Abstract
Ethylene is a hormone involved in numerous aspects of growth, development, and responses to biotic and abiotic stresses in plants. Ethylene is perceived through its binding to endoplasmic reticulum-localized receptors that function as negative regulators of ethylene signaling in the absence of the hormone. In Arabidopsis thaliana, five structurally and functionally different ethylene receptors are present. These differ in their primary sequence, in the domains present, and in the type of kinase activity exhibited, which may suggest functional differences among the receptors. Whereas ethylene receptors functionally overlap to suppress ethylene signaling, certain other responses are controlled by specific receptors. In this review, I examine the nature of these receptor differences, how the evolution of the ethylene receptor gene family may provide insight into their differences, and how expression of receptors or their accessory proteins may underlie receptor-specific responses.
Introduction
The effects of ethylene on plant growth, development, and responses to biotic and abiotic stresses are wide-ranging, and the diversity of those processes affected is perhaps greater than for many other phytohormones [1–6]. Ethylene is perceived through its binding to receptors [7], which are localized to the endoplasmic reticulum (ER) and Golgi apparatus and which function to negatively regulate ethylene responses [5,6,8–16]. In the absence of the hormone, ethylene receptors activate constitutive triple response 1 (CTR1), a Raf-like serine/threonine (Ser/Thr) kinase [17,18]. CTR1 phosphorylates ethylene insensitive (EIN)-2, an ER-bound, Nramp-like transmembrane protein, to repress its ability to induce ethylene responses [19–22] (Figure 1). After the binding of ethylene to a receptor, CTR1 activity is repressed, and in the absence of CTR1 phosphorylation, EIN2 undergoes proteolytic processing to release its C-terminal domain, which migrates to the nucleus to activate a transcriptional cascade involving EIN3/EIN3-like and ethylene response factor (ERF) transcription factors [20–22] (Figure 1).
Figure 1. The ethylene signaling pathway.

An N-terminal transmembrane domain of ethylene receptors targets the receptors to the endoplasmic reticulum (ER) membrane, where they form homodimers. CTR1 associates with the His kinase and receiver domains of a receptor, and, in the absence of the hormone, ethylene receptors maintain CTR1 in an active state to phosphorylate EIN2, which inhibits the ability of the latter to induce ethylene responses. EIN2 also interacts directly with ethylene receptors. When present, ethylene binds to the copper bound in the N-terminal transmembrane domain, resulting in the inactivation of the ethylene receptor-CTR complex, perhaps through conformational changes. Loss of EIN2 phosphorylation results in the proteolytic release of its C-terminal domain, which is targeted to the nucleus. The presence of the EIN2 C-terminal domain in the nucleus prevents further degradation of the ethylene response transcription factors EIN3/EIL1 by two F-box proteins (EBF1/2) and induces EBF1/2 degradation. Dimerization of EIN3/EIL1 and their binding to the promoter region of ethylene response factor (ERF) transcription factor genes activate the gene expression of the latter, resulting in the activation of downstream ethylene response genes. CTR, constitutive triple response; EIL, ethylene insensitive 3-like; EIN, ethylene insensitive.
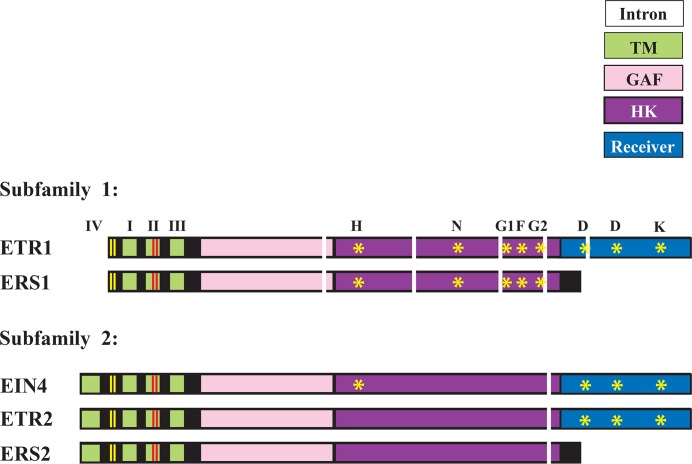
Five different types of ethylene receptors are present in Arabidopsis thaliana: ETR1, ERS1, EIN4, ETR2, and ERS2. Each contain N-terminal transmembrane, GAF, and histidine (His) kinase domains, and ETR1, EIN4, and ETR2 also contain a receiver domain (Figure 2) [23]. Ethylene receptors likely evolved from bacterial and yeast two-component regulators which have similar signal input and output domains with His kinase activity [9,24–26]. The receptors are divided into two subfamilies based on the presence or absence of conserved elements in the His kinase domain. Arabidopsis thaliana subfamily 1 receptors (ETR1 and ERS1) contain three membrane-spanning regions in their transmembrane domain and possess the motifs necessary for their His kinase activity, whereas subfamily 2 receptors (ETR2, ERS2, and EIN4) contain four transmembrane regions, lack most or all of the His kinase motifs, and exhibit Ser/Thr kinase activity which ERS1 also exhibits [23,27–33]. The role of phosphorylation in receptor function has been reviewed recently [6,14–16]. Thus, ethylene receptors exhibit a surprising level of structural and functional diversity that may be greater than that observed among receptors for other plant hormones.
Figure 2. Five ethylene receptors present in Arabidopsis thaliana.
The domain organization of subfamily 1 receptors (ETR1 and ERS1) and subfamily 2 receptors (ETR2, EIN4, and ERS2) in A. thaliana is shown. The N-terminal hydrophobic transmembrane domains (TMs) are indicated by green boxes. The N-proximal Cys residues that are required for the dimerization of receptors are indicated by the vertical yellow bars. The Cys and His residues of TM II required for ethylene binding are indicated by the vertical red bars. The GAF, His kinase (HK), and receiver domains are indicated according to the key. Yellow asterisks indicate the conserved motifs of the HK domain (H, N, G1, F, and G2) and the conserved aspartate and lysine residues of the receiver domain. The vertical white bars indicate intron position but not intron length. EIN, ethylene insensitive; ERS, ethylene response sensor; ETR, ethylene response; GAF; cGMP-specific phosphodiesterases, adenylyl cyclases and FhlA; His, histidine.
Loss of subfamily 1 receptor expression in Arabidopsis induces a more severe constitutive ethylene response than does the loss of subfamily 2 receptor expression [34,35]. Expression of ETR1 as the only receptor suppresses ethylene responses to a greater extent than any other receptor [35,36]. Moreover, ectopic expression of subfamily 2 receptors cannot rescue subfamily 1 null mutants [34,37–40]. Although loss-of-function (LOF) mutant analysis suggested functional overlap among the receptors in suppressing ethylene responses [7] (with ETR1 being predominant), such analyses also indicated functional differences between the subfamilies.
Ethylene receptors form homodimers through a disulfide bond between Cys-4 and Cys-6, and the receptor dimer binds one molecule of ethylene [41–43]. Receptor heterodimers also occur, perhaps mediated through the GAF domain, suggesting receptor clustering for signal amplification and possible crosstalk between receptor types in a heterodimer [13,44,45]. Together, the receptors CTR1 and EIN2 interact to form a signaling complex at the ER [20].
