Abstract
Background
We previously demonstrated that patients with metastatic KRAS mutant lung cancers have a shorter survival compared to patients with KRAS wild type cancers. Recent reports have suggested different clinical outcomes and distinct activated signaling pathways depending on KRAS mutation subtype. To better understand the impact of KRAS mutation subtype, we analyzed data from 677 patients with KRAS mutant metastatic lung cancer.
Methods
We reviewed all patients with metastatic or recurrent lung cancers found to have KRAS mutations over a 6 year time period. We evaluated the associations between KRAS mutation type, clinical factors, and overall survival in univariate and multivariate analyses. Any significant findings were validated in an external multi-institution patient data set.
Results
Among 677 patients with KRAS mutant lung cancers (53 at codon 13, 624 at codon 12), there was no difference in overall survival for patients when comparing KRAS transition versus transversion mutations (p=0.99), smoking status (p=0.33) or when comparing specific amino acid substitutions (p=0.20). In our data set, patients with KRAS codon 13 mutant tumors (n=53) had shorter overall survival compared to patients with codon 12 mutant tumors (n=624)( 1.1 vs 1.3 years, respectively, p=0.009), and the findings were confirmed in a multivariate Cox model controlling for age, sex and smoking status (HR 1.52 95% CI 1.11-2.08, p=0.008). In an independent validation set of tumors from 682 patients with stage IV KRAS mutant lung cancers, there was no difference in survival between patients with KRAS codon 13 versus codon 12 mutations (1.0 vs 1.1 years respectively, p=0.41).
Conclusions
Among individuals with KRAS mutant metastatic lung cancers treated with conventional therapy, there are apparent differences in outcome based on KRAS mutation subtype
Introduction
RAS mutations are identified in 25-30% of lung adenocarcinomas; the vast majority are KRAS mutations occurring at codon 12 or 13. In small cohorts, specific point mutations such as G12V and G12R and KRAS codon 12 have been associated with trends towards poorer outcomes.1-3 It is difficult to make definitive conclusions about KRAS mutations as a prognostic marker as published studies use differing molecular diagnostic techniques, comparison populations and endpoints.1-9 Moreover, the discovery of other oncogenes such as EGFR and ALK with potent targeted therapy available10,11, must be taken into account when evaluating the prognostic significance of any biomarker as these discoveries highlight the heterogeneity of the KRAS wild type designation.
Studies report superior survival for never-smokers compared to current or former smokers12, which may be due to a differing distribution of oncogenic drivers with KRAS more frequent in smokers and EGFR and ALK alterations more common in never smokers.13 There are no differences in survival between current/former smokers and never smokers when their tumors harbor the same driver oncogene.13 Transversion mutations refer to the substitution of a purine nucleotide to a pyrimidine, or vice versa. Transitions refer to a purine to purine or pyrimidine to pyrimidine nucleotide change. Within KRAS, transversion mutations are more common in current or former smokers, and never-smokers have a higher frequency of transition mutations.14,15 Data with regard to outcomes for KRAS transition versus transversion mutations are conflicting.2,16
In colorectal cancer, the specific KRAS point mutation present may be a prognostic and predictive marker with G12V conveying an increased risk of disease recurrence and death.17-19 Patients with KRAS codon 12 mutant colorectal tumors had shorter overall survival compared to patients with KRAS codon 13 mutant tumors.19 The biologic basis to these findings is not fully understood but may be related to differences in downstream signaling or protein expression.20,21 In colorectal cancer, KRAS mutation status has been validated as a predictive marker of response to EGFR targeted therapies with the presence of a KRAS mutation predicting a lack of response to cetuximab or panitumumab22, and newer data raise the possibility that specific KRAS point mutations may induce differential responses to EGFR directed therapies.23
In lung cancers, the predictive utility of KRAS mutations as a marker of response to both targeted therapy and standard cytotoxic chemotherapy has been of great interest (Table 1). The presence of a KRAS mutation suggests a lack of response to EGFR tyrosine kinase inhibitors24,25, but has not been helpful in selecting patients for treatment with EGFR monoclonal antibodies.26,27 In the BATTLE trial, patients with KRAS G12C and G12V mutant lung cancers were found to have a shorter progression free survival than other KRAS genotypes with certain targeted therapies such as erlotinib, vandetanib, bexarotene and sorafenib.28 Recent data suggest that the specific KRAS mutation present may predict response to adjuvant chemotherapy for patients with resected NSCLC. Patients harboring KRAS codon 13 mutations appear to have poorer outcomes with adjuvant cisplatin-based chemotherapy.29 In the metastatic setting, KRAS mutations do not appear to independently predict response or resistance to chemotherapy treatments.30-33
Table 1.
