38–48 (2008). 10.1016/j.jalz.2007.08.006 [DOI] [PubMed] [Google Scholar]
- 27.Roses A. D., et al. , “A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer’s disease,” Pharmacogenomics J. 10, 375–384 (2009). 10.1038/tpj.2009.69 [DOI] [PMC free article] [PubMed] [Google Scholar]