Not all ethylene receptors are created equal
Although ethylene receptors functionally overlap in their ethylene signaling suppression activity [7], they also exhibit functional specialization and a surprising degree of complexity in their interactions with one another and with partner proteins. Specialized roles for receptor family members are supported by the observation that Arabidopsis receptor LOF mutants exhibit elevated expression of the remaining receptor isoforms at the level of mRNA expression and ethylene-binding activity but not at the level of receptor output [46]. Moreover, the dominance of ETR1 in growth responses to ethylene in Arabidopsis is consistent with the notion of functional differences among receptors, a conclusion supported by the observations that specific receptors exhibit a disproportionate effect on fruit ripening in tomato, on salt stress in tobacco, and on the growth and development of rice [32,33,47,48].
Despite the role of ethylene receptors as negative regulators of ethylene responses, loss of ERS1 expression in Arabidopsis partially reverses the constitutive ethylene-response phenotype exhibited by any receptor mutant or combination of mutants in which wildtype ETR1 was present, suggesting that ERS1 modifies ETR1 activity or positively regulates ethylene responses under certain circumstances [35]. Arabidopsis mutants expressing only receptors without a receiver domain (that is, ERS1 and ERS2) recover from ethylene exposure more slowly than do mutants expressing receptors with a receiver domain (that is, ETR1, ETR2, and EIN4) [49], indicating that the receiver domain is important in growth recovery from ethylene.
Receptors may also regulate development differentially. Loss of ETR2 expression specifically resulted in an increase in branched root hairs and two-branch trichomes which was accompanied by altered expression of microtubule-associated protein 4 [50,51], suggesting that microtubule assembly is regulated specifically by ETR2. Moreover, EIN4 but not ETR1 is required for the ethylene suppression of defects in the development of root hairs in the Arabidopsis mutant root hair defective 1 (rhd1) [52].
Loss of ETR1 expression specifically resulted in a loss of the ethylene-mediated oscillatory movements of the apical hook known as nutational bending, whereas loss of expression of the other four receptors in Arabidopsis resulted in constitutive nutations [49,53], indicating that ETR1 is functionally distinct in this developmental response. ETR1 may also be involved in hydrogen peroxide signaling in stomatal guard cells that other receptors are not involved in [54].
Responses to pathogens may be receptor-specific. Gain-of-function mutants expressing etr1-1, which confers ethylene insensitivity, exhibited increased susceptibility to fumonisin B1, a fungal toxin that induces cell death, whereas ein4-1, which is also ethylene-insensitive, was less susceptible [50], suggesting that ETR1 and EIN4 have opposite roles in regulating susceptibility to fumonisin B1 toxin.
Receptor-specific interactions were observed between ERS1 and tetratricopeptide repeat protein 1 (AtTRP1) [55]. In addition to altering some auxin responses, AtTRP1 overexpression increased ethylene sensitivity, suggesting that AtTRP1 may compete with CTR1 for receptor binding or may promote receptor degradation [55].
ETR1 specifically interacts with RTE1, a transmembrane protein that stabilizes or promotes a stable signaling conformation [56–60]. RTE1 physically associates with cytochrome b5 (Cb5), which functions similarly to RTE1 in promoting ETR1 signaling, as loss of either partially suppresses the ethylene insensitivity conferred by etr1-2 and overexpression of either reduces ethylene sensitivity [61]. However, Cb5 overexpression phenotypes are RTE1-dependent, whereas RTE1 overexpression phenotypes do not require Cb5, suggesting that Cb5 functions upstream of RTE1. The specific interaction of RTE1 with ETR1 to promote its signaling may contribute to the dominance of ETR1.
ETR1 and EIN4 inhibit germination during salt stress, whereas ETR2 has the opposite effect [62]. The receiver domain of ETR1 was required for its inhibitory effect. As exposure to mannitol had a smaller effect, the receptors likely were mediating the effects of ion toxicity associated with salt rather than osmotic stress. Moreover, these effects were independent of ethylene signaling as they were lost under conditions of higher ethylene. Loss of ETR1 resulted in reduced sensitivity to ABA, whereas loss of ETR2 resulted in increased ABA sensitivity [62].
These results suggest that ethylene receptors differentially regulate an ethylene signaling component in addition to CTR1. One possible target might be EIN2, which is supported by the observation that ethylene receptors interact directly with EIN2 [63,64]. Whether an ethylene receptor possesses His kinase or Ser/Thr kinase activity may result in the phosphorylation of different sites in EIN2 and yield different signaling outcomes. The regulation of EIN2 activity through phosphorylation by CTR1 demonstrates that EIN2 is subject to such a mechanism.
Another, not mutually exclusive, possibility is that ethylene receptors differentially interact with His-containing phosphotransfer proteins (authentic histidine-containing phosphotransfer proteins [AHPs]), which receive a phosphate from the receiver domain of cytokinin receptors, or with response regulatory proteins (Arabidopsis thaliana response regulator [ARRs]), which receive the phosphate from AHPs [65–69]. The former is supported by the observation that ETR1, but not ERS1, interacts with AHPs [65], whereas the latter is supported by the observation that some ARRs exhibit opposing effects on germination during salt stress [70].
Need as a creative force driving complexity?
The complexity of ethylene receptors may have been a result of the diverse regulatory roles the hormone has come to play during development and during responses to biotic and abiotic stimuli. The structural differences in receptors likely enable the association of isoform-specific factors (for example, RTE1) or binding of general signaling factors with different affinities that may be needed for these individual roles. Differences in general signaling factor binding affinity were observed with CTR1, which interacts more strongly with subfamily 1 receptors than with ETR2 and interacts with the receiver domain in addition to the kinase domain of receptors [18,71]. This may contribute to the dominance of subfamily 1 receptors and ETR1 in particular. However, studies examining the amount of membrane-associated CTR1, presumably through receptor association, in subfamily 1-deficient Arabidopsis expressing kinase-defective ETR1 revealed little correlation between the amount of bound CTR1 and ethylene sensitivity [72]. Moreover, loss of either ETR1 or ERS1, which increases ethylene sensitivity, was accompanied by increased levels of membrane-associated CTR1 [34,73]. Therefore, it may be the extent to which a receptor activates CTR1 that determines signal output intensity, and subfamily 1 receptors may activate CTR1 more effectively than subfamily 2 receptors.
Although ethylene receptors are thought to signal through CTR1 and EIN2 [74], recent evidence suggests that signaling independent of CTR1 or EIN2 can occur as well. The ctr1 mutant retains a degree of ethylene responsiveness [75], and expression of the N-terminal half of ETR1 partially suppresses the ctr1 phenotype [76], observations supporting receptor function through a CTR1-independent pathway. As the latter observation required RTE1, RTE1 may assist ETR1 signaling or ETR1 may function in an alternative pathway through RTE1.
The observation that ethylene receptors can also interact with AHP and ARR proteins [65,66,77] suggests the possibility of CTR1- or EIN2-independent signaling crosstalk with other hormone pathways. Collectively, these findings suggest that, despite their functional overlap in ethylene signaling suppression, there are also ethylene receptor-specific functions that argue for accessory signaling factors or involvement of additional signaling pathways (or both) that would help explain the diverse responses elicited by ethylene.