KRAS as a predictive and prognostic marker
| KRAS as a Predictive Marker | ||||
|---|---|---|---|---|
| Author | Trial Design | Treatment | % KRAS mutant | Findings |
| Rodenhuis30 | Single arm phase II of patients with metastatic disease | Carboplatin, Etoposide, Mesna, Ifosfamide | 26% (n=16) | No difference Similar OS and PFS among KRAS and KRAS wild-type |
| Schiller5 | E4592, Completely resected pts randomized post-operative RT alone or with chemo | RT +/− chemo | 24% (n=44) | No difference Similar OS and PFS among KRAS and KRAS wild-type |
| Eberhard44 | TRIBUTE, Untreated patients with metastatic disease randomized to chemo alone or with erlotinib | Carboplatin, paclitaxel +/− erlotinib | 21% (n=55) | No difference Similar OS, TTP and ORR among KRAS and KRAS wild-type |
| Tsao4 | JBR.10, Completely resected pts randomized to observation or chemo | Cisplatin, vinorelbine | 26% (n=117) | No apparent benefit with adjuvant chemo in RAS mt (HR=1.02), but chemo, RAS interaction test non significant. |
| Massarelli45 | Metastatic pts treated with EGFR TKI | Gefitinib or erlotinib | 16% (n=23) | Non significant association with poor response in RAS mt (p=0.06) |
| Shepherd29 | ANITE, JBR.10, IALT, CALGB-9633 | Platinum based chemotherapy | 19% (n=300) | No difference in OS or DFS when comparing RAS mt and wild-type, possible worse OS with chemo in KRAS codon 13 mt pateints |
| Zhu46 | BR.21, Metastatic pts randomized to erlotinib or placebo | Erlotinib | 15% (n=30) | Lack of response in RAS mt, HR 1.67 for RAS mt treated with erlotinib compared to HR 0.69 for RAS wild-type |
| Douillard47 | INTEREST, Metastatic pts randomized to gefitinib or docetaxel | Gefitinib or docetaxel | 18% (n=49) | No difference between treatments in RAS mt |
| KRAS as a prognostic marker | ||||
|---|---|---|---|---|
| Author | Endpoint | Comparison | Number of pts | Findings |
| Slebos1 | Overall survival, disease-free survival | KRAS wild-type early stage adenocarcinomas | 28% (n=19) | Shorter survival (p=0.002), DFS (p=0.038) in KRAS mt |
| Keohavong2 | Overall survival | KRAS wild-type early stage adenocarcinomas | 32% (n=41) | No difference, but with KRAS G12C/V having a trend (p=0.07) towards poorer prognosis compared to wild-type or other KRAS mts |
| Villaruz3 | Overall survival, recurrence free survival | KRAS wild-type predominantly early stage (79%) adenocarcinomas | 32% (n=318) | No difference by KRAS point mutation in overall survival (p=0.612) or RFS (p=0.089), trend towards better OS for KRAS codon 13 mts (p=0.052) |
| Tsao4 | Overall survival | KRAS wild-type early stage NSCLC | 26% (n=117) | No difference in overall survival (p=0.40) |
| Schiller5 | Overall survival, progression free survival | KRAS wild-type early stage NSCLC | 24% (n=44) | No difference in overall survival (p=0.38)or progression free survival |
| Graziano6 | Overall survival | KRAS wild-type early stage NSCLC | 16% (n=35) | No difference in overall survival (p=0.33) |
| Johnson7 | Overall survival | KRAS, EGFR wild-type advanced adenocacinomas | 23% (n=241) | KRAS mutant had shorter survival compared to KRAS/EGFR wild-type (p=0.048) |
| Mascaux8 | Overall survival, meta-analysis | Not consistent over studies | 28 total studies | KRAS mutants with shorter survival (HR 1.35 95% CI 1.16-1.56) |
We hypothesized that with a large number of patients with KRAS mutant advanced lung cancers, we would have sufficient power to identify any clinically meaningful differences in outcome related to specific KRAS mutation subtypes. To evaluate this hypothesis, we reviewed specific KRAS point mutation status, clinical characteristics and survival of patients with metastatic KRAS mutant lung cancers identified at our institution and then evaluated key findings in an independent group of patients from other institutions.
Methods
Patients
Consecutive patients with metastatic or recurrent lung cancers found to have a KRAS-mutation by routine molecular testing performed between January 2005 and January 2011 were included in this analysis. An electronic medical record search was used to identify individuals seen at Memorial Sloan-Kettering with a primary tumor diagnosis of lung cancer by ICD-0 code with available diagnostic molecular pathology reports that indicated the presence of a KRAS mutation. The list was then manually reviewed to exclude patients who did not have metastatic or recurrent disease or a tumor diagnosis of a primary lung cancer. Data collection was approved by the MSKCC Institutional Review Board/Privacy Board. We collected clinical characteristics and treatment course for all patients. Overall survival was defined as the time from date of advanced disease (stage IV or recurrent) until date of death or last follow up. KRAS mutation analysis was performed prior to this retrospective review on available tissue by standard Sanger sequencing or by a mass spectrometry based mutation profiling assay.34,35
Information on mutation status and outcomes for patients with stage IV KRAS-mutant lung cancers identified by routine molecular sequencing during the same time period were collected from Dana Farber Cancer Institute, Massachusetts General Hospital Cancer Center and Vanderbilt Ingram Cancer Center. This data set was compiled with the intent to verify any significant findings in our institutional dataset.
Statistical methods
We compared characteristics of patients with KRAS codon 12 and codon 13 mutant tumors using t test (for continuous variables) and chi-square test (for categorical variables). Overall survival following diagnosis of stage IV lung cancer was estimated using Kaplan-Meier methodology. Patients were followed until death; patients alive at the end of the study