Can evolution inform function?
The presence of ethylene receptors in plants raises questions regarding when such receptors first appeared, which receptor was the foundational member, when other receptors evolved, and receptor distribution among species. Proteins in Synechocystis (slr1212) and Anabaena share limited similarity to ETR1, and slr1212 binds ethylene [43,78,79]. As chloroplasts and cyanobacteria share a common ancestor, these data might suggest that the ancestral ethylene receptor was introduced into evolving plants through the endosymbiosis of the ancestral cyanobacterium and subsequent migration of the gene to the nucleus accompanied by re-targeting the receptors to the ER. However, these cyanobacterial proteins differ structurally from ethylene receptors in that the former lack the GAF domain, some conserved sequence motifs of the His kinase domain, or the receiver domain entirely while at the same time contain domains not present in plant ethylene receptors [79]. Moreover, though not reported for Anabaena, Synechocystis produces no detectable ethylene, and the hormone elicits no obvious response [78], raising the question of what function its ethylene binding serves. Neither species contains homologs for the other components of ethylene signaling, suggesting that any ethylene signaling output from these putative receptors would require novel interactions. The observation that marine algae lack any clear ethylene receptor homolog supports the possibility that ethylene receptors had not evolved in early plants.
In contrast to this, proteins containing the hallmarks of an ETR1 receptor are present in the charophyte Klebsormidium flaccidum (Figure 3), a freshwater algae whose genome sequence was reported recently [80]. Moreover, homologs for CTR1, EIN3, and EBF1 are present in K. flaccidum, Spirogyra pratensis, and Coleochaete orbicularis [80,81]. The S. pratensis ETR1-like and EIN3-like homologs complement ethylene receptor and ein3 mutants, respectively, in Arabidopsis thaliana [81], demonstrating remarkable conservation in the ethylene signaling pathway between freshwater algae and higher plants. K. flaccidum, which represents an older plant lineage than S. pratensis, appears to lack ERF1, and its homolog for EIN2 lacks the C-terminal domain involved in signaling, whereas ethylene receptors may be absent altogether in Mesostigma viride, an even older charophyte [81]. These observations suggest that the ethylene signaling pathway may have been evolving concomitant with charophytes. As land plants evolved from aquatic ancestors that are sister groups to charophycean algae, these observations demonstrate that ethylene receptor evolution predates the appearance of land plants and may have arisen in response to the terrestrial aspects of charophycean habitats, which include growth in moist soils that might expose them to abiotic stresses.
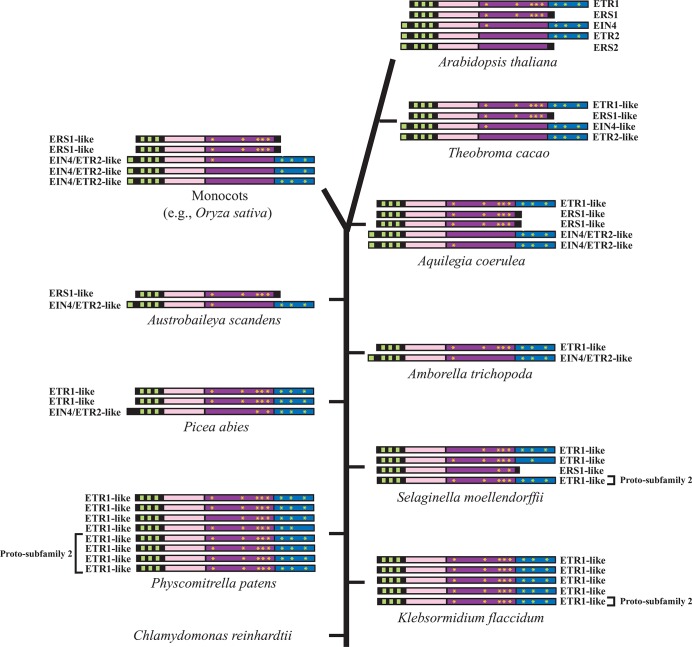
Figure 3. Timeline of appearance of ethylene receptor gene family members in plants.
The ethylene receptor genes in the lower and higher plant species indicated are shown along a timeline of plant evolution from bottom to top. Transmembrane domains (green), GAF domain (pink), His kinase domain (purple), and receiver domain (blue) are indicated. The number of genes represents the size of the receptor gene family for each species. Yellow asterisks indicate the conserved motifs of the His kinase and receiver domains. ETR1-like receptors contain the conserved motifs of the His kinase domain and a receiver domain. ERS1-like receptors contain the conserved motifs of the His kinase domain but lack a receiver domain. EIN4-like and ETR2-like receptors lack most or all of the conserved motifs of the His kinase domain but contain a receiver domain. EIN, ethylene insensitive; ERS, ethylene response sensor; ETR, ethylene response; GAF, cGMP-specific phosphodiesterases, adenylyl cyclases and FhlA; His, histidine.
Despite possessing only ETR1-like receptors, the first indication of subfamily 2 gene evolution is revealed in the intron structure of one of the five K. flaccidum ETR1-like genes in which the single intron diagnostic of subfamily 2 receptor genes first appears [82]. A receptor lacking some of the conserved motifs of the His kinase domain first appears in gymnosperms followed by the appearance of an EIN4/ETR2-like receptor in Amborella trichopoda (Figure 3), the basal-most angiosperm species, with an intron structure characteristic of subfamily 2 genes [82]. Distinct EIN4-like and ETR2-like receptors appear only in core eudicots, and the occurrence of ERS2, the most recent subfamily 2 member to evolve, is limited to the Brassicaceae [82]. ERS1-like receptors did not appear until after A. trichopoda, and in these basal angiosperm species (as in monocots in general), only ERS1-like and EIN4/ETR2-like receptors are present [82]. Thus, despite being a foundational member, ETR1-like receptors are not present in all plants.
From these observations, we can conclude that an ETR1-like receptor evolved first and did so perhaps during charophycean evolution. The five- and eight-member gene families present in K. flaccidum and the bryophyte Physcomitrella patens suggest that ethylene responses were likely important during the transition to land environments. The differentiation of a charophycean receptor gene into a proto-subfamily 2 receptor gene followed by the appearance of a transitional subfamily 2 receptor in gymnosperms and then a true subfamily 2 receptor appearing in the basal-most angiosperm species suggests a need for this second subfamily receptor type as the colonization of land environments progressed, perhaps in response to drier and warmer conditions. The presence of an ERS1-like receptor as the only subfamily 1 receptor in some basal and early angiosperms as well as monocots demonstrates that ETR1-like receptors are not essential for plant competitiveness. However, at least one receiver domain-containing receptor is expressed in every land plant species examined. The recent evolution of ERS2 in the Brassicaceae indicates that the gene family continues to evolve in a dynamic and specific manner.
Is ethylene receptor expression regulated by small upstream open reading frames?
The amount of receptor expression influences ethylene responses as shown through the regulated expression of etr1-1 where the degree of ethylene insensitivity conferred was controlled by the level of mutant receptor expression [83]. Exposure to ethylene also results in the induction of specific receptors (for example, ERS1, ETR2, and ERS2 in Arabidopsis thaliana) perhaps to prevent an excessive response to ethylene [7,84–86].