were censored at the time of the last available follow-up. Univariate group comparisons were performed using log-rank tests. A multivariate Cox proportional hazards model was used to assess the independent effect of KRAS mutation type, controlling for potential confounding factors associated with overall survival in univariate analysis. All associations found significant were validated using the external validation set.
RESULTS
Clinical characteristics
We evaluated tumor specimens from 3,357 unique patients with lung cancers for KRAS mutations. During that period, 677 patients were identified to have metastatic lung cancer that harbored a KRAS mutation. Patient demographics are noted in Table 2. The majority (59%) had chemotherapy treatment details available. Of those with available data, there was no difference in the frequency of platinum-based chemotherapy, pemetrexed and/or bevacizumab among patients with KRAS codon 12 versus codon 13 mutations. The median lines of treatment in those who received chemotherapy at MSKCC was 2 (range 1-9).
Table 2.
Patient Characteristics in original KRAS dataset
| Total | KRAS Codon 12 | KRAS Codon 13 | |
|---|---|---|---|
| Characteristic | N=677 | N=624 | N=53 |
| Age at Diagnosis (Stage IV) | |||
| Median | 66 | 66 | 64 |
| Range | 31-89 | 31-89 | 44-87 |
| Sex-(%) | |||
| Men | 260 (38) | 240 (38) | 20 (38) |
| Women | 417 (62) | 384 (62) | 33 (62) |
| Smoking history- (%) | |||
| Never-smoker | 48 (7) | 46 (7) | 2 (4) |
| Former/current smoker | 625 (92) | 576 (92) | 49 (93) |
| Median pack year | 38 | 35 | 40 |
| Range | 1-245 | 1-245 | 7-120 |
| Unknown | 4 | 2 | 2 |
KRAS mutation subtype
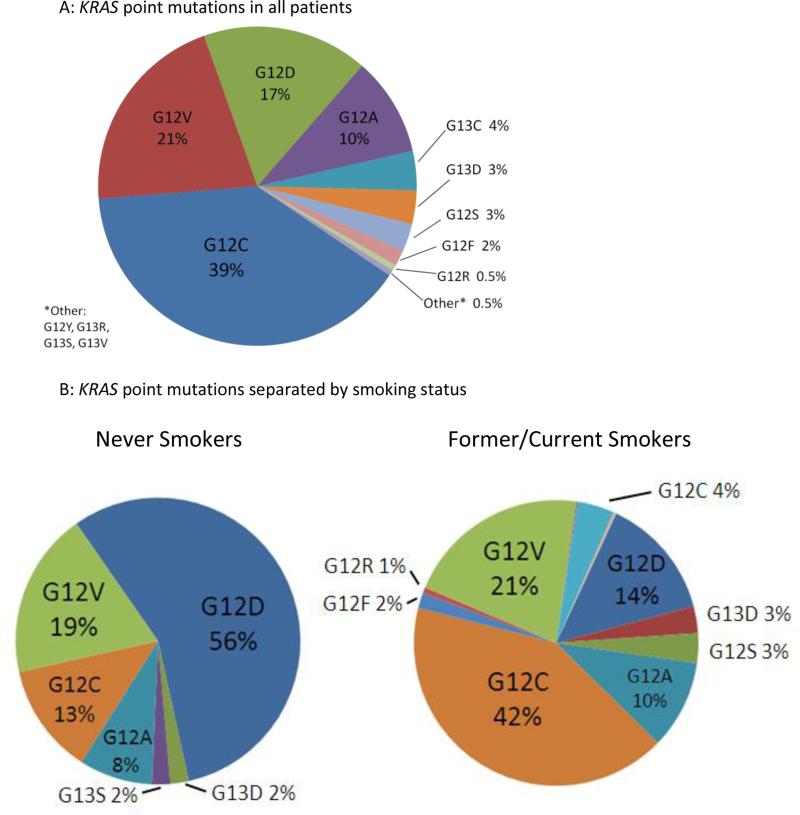