Not all receptors function equally, however, in that subfamily 1 receptors appear to be more important than subfamily 2 receptors. For example, loss of ETR1 specifically results in greater ethylene responsiveness than does the loss of other receptors, supporting the notion that ETR1 is dominant [34,35]. This has been ascribed to its function and not to its expression level as ETR1 and ERS1 transcript abundance is similar [85]. Gene expression, however, is not controlled at the transcriptional level alone. Like many genes encoding regulators, ethylene receptor genes contain upstream open reading frames (uORFs) in their 5′-leader sequence (Figure 4). Translation of uORFs typically reduces expression from the main ORF, and uORFs can serve a regulatory role in response to internal or external cues.
Figure 4. Upstream open reading frames (uORFs) present in Arabidopsis thaliana ethylene receptor genes.
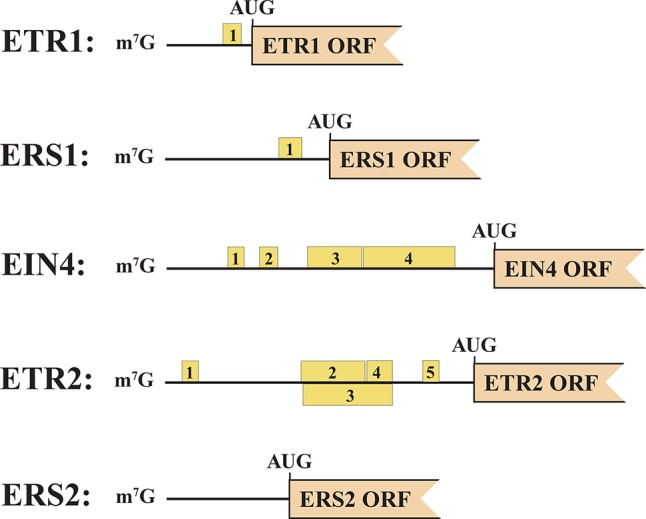
The presence of uORFs in the 5′-untranslated sequence of Arabidopsis thaliana ETR1, ERS1, ETR2, EIN4, and ERS2 mRNAs is shown. uORFs are indicated by yellow and numbered boxes. The main ORF encoding the receptor is indicated in orange. The context of most of the uORF AUGs (that is, the sequence surrounding a start codon that determines the strength of initiation) is not optimal, suggesting leaky 40S subunit scanning (inefficient initiation of protein synthesis) under normal conditions. EIN, ethylene insensitive; ERS, ethylene response sensor; ETR, ethylene response.
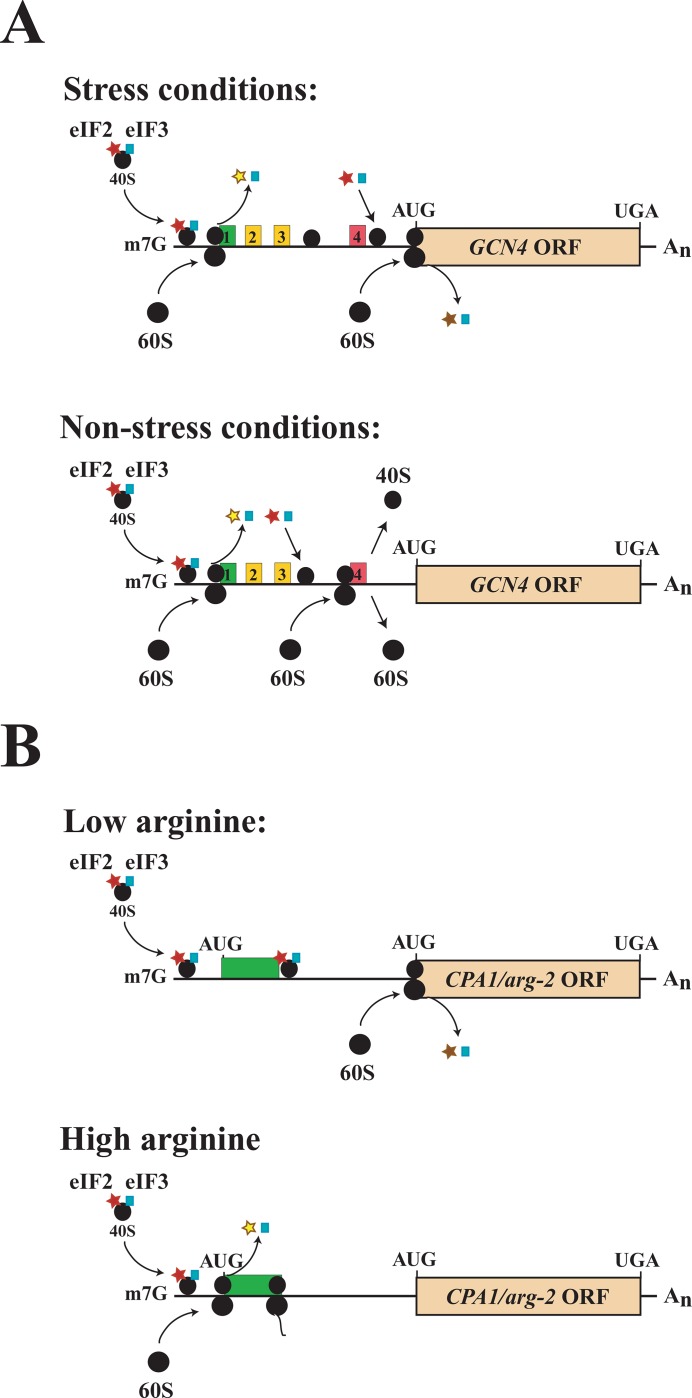
An example of uORF-mediated translational regulation is yeast GCN4, which contains four uORFs upstream of the GCN4 ORF, which encodes an amino acid biosynthesis transcriptional activator (Figure 5A). The degree to which each uORF is translated is dependent on amino acid availability, which determines whether translation re-initiates at the GCN4 ORF (Figure 5A) [87,88]. For this type of regulation, the uORFs are typically no more than a few codons long and their encoded proteins have no function. Translation of such uORFs can cause up to 100% of ribosomes to dissociate after uORF translation, thus preventing translation from the main ORF. In a second type of uORF-mediated regulation, translation of the uORF results in ribosome stalling mediated by the nascent uORF-encoded polypeptide, whose activity is determined by the availability of a co-factor [88] (Figure 5B). For this type, the uORFs are longer, as the regulation is mediated by the encoded protein.
Figure 5. Upstream open reading frame-mediated regulation of translation.