The most frequent nucleotide change in tumor specimens was a guanine to thymidine (G>T) seen in 366 patients. The prevalence of specific KRAS point mutations is summarized in Figure 1. Mutations were found at codon 12 in 624 (92%) patients, and codon 13 in 53 (8%) patients. Twenty-three percent (157/677) of patients had transition mutations (G12D, G12S, G13D, G13S). The prevalence of specific point mutations differed between former/current smokers and never-smokers (Figure 1). Patients with transition mutations were more likely to be never-smokers, compared to patients with transversion mutations (p<0.001). Clinical characteristics were similar between patients with KRAS codon 12 versus 13 mutations (Table 2). No concurrent EGFR mutations or ALK rearrangements were found in any patients.
Figure 1.
KRAS point mutations in the original KRAS dataset
KRAS mutation and survival
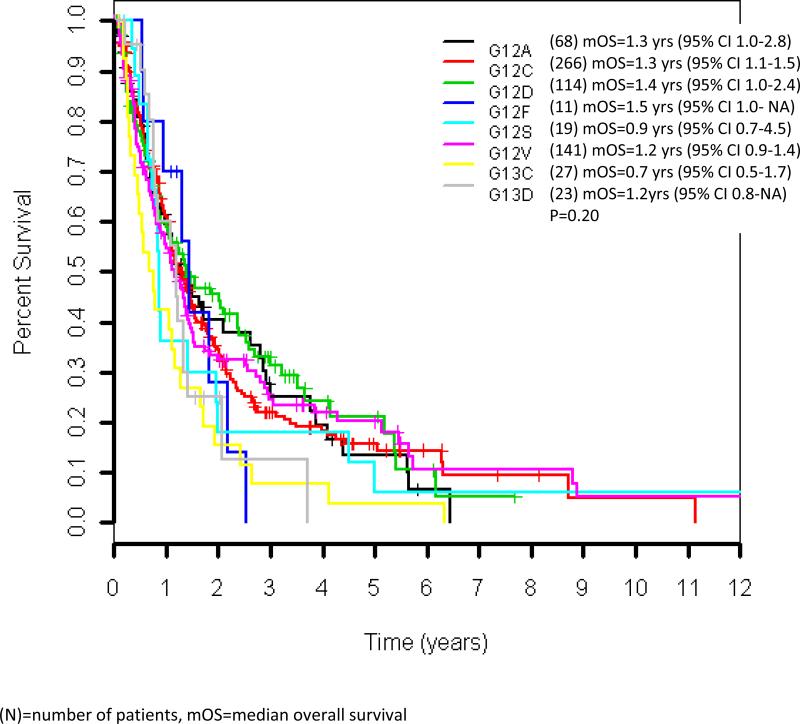
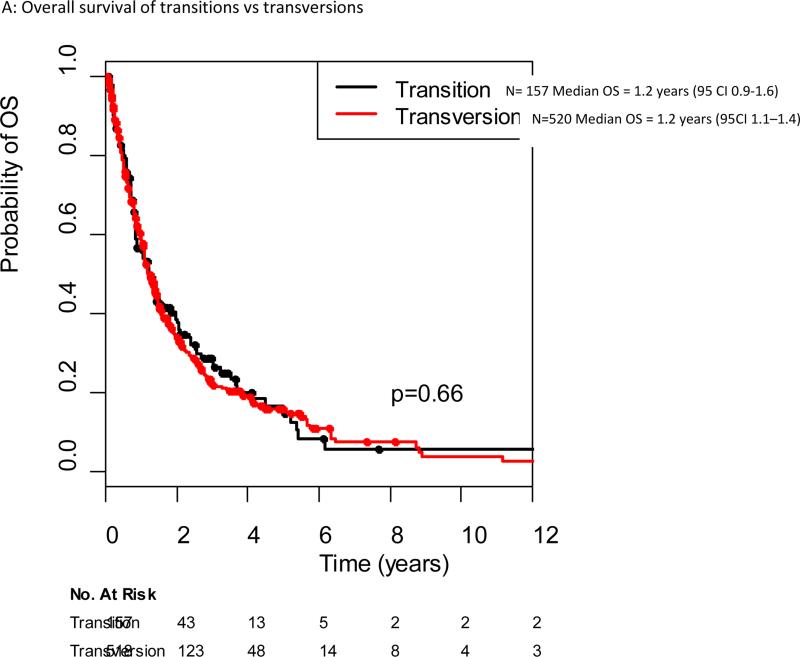
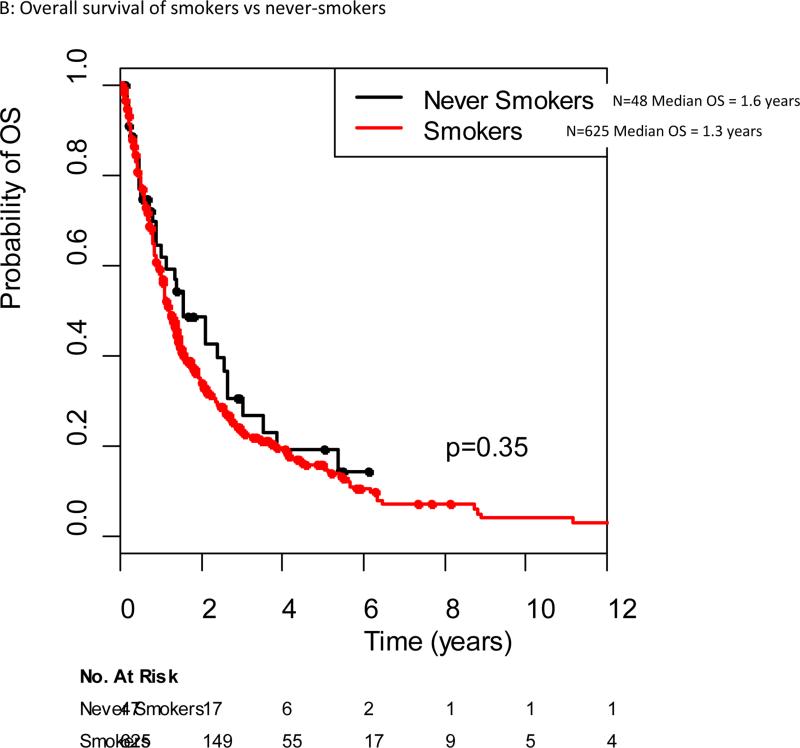
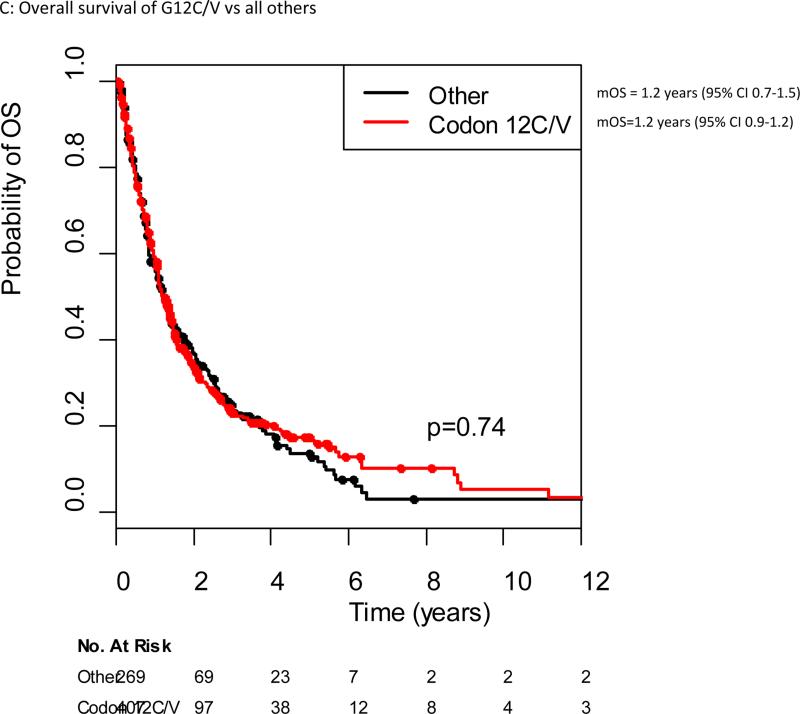
The median follow-up among the 197 patients alive at the data cut off of June 2012 was 17 months (range 1-207 months). The median overall survival for all patients with KRAS mutant advanced lung cancers in our cohort was 1.2 years (95% CI 1.2-1.4 years). Median overall survival for specific KRAS point mutations ranged from 0.7 years (G13C) to 1.5 years (G12F), although no significant difference in survival was seen when comparing different KRAS point mutations (Figure 2). There was no difference in outcome for patients with KRAS transition versus transversion mutations, with a median survival of 1.2 years for both (p=0.66) (Figure 3A). No difference in overall survival was seen in patients with KRAS mutant lung cancers who are current or former smokers (n=625) compared to never-smokers (n=48), median overall survival 1.2 years (95% CI 1.1-1.4 years) and 1.6 years (95% CI 0.9-2.6), respectively (p=0.34)(Figure 3B). The median overall survival of patients with G12C/G12V mutant tumors was not different from patients with all other point mutations, 1.2 years for both (p=0.74)(Figure 3C).