(A) The upstream open reading frames (uORFs) in yeast GCN4 mRNA regulate translation of the GCN4 ORF through a re-initiation mechanism controlled by amino acid biosynthesis availability. The GCN4 uORFs are no more than a few codons long and their encoded proteins have no function. Under non-starvation conditions, translation initiates at uORF1, but, upon termination, up to 50% of the 40S ribosomal subunits remain associated with the mRNA. Retention of the 40S subunits is due to sequences immediately upstream and downstream of the uORF1 start and stop codons, respectively, which interact with the translation initiation factor, eIF3a, which normally is released from the 40S subunit after initiation. Retention of eIF3 enables the 40S subunit to resume scanning and re-initiate translation. The 40S subunit regains competency to recognize initiation codons quickly because of the binding of a new eIF2-tRNAi-GTP ternary complex. Translation of uORF4 leads to full ribosome dissociation at its termination codon, preventing re-initiation at the GCN4 ORF [85]. Amino acid starvation results in reduced levels of ternary complex through the GCN2-mediated phosphorylation of the eIF2α subunit, which inhibits its recycling into a new ternary complex. As a result of the reduced availability of ternary complex, a 40S subunit that has resumed scanning after uORF1 translation will scan past uORF4 before a ternary complex binds, thus enabling the 40S subunit to reach the GCN4 ORF to initiate protein synthesis. (B) The single uORF in yeast CPA1/Neurospora crassa arg-2 mRNAs regulates translation through a ribosome-stalling mechanism controlled by arginine availability. Translation of the uORF-encoded, nascent arginine attenuator peptide (AAP) in response to arginine causes ribosome stalling at the uORF termination codon. This in turn inhibits leaky scanning of any 40S ribosomal subunits through the uORF and thus prevents them from reaching the main ORF. The amino acid sequence of the AAP peptide is key to the negative cis-acting regulation, which in the presence of arginine may interact with the ribosome tunnel near the large ribosomal protein L22 to cause stalling [88]. For peptide-mediated regulators, the uORFs that encode them are typically longer, as the regulation is mediated by the encoded protein. Translation of uORFs may also trigger nonsense-mediated decay (NMD) of the mRNA in addition to the translational regulation, thus serving as another means to regulate expression. For example, translation of the CPA1 uORF induces NMD whereas translation of the GCN4 ORFs does not [88].
uORFs whose length is consistent with both of these uORF types are present in four of the five ethylene receptors in Arabidopsis (Figure 4). ETR1 and ERS1 contain a single uORF, EIN4 and ETR2 show a more complex uORF arrangement, and ERS2 lacks uORFs. Although the extent to which any of these uORFs is translated has not been investigated, the frequency of initiation at an upstream AUG is determined by the sequence context surrounding it and the presence of downstream proximal secondary structure [89]. Therefore, the extent to which expression of each receptor type is affected will be determined by the presence and number of uORFs and the extent to which each is translated. As in the cases above, one or more of the uORFs may regulate receptor expression in response to growth conditions which could result in relative changes in specific receptors in different tissues or after receipt of internal or external cues. Therefore, whether ETR1 is truly the dominant receptor would require measuring the steady-state protein level of this receptor relative to ERS1 or other receptors and in specific tissues or growth conditions. Moreover, the potentially repressive effect that multiple uORFs in EIN4 and ETR2 could exert necessitates comparing receptor expression at the protein level in addition to transcript abundance in order to draw accurate conclusions regarding dominance of any individual receptor.
Future challenges – in pursuit of the known unknowns
Given the diversity of ethylene responses, we are just beginning to understand that ethylene signaling is considerably more complex than initially thought. The notion that multiple receptor types differ only in the level of their sensitivity to ethylene and signal output is likely insufficient to explain the many processes the hormone controls. Receptors of different types can form heterodimers, providing yet another level of complexity if receptor crosstalk within a heterodimer can alter the strength or type of signaling activity of a receptor. The observation that ethylene receptor expression exhibits an overlapping but distinct developmental pattern among the isoforms in Arabidopsis increases the potential for complexity, particularly when possible receptor crosstalk in heterodimer complexes affecting the strength and type of signal output is also considered [13]. The observation that ETR1 suppresses ethylene responses to a greater extent than does ERS1 when either is expressed as the only receptor in Arabidopsis [35,36] is illustrative of the need to determine whether receptors are inherently different in their signaling activity, whether receptors modify one another's output through the formation of heterodimers or higher-order complexes, or whether they differ in their strength or type of interactions with partner proteins.
Central to unraveling this complexity will be the identification of all components of the receptorsome, both those shared by all receptor members and those that are receptor-specific. This will require isolation of receptor complexes followed by proteomic identification of each associated protein. Identifying how the partner proteins present in a homodimeric receptorsome differ from those present in a heterodimeric receptorsome will be critical in determining how receptor interactions might alter their signaling behavior. Already, initial isolation has revealed considerable heterogeneity in receptor complexes, which may be regulated by ethylene in a receptor-specific manner [90].
As the ethylene signaling of receptors is dependent on their interactions with partner proteins (for example, CTR1), proteomic profiling of the receptorsome in different organs, different developmental stages, or under different growth conditions is needed to understand how changes in receptorsome components might explain the complexity of receptor-mediated responses to internal and external signals.
Whether such factors also alter receptor association with other receptors or with CTR1 or EIN2 could also be determined from this approach. Comparative proteomic profiling of receptorsomes in different species would be of particular interest, especially between Arabidopsis (which has five receptors) and P. patens (which possesses only ETR1-like receptors) or monocots (which express only ERS1-like and EIN4/ETR2-like receptors). How might the composition of the corresponding receptorsome differ among species?
Identification of receptorsome components may also reveal receptor signaling through CTR1- or EIN2-independent pathways or both. Ethylene receptor interaction with AHP proteins [65,66] is an example of how receptor signaling to other factors may work independently or together with EIN2 to tailor a response appropriately. This could involve the induction or repression of other genes which are not controlled by EIN2 but which alter or complement EIN2-mediated responses.
Given the potential for uORF-mediated regulation, the diversity of ethylene responses may involve differential changes in receptor mRNA translation which would alter the relative amounts of each receptor present. Profiling ethylene receptor expression at the level of transcription, translation, and mRNA and protein turnover will be needed to determine how expression contributes to the functional effects of each receptor and to resolve whether the dominance of ETR1 or subfamily 1 receptors is inherent or a result of expression differences.
Acknowledgments
Work cited from Daniel Gallie's laboratory was funded by the University of California Agricultural Experiment Station.
Abbreviations
- AHP
authentic histidine-containing phosphotransfer protein
- ARR
Arabidopsis thaliana response regulator
- AtTRP1
a tomato tetratricopeptide repeat protein 1
- Cb5
cytochrome b5
- CTR1
constitutive triple response 1
- Cys
cysteine
- EIL
ethylene insensitive 3-like
- EIN
ethylene insensitive
- ER
endoplasmic reticulum
- ERS
ethylene response sensor
- ETR
ethylene response
- GAF
cGMP-specific phosphodiesterases, adenylyl cyclases, and FhlA
- His
histidine
- LOF
loss-of-function
- ORF
open reading frame
- Ser
serine
- Thr
threonine
- uORF
upstream open reading frame
Disclosures
The author declares that he has no disclosures.