Figure 2.
Overall survival by different KRAS point mutations in the original KRAS dataset
Figure 3.
Overall survival from diagnosis of stage IV cancer in the original KRAS dataset
Patients with KRAS codon 13 mutant tumors (n=53) had inferior survival compared to patients with codon 12 mutant tumors (n=624), median 1.1 years (95% CI 0.8-1.3) and 1.3 years (95% 1.1-2.4), respectively (p=0.008)(Figure 3D). When comparing patients with G12D versus G13D mutant tumors, there was no difference in overall survival, 1.4 years and 1.2 years respectively (p=0.16). Among patients with KRAS transition mutations, there is no difference in overall survival when comparing smokers versus never smokers, 1.2 years and 1.4 years (p=0.95). Among patients with KRAS G12D mutations, there is no difference in outcome when comparing current/former smokers to never smokers (p=0.66).
Multivariate Analysis
We evaluated sex, age, smoking history, and KRAS codon to determine impact on survival (Table 3). Sex, age and KRAS codon were associated with survival in univariate analysis. Men had increased risk of death, compared to women (HR 1.28 95% CI 1.07-1.55 p=0.008). Older age was significantly associated with increased risk of death, with each 5 additional years at diagnosis increasing the risk of death by 5% (HR 1.05 for each 5 added yrs, 95% CI 1.01-1.10 p=0.026). Smoking history did not affect outcome among those with KRAS mutations (HR = 1.2, 95% CI 0.83 – 1.72, p=0.34). KRAS codon was associated with overall survival, with KRAS codon 13 mutant tumors having an increased risk of death, compared to patients with KRAS codon 12 (HR 1.50 95% CI 1.11-2.04 p=0.009). In a multivariate analysis controlling for age, sex and smoking history, KRAS codon 13 was associated with shorter overall survival (HR 1.52 95% CI 1.11-2.08 p=0.008)(Table 3).
Table 3.