The electronic version of this article is the complete one and can be found at: http://f1000.com/prime/reports/b/7/39
References
- 1.Mattoo AK, Suttle JC. The Plant Hormone Ethylene. Boca Raton: CRC Press; 1991. [Google Scholar]
- 2.Abeles FB, Morgan PW, Saltveit ME., Jr . Ethylene in Plant Biology. 2. San Diego: Academic Press; 1992. [Google Scholar]
- 3.Bleecker AB, Kende H. Ethylene: a gaseous signal molecule in plants. Ann Rev Cell Dev Biol. 2000;16:1–18. doi: 10.1146/annurev.cellbio.16.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Klee HJ. Ethylene signal transduction. Moving beyond Arabidopsis. Plant Physiol. 2004;135:660–7. doi: 10.1104/pp.104.040998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin Z, Zhong S, Grierson D. Recent advances in ethylene research. J Exp Bot. 2009;60:3311–36. doi: 10.1093/jxb/erp204. [DOI] [PubMed] [Google Scholar]
- 6.Shakeel SN, Wang X, Binder BM, Schaller GE. Mechanisms of signal transduction by ethylene: overlapping and non-overlapping signalling roles in a receptor family. AoB Plants. 2013;5:plt010. doi: 10.1093/aobpla/plt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hua J, Meyerowitz EM. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;94:261–71. doi: 10.1016/S0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- 8.Chang C, Shockey JA. The ethylene-response pathway: signal perception to gene regulation. Curr Opin Plant Biol. 1999;2:352–8. doi: 10.1016/S1369-5266(99)00004-7. [DOI] [PubMed] [Google Scholar]
- 9.Chang C, Stadler R. Ethylene hormone receptor action in Arabidopsis. Bioessays. 2001;23:619–27. doi: 10.1002/bies.1087. [DOI] [PubMed] [Google Scholar]
- 10.Wang KL, Li H, Ecker JR. Ethylene biosynthesis and signaling networks. Plant Cell. 2002;14:S131–51. doi: 10.1105/tpc.001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen YF, Randlett MD, Findell JL, Schaller GE. Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. J Biol Chem. 2002;277:19861–6. doi: 10.1074/jbc.M201286200. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1006557
- 12.Stepanova AN, Alonso JM. Arabidopsis ethylene signaling pathway. Sci STKE. 2005;2005:cm4. doi: 10.1126/stke.2762005cm4. [DOI] [PubMed] [Google Scholar]
- 13.Grefen C, Städele K, Růzicka K, Obrdlik P, Harter K, Horák J. Subcellular localization and in vivo interaction of the Arabidopsis thaliana ethylene receptor family members. Mol Plant. 2008;1:308–20. doi: 10.1093/mp/ssm015. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/720316993
- 14.Schaller GE. Ethylene and the regulation of plant development. BMC Biol. 2012;10:9. doi: 10.1186/1741-7007-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Binder BM, Chang C, Schaller GE. Annual Plant Reviews, The Plant Hormone Ethylene. Vol. 44. Hoboken: Wiley-Blackwell; 2012. Perception of ethylene by plants - ethylene receptors; pp. 117–145. [DOI] [Google Scholar]
- 16.Merchante C, Alonso JM, Stepanova AN. Ethylene signaling: simple ligand, complex regulation. Curr Opin Plant Biol. 2013;16:554–60. doi: 10.1016/j.pbi.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell. 1993;72:427–41. doi: 10.1016/0092-8674(93)90119-B. [DOI] [PubMed] [Google Scholar]
- 18.Clark KL, Larsen PB, Wang X, Chang C. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci USA. 1998;95:5401–6. doi: 10.1073/pnas.95.9.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284:2148–52. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- 20.Ju C, Yoon GM, Shemansky JM, Lin DY, Ying ZI, Chang J, Garrett WM, Kessenbrock M, Groth G, Tucker ML, Cooper B, Kieber JJ, Chang C. CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signalling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci USA. 2012;109:19486–91. doi: 10.1073/pnas.1214848109. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/717963586
- 21.Qiao H, Shen ZS, Huang C, Schmitz RJ, Urich MA, Briggs SP, Ecker JR. Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science. 2012;338:390–3. doi: 10.1126/science.1225974. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/717961859
- 22.Wen X, Zhang C, Ji Y, Zhao Q, He W, An F, Jiang L, Guo H. Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res. 2012;22:1613–6. doi: 10.1038/cr.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/725361473
- 23.Hua J, Sakai H, Nourizadeh S, Chen QG, Bleecker AB, Ecker JR, Meyerowitz EM. EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell. 1998;10:1321–32. doi: 10.1105/tpc.10.8.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaller GE. Ethylene and cytokinin signalling in plants: the role of two-component systems. Essays in Biochemistry. 1997;32:101–11. [PubMed] [Google Scholar]
- 25.Chang C, Stewart RC. The two-component system. Regulation of diverse signaling pathways in prokaryotes and eukaryotes. Plant Physiol. 1998;117:723–31. doi: 10.1104/pp.117.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lohrmann J, Harter K. Plant two-component signaling systems and the role of response regulators. Plant Physiol. 2002;128:363–9. doi: 10.1104/pp.010907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science. 1993;262:539–44. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- 28.Hua J, Chang C, Sun Q, Meyerowitz EM. Ethylene insensitivity conferred by Arabidopsis ERS gene. Science. 1995;269:1712–4. doi: 10.1126/science.7569898. [DOI] [PubMed] [Google Scholar]
- 29.Gamble RL, Coonfield ML, Schaller GE. Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc Natl Acad Sci USA. 1998;95:7825–9. doi: 10.1073/pnas.95.13.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakai H, Hua J, Chen QG, Chang C, Medrano LJ, Bleecker AB, Meyerowitz EM. ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:5812–7. doi: 10.1073/pnas.95.10.5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moussatche P, Klee HJ. Autophosphorylation activity of the Arabidopsis ethylene receptor multigene family. J Biol Chem. 2004;279:48734–41. doi: 10.1074/jbc.M403100200. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1021145
- 32.Chen T, Liu J, Lei G, Liu YF, Li ZG, Tao JJ, Hao YJ, Cao YR, Lin Q, Zhang WK, Ma B, Chen SY, Zhang JS. Effects of tobacco ethylene receptor mutations on receptor kinase activity, plant growth and stress responses. Plant Cell Physiol. 2009;50:1636–50. doi: 10.1093/pcp/pcp107. [DOI] [PubMed] [Google Scholar]
- 33.Wuriyanghan H, Zhang B, Cao WH, Ma B, Lei G, Liu YF, Wei W, Wu HJ, Chen LJ, Chen HW, Cao YR, He SJ, Zhang WK, Wang XJ, Chen SY, Zhang JS. The ethylene receptor ETR2 delays floral transition and affects starch accumulation in rice. Plant Cell. 2009;21:1473–94. doi: 10.1105/tpc.108.065391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qu X, Hall B, Gao Z, Schaller GE. A strong constitutive ethylene-response phenotype conferred on Arabidopsis plants containing null mutations in the ethylene receptors ETR1 and ERS1. BMC Plant Biol. 2007;7:3. doi: 10.1186/1471-2229-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/722336883