Univariate and multivariate analysis of original KRAS dataset
| Table 3A: Univariate analysis | |||
|---|---|---|---|
| Variable | Survival (yrs) | HR | P value |
| KRAS (G13 vs G12) | 1.1 vs 1.3 | 1.50 (1.11-2.04 | 0.009 |
| Sex (M vs F) | 1.0 vs 1.4 | 1.28 (1.07-1.54) | 0.008 |
| Age | 1.05* | 0.026 | |
| Smoking (Y vs N) | 1.2 vs 1.6 | 1.20 (0.83-1.72) | 0.34 |
| Table 3B: Multivariate analysis | ||
|---|---|---|
| Variable | HR | P value |
| KRAS (G13 vs G12) | 1.52 (95% CI 1.11-2.08) | 0.008 |
| Sex (M vs F) | 1.29 (95% CI 1.07-1.55) | 0.007 |
| Age | 1.01 (95% CI 1.001-1.02) | 0.025 |
| Smoking (Y vs N) | 1.18 (95% CI 0.82-1.69) | 0.39 |
External validation set
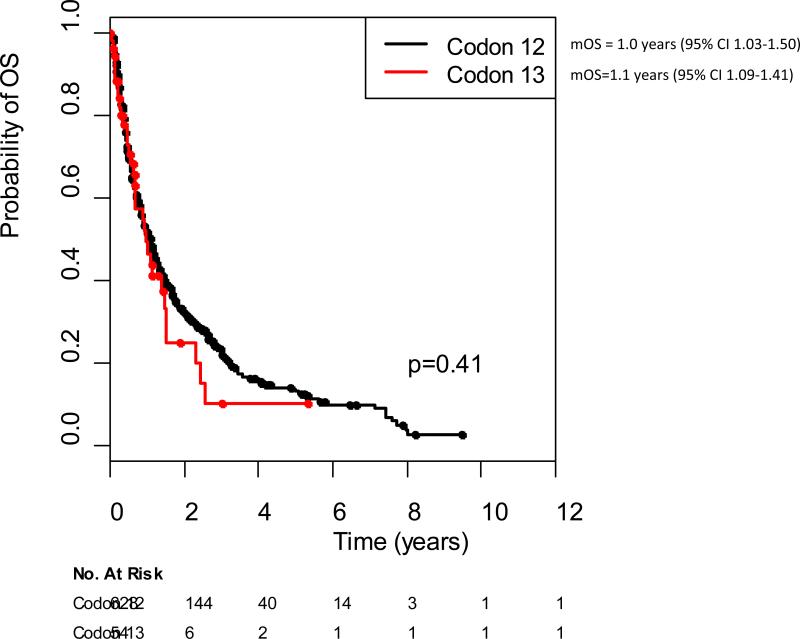
To verify the survival difference in patients with KRAS codon 13 versus codon 12 mutant lung cancer, we used an external validation set consisting of patients with KRAS mutant lung cancers treated at other institutions. In total, 682 patients were analyzed: 354 from Dana Farber Cancer Institute, 242 patients from Massachusetts General Hospital and 86 from Vanderbilt Ingram Cancer Center. In this collected set, there was no difference in overall survival from time of advanced disease in patients with KRAS codon 13 versus codon 12 mutant metastatic lung cancers, 1 year (95% CI 0.7-1.5 yrs) vs 1.1 (95% CI 0.9-1.2 yrs), respectively. Median follow-up was similar in our dataset and the validation dataset. Median overall survival was longer in our dataset compared to the validation set, 15 vs 13 months, respectively (p=0.05).
Discussion
In this series of patients, the largest reported series of patients with KRAS mutant lung cancers, we validated clinical prognostic factors known to be relevant in advanced lung cancers, but did not observe any difference in outcomes based on individual KRAS genotype (specific point mutation, transition vs transversion, G12 vs 13). While others have noted that the specific KRAS point mutations present is associated with outcomes in smaller series2, such differences were not observed in our group of patients. The importance of independent validation of observed prognostic differences is underscored by our identification of a difference in outcomes for KRAS G13 vs G12, although this finding was not confirmed in a cohort of similar patients seen at three other cancer centers.
Similar to our findings, Shepherd and colleagues found that KRAS mutation was not prognostic in patients with early stage lung cancer.29 Other factors besides mutation subtype might be influencing survival. The presence of concurrent mutations may also influence the clinical phenotype seen with different KRAS mutation subtypes. LKB1 (STK11) and p53 mutations are seen concurrently with KRAS mutations and portend a poorer prognosis in patients, although the prevalence of concurrent mutations has only been assessed in small series.36-40 Preclinical data indicate that loss of LKB1 leads to a more aggressive tumor phenotype, with short tumor latency and greater rate of metastasis.37 The presence of concurrent mutations may also have treatment implications as preclinical data suggest concurrent LKB1 inactivation alters downstream signaling and sensitivity to mTOR and MEK inhibition.41-43 More clearly characterizing the frequency of concurrent mutations and understanding their correlation with clinical behavior will be helpful in further defining the prognosis of patients with KRAS mutant lung cancers.
There are limitations to our analysis. All patients were identified based upon molecular testing at a single institution during a time (beginning in 2005) in which molecular analysis was not broadly performed, and therefore these patients may not be representative of a general population. They received diverse treatment and the vast majority did not receive any KRAS directed targeted therapy. Patients with different KRAS mutation subtypes may respond differently to chemotherapy, and our analysis was not powered to identify any predictive effects of KRAS mutation subtype. Performance status, a powerful prognostic factor, was not included in our analysis. In addition, even with a larger dataset, we are limited in our ability to make conclusions regarding KRAS mutation subsets that are rare. Due to these limitations, we attempted to validate our findings in an external dataset, in which we did not find a difference in survival when comparing patients with KRAS G12 vs G13 mutant tumors. Further validation would be needed to draw definitive conclusions regarding the prognostic value of KRAS mutation subtype.