- 35.Liu Q, Xu C, Wen CK. Genetic and transformation studies reveal negative regulation of ERS1 ethylene receptor signaling in Arabidopsis. BMC Plant Biol. 2010;10:60. doi: 10.1186/1471-2229-10-60. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/722336486
- 36.Liu Q, Wen CK. Arabidopsis ETR1 and ERS1 differentially repress the ethylene response in combination with other ethylene receptor genes. Plant Physiol. 2012;158:1193–207. doi: 10.1104/pp.111.187757. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/723350906
- 37.Zhao XC, Qu X, Mathews DE, Schaller GE. Effect of ethylene pathway mutations upon expression of the ethylene receptor ETR1 from Arabidopsis. Plant Physiol. 2002;130:1983–91. doi: 10.1104/pp.011635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hall AE, Bleecker AB. Analysis of combinatorial loss-of-function mutants in the Arabidopsis ethylene receptors reveals that the ers1 etr1 double mutant has severe developmental defects that are EIN2 dependent. Plant Cell. 2003;15:2032–41. doi: 10.1105/tpc.013060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang W, Hall AE, O'Malley R, Bleecker AB. Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proc Natl Acad Sci USA. 2003;100:352–7. doi: 10.1073/pnas.0237085100. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1011611
- 40.Xie F, Liu Q, Wen C-K. Receptor signal output mediated by the ETR1 N-terminus is primarily subfamily I receptor dependent. Plant Physiol. 2006;142:492–508. doi: 10.1104/pp.106.082628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaller GE, Bleecker AB. Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science. 1995;270:1809–11. doi: 10.1126/science.270.5243.1809. [DOI] [PubMed] [Google Scholar]
- 42.Schaller GE, Ladd AN, Lanahan MB, Spanbauer JM, Bleecker AB. The ethylene response mediator ETR1 from Arabidopsis forms a disulfide-linked dimer. J Biol Chem. 1995;270:12526–30. doi: 10.1074/jbc.270.21.12526. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez FI, Esch JJ, Hall AE, Binder BM, Schaller GE, Bleecker AB. A copper co-factor for the ethylene receptor ETR1 from Arabidopsis. Science. 1999;283:996–8. doi: 10.1126/science.283.5404.996. [DOI] [PubMed] [Google Scholar]
- 44.Gao Z, Wen CK, Binder BM, Chen YF, Chang J, Chiang YH, Kerris RJ, 3rd, Chang C, Schaller GE. Heteromeric interactions among ethylene receptors mediate signaling in Arabidopsis. J Biol Chem. 2008;283:23801–10. doi: 10.1074/jbc.M800641200. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718931443
- 45.Gao Z, Schaller GE. The role of receptor interactions in regulating ethylene signal transduction. Plant Signaling Behavior. 2009;4:1152–3. doi: 10.4161/psb.4.12.9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Malley RC, Rodriguez FI, Esch JJ, Binder BM, O'Donnell P, Klee HJ, Bleecker AB. Ethylene-binding activity, gene expression levels, and receptor system output for ethylene receptor family members from Arabidopsis and tomato. Plant J. 2005;41:651–9. doi: 10.1111/j.1365-313X.2004.02331.x. [DOI] [PubMed] [Google Scholar]
- 47.Tieman DM, Taylor MG, Ciardi JA, Klee HJ. The tomato ethylene receptors NR and LeETR4 are negative regulators of ethylene response and exhibit functional compensation within a multigene family. Proc Natl Acad Sci USA. 2000;97:5663–8. doi: 10.1073/pnas.090550597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kevany BM, Tieman DM, Taylor MG, Cin VD, Klee HJ. Ethylene receptor degradation controls the timing of ripening in tomato fruit. Plant J. 2007;51:458–67. doi: 10.1111/j.1365-313X.2007.03170.x. [DOI] [PubMed] [Google Scholar]
- 49.Kim H, Helmbrecht EE, Stalans MB, Schmitt C, Patel N, Wen CK, Wang W, Binder BM. 2007. Ethylene receptor ETHYLENE RECEPTOR1 domain requirements for ethylene responses in Arabidopsis seedlings. Plant Physiol. 2011;156:417–29. doi: 10.1104/pp.110.170621. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/12078956
- 50.Plett JM, Cvetkovska M, Makenson P, Xing T, Regan S. Arabidopsis ethylene receptors have different roles in Fumonisin B1-induced cell death. Physiol Mol Plant Pathol. 2009a;74:18–26. doi: 10.1016/j.pmpp.2009.08.004. [DOI] [Google Scholar]
- 51.Plett JM, Mathur J, Regan S. Ethylene receptor ETR2 controls trichome branching by regulating microtubule assembly in Arabidopsis thaliana. J Exp Bot. 2009b;60:3923–33. doi: 10.1093/jxb/erp228. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/720758865
- 52.Seifert GJ, Barber C, Wells B, Roberts K. Growth regulators and the control of nucleotide sugar flux. Plant Cell. 2004;16:723–30. doi: 10.1105/tpc.019661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Binder BM, O'Malley RC, Wang W, Zutz TC, Bleecker AB. Ethylene stimulates nutations that are dependent on the ETR1 receptor. Plant Physiol. 2006;142:1690–1700. doi: 10.1104/pp.106.087858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Desikan R, Hancock JT, Bright J, Harrison J, Weir I, Hooley R, Neill SJ. A role for ETR1 in hydrogen peroxide signaling in stomatal guard cells. Plant Physiol. 2005;137:831–4. doi: 10.1104/pp.104.056994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin Z, Ho CW, Grierson D. AtTRP1 encodes a novel TPR protein that interacts with the ethylene receptor ERS1 and modulates development in Arabidopsis. J Exp Bot. 2009b;60:3697–714. doi: 10.1093/jxb/erp209. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/720758896
- 56.Resnick JS, Wen CK, Shockey JA, Chang C. REVERSION-TO-ETHYLENE SENSITIVITY1, a conserved gene that regulates ethylene receptor function in Arabidopsis. Proc Natl Acad Sci USA. 2006;103:7917–22. doi: 10.1073/pnas.0602239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou X, Liu Q, Xie F, Wen CK. RTE1 is a Golgi-associated and ETR1-dependent negative regulator of ethylene responses. Plant Physiol. 2007;145:75–86. doi: 10.1104/pp.107.104299. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1098646
- 58.Dong CH, Rivarola M, Resnick JS, Maggin BD, Chang C. Subcellular co-localization of Arabidopsis RTE1 and ETR1 supports a regulatory role for RTE1 in ETR1 ethylene signaling. Plant J. 2008;53:275–86. doi: 10.1111/j.1365-313X.2007.03339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dong CH, Jang M, Scharein B, Malach A, Rivarola M, Liesch J, Groth G, Hwang I, Chang C. Molecular association of the Arabidopsis ETR1 ethylene receptor and a regulator of ethylene signaling, RTE1. J Biol Chem. 2010;285:40706–13. doi: 10.1074/jbc.M110.146605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rivarola M, McClellan CA, Resnick JS, Chang C. ETR1-specific mutations distinguish ETR1 from other Arabidopsis ethylene receptors as revealed by genetic interaction with RTE1. Plant Physiol. 2009;150:547–51. doi: 10.1104/pp.109.138461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang J, Clay JM, Chang C. Association of cytochrome b5 with ETR1 ethylene receptor signaling through RTE1 in Arabidopsis. Plant J. 2014;77:558–67. doi: 10.1111/tpj.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718313406
- 62.Wilson RL, Kim H, Bakshi A, Binder BM. The ethylene receptors ETHYLENE RESPONSE1 and ETHYLENE RESPONSE2 have contrasting roles in seed germination of Arabidopsis during salt stress. Plant Physiol. 2014;165:1353–66. doi: 10.1104/pp.114.241695. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718384802
- 63.Bisson MM, Bleckmann A, Allekotte S, Groth G. EIN2, the central regulator of ethylene signalling, is localized at the ER membrane where it interacts with the ethylene receptor ETR1. Biochem J. 2009;424:1–6. doi: 10.1042/BJ20091102. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/719241648
- 64.Bisson MM, Groth G. New insight in ethylene signaling: autokinase activity of ETR1 modulates the interaction of receptors and EIN2. Mol Plant. 2010;3:882–9. doi: 10.1093/mp/ssq036. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/720316747
- 65.Urao T, Miyata S, Yamaguchi-Shinozaki K, Shinozaki K. Possible His to Asp phosphorelay signaling in an Arabidopsis two-component system. FEBS Lett. 2000;478:227–32. doi: 10.1016/S0014-5793(00)01860-3. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/719990509
- 66.Scharein B, Voet-van-Vormizeele J, Harter K, Groth G. Ethylene signaling: identification of a putative ETR1-AHP1 phosphorelay complex by fluorescence spectroscopy. Anal Biochem. 2008;377:72–6. doi: 10.1016/j.ab.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 67.Argyros RD, Mathews DE, Chiang YH, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, Schaller GE. Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell. 2008;20:2102–16. doi: 10.1105/tpc.108.059584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schaller GE, Kieber JJ, Shiu SH. Two-component signaling elements and histidyl-aspartyl phosphorelays. The Arabidopsis Book. 2008;6:e0112. doi: 10.1199/tab.0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.To JPC, Kieber JJ. Cytokinin signaling: two-components and more. TIPS. 2008;13:85–92. doi: 10.1016/j.tplants.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 70.Mason MG, Jha D, Salt DE, Tester M, Hill K, Kieber JJ, Schaller GE. Type-B response regulators ARR1 and ARR12 regulate expression of AtHKT1;1 and accumulation of sodium in Arabidopsis shoots. Plant J. 2010;64:753–63. doi: 10.1111/j.1365-313X.2010.04366.x. [DOI] [PubMed] [Google Scholar]
- 71.Cancel JD, Larsen PB. Loss-of-function mutations in the ethylene receptor ETR1 cause enhanced sensitivity and exaggerated response to ethylene in Arabidopsis. Plant Physiol. 2002;129:1557–67. doi: 10.1104/pp.003780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hall BP, Shakeel SN, Amir M, Haq NU, Qu X, Schaller GE. Histidine kinase activity of the ethylene receptor ETR1 facilitates the ethylene response in Arabidopsis. Plant Physiol. 2012;159:682–95. doi: 10.1104/pp.112.196790. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/723350815
- 73.Gao Z, Chen YF, Randlett MD, Zhao XC, Findell JL, Kieber JJ, Schaller GE. Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signaling complexes. J Biol Chem. 2003;278:34725–32. doi: 10.1074/jbc.M305548200. [DOI] [PubMed] [Google Scholar]
- 74.Bleecker AB, Esch JJ, Hall AE, Rodríguez FI, Binder BM. The ethylene-receptor family from Arabidopsis: structure and function. Philos Trans R Soc Lond B Biol Sci. 1998;353:1405–12. doi: 10.1098/rstb.1998.0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guo H, Ecker JR. The ethylene-signaling pathway: new insights. Curr Opin Plant Biol. 2004;7:40–9. doi: 10.1016/j.pbi.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 76.Qiu L, Xie F, Yu J, Wen CK. Arabidopsis RTE1 is essential to ethylene receptor ETR1 N-terminal signaling independent of CTR1. Plant Physiol. 2012;159:1263–76. doi: 10.1104/pp.112.193979. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/717953741
- 77.Hass C, Lohrmann J, Albrecht V, Sweere U, Hummel F, Yoo SD, Hwang I, Zhu T, Schäfer E, Kudla J, Harter K. The response regulator 2 mediates ethylene signalling and hormone integration in Arabidopsis. EMBO J. 2004;23:3290–302. doi: 10.1038/sj.emboj.7600337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bleecker AB. Ethylene signaling: an evolutionary perspective. TIPS. 1999;4:269–274. doi: 10.1016/S1360-1385(99)01427-2. [DOI] [PubMed] [Google Scholar]
- 79.Mount SM, Chang C. Evidence for a plastid origin of plant ethylene receptor genes. Plant Physiol. 2002;130:10–4. doi: 10.1104/pp.005397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hori K, Maruyama F, Fujisawa T, Togashi T, Yamamoto N, Seo M, Sato S, Yamada T, Mori H, Tajima N, Moriyama T, Ikeuchi M, Watanabe M, Wada H, Kobayashi K, Saito M, Masuda T, Sasaki-Sekimoto Y, Mashiguchi K, Awai K, Shimojima M, Masuda S, Iwai M, Nobusawa T, Narise T, Kondo S, Saito H, Sato R, Murakawa M, Ihara Y, et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat Commun. 2014;5:3978. doi: 10.1038/ncomms4978. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718421764
- 81.Ju C, Van de Poel Bram, Cooper ED, Thierer JH, Gibbons TR, Delwiche CF, Chang C. Conservation of ethylene as a plant hormone over 450 million years of evolution. NPLANTS. 2015;1:14004. doi: 10.1038/nplants.2014.4. [DOI] [PubMed] [Google Scholar]
- 82.Gallie DR. Appearance and elaboration of the ethylene receptor family during land plant evolution. Plant molecular biology. 2015;87:521–39. doi: 10.1007/s11103-015-0296-z. [DOI] [PubMed] [Google Scholar]
- 83.Gallie DR. Regulated ethylene insensitivity through the inducible expression of the Arabidopsis etr1-1 mutant ethylene receptor in tomato. Plant Physiol. 2010;152:1928–39. doi: 10.1104/pp.109.151688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wilkinson JQ, Lanahan MB, Yen H-C, Giovannoni JJ, Klee HJ. An ethylene-inducible component of signal transduction encoded by Never-ripe. Science. 1995;270:1807–9. doi: 10.1126/science.270.5243.1807. [DOI] [PubMed] [Google Scholar]
- 85.Binder BM, O'Malley RC, Wang W, Moore JM, Parks BM, Spalding EP, Bleecker AB. Arabidopsis seedling growth response and recovery to ethylene. A kinetic analysis. Plant Physiol. 2004;136:2913–20. doi: 10.1104/pp.104.050369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ma N, Tan H, Liu X, Xue J, Li Y, Gao J. Transcriptional regulation of ethylene receptor and CTR genes involved in ethylene-induced flower opening in cut rose (Rosa hybrida) cv. Samantha. J Exp Bot. 2006;57:2763–73. doi: 10.1093/jxb/erl033. [DOI] [PubMed] [Google Scholar]
- 87.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–50. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 88.Hood HM, Neafsey DE, Galagan J, Sachs MS. Evolutionary roles of upstream open reading frames in mediating gene regulation in fungi. Annu Rev Microbiol. 2009;63:385–409. doi: 10.1146/annurev.micro.62.081307.162835. [DOI] [PubMed] [Google Scholar]
- 89.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–70. [PubMed] [Google Scholar]
- 90.Chen JF, Gallie DR. Analysis of the functional conservation of ethylene receptors between maize and Arabidopsis. Plant Mol Biol. 2010;74:405–21. doi: 10.1007/s11103-010-9686-4. [DOI] [PMC free article] [PubMed] [Google Scholar]