KRAS mutation subtype does not appear to be associated with overall survival from the diagnosis of lung cancer. Investigation into other areas such as variable gene expression and identifying concurrent mutations may identify potential molecular prognostic markers in patients with KRAS mutant lung cancers.
Figure 4.
Overall survival of codon 12 vs codon 13 in external validation dataset
Table 4.
Overall survival by KRAS subgroup in original KRAS dataset
| Table 4: Overall Survival by KRAS subgroup | ||
|---|---|---|
| Subset | Median survival (yrs) | P value |
| Specific point mutations | 0.5 to 1.5 | |
| Transition vs transversion | 1.2 vs 1.2 | 0.66 |
| Smoker vs never-smoker | 1.2 vs 1.6 | 0.34 |
| G12C/V vs other | 1.2 vs 1.2 | 0.74 |
| KRAS (G13 vs G12) | 1.1 vs 1.3 | 0.009 |
| G12D vs G13D | 1.4 vs 1.2 | 0.16 |
| Transition: smoker vs never-smoker | 1.2 vs 1.4 | 0.95 |
| KRAS G12D: smoker vs never smoker | 0.66 | |
Acknowledgments
Funding: This study was supported by NIH funding, P01CA129243 (Mark Kris). The sponsor played no role in data collection, analysis or interpretation.
Works Cited
- 1.Slebos RJ, Kibbelaar RE, Dalesio O, et al. K-ras oncogene activation as a prognostic marker in adenocarcinoma of the lung. The New England journal of medicine. 1990;323:561–5. doi: 10.1056/NEJM199008303230902. [DOI] [PubMed] [Google Scholar]
- 2.Keohavong P, DeMichele MA, Melacrinos AC, Landreneau RJ, Weyant RJ, Siegfried JM. Detection of K-ras mutations in lung carcinomas: relationship to prognosis. Clinical cancer research. 1996;2:411–8. [PubMed] [Google Scholar]
- 3.Villaruz LC, Socinski MA, Cunningham DE, et al. The prognostic and predictive value of KRAS oncogene substitutions in lung adenocarcinoma. Cancer. 2013;119(12):2268–74. doi: 10.1002/cncr.28039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsao MS, Aviel-Ronen S, Ding K, et al. Prognostic and predictive importance of p53 and RAS for adjuvant chemotherapy in non small-cell lung cancer. Journal of clinical oncology. 2007;25:5240–7. doi: 10.1200/JCO.2007.12.6953. [DOI] [PubMed] [Google Scholar]
- 5.Schiller JH, Adak S, Feins RH, et al. Lack of prognostic significance of p53 and K-ras mutations in primary resected non-small-cell lung cancer on E4592: a Laboratory Ancillary Study on an Eastern Cooperative Oncology Group Prospective Randomized Trial of Postoperative Adjuvant Therapy. Journal of clinical oncology. 2001;19:448–57. doi: 10.1200/JCO.2001.19.2.448. [DOI] [PubMed] [Google Scholar]
- 6.Graziano SL, Gamble GP, Newman NB, et al. Prognostic significance of K-ras codon 12 mutations in patients with resected stage I and II non-small-cell lung cancer. J Clin Oncol. 1999;17:668–75. doi: 10.1200/JCO.1999.17.2.668. [DOI] [PubMed] [Google Scholar]
- 7.Johnson ML, Sima CS, Chaft J, et al. Association of KRAS and EGFR mutations with survival in patients with advanced lung adenocarcinomas. Cancer. 2013;119(2):356–62. doi: 10.1002/cncr.27730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mascaux C, Iannino N, Martin B, et al. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. British journal of cancer. 2005;92:131–9. doi: 10.1038/sj.bjc.6602258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitsudomi T, Steinberg SM, Oie HK, et al. ras gene mutations in non-small cell lung cancers are associated with shortened survival irrespective of treatment intent. Cancer research. 1991;51:4999–5002. [PubMed] [Google Scholar]
- 10.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mok TS, Wu Y-L, Thongprasert S, et al. Gefitinib or Carboplatin-Paclitaxel in Pulmonary Adenocarcinoma. N Engl J Med. 2009;361:947–57. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 12.Janjigian YY, McDonnell K, Kris MG, et al. Pack-years of cigarette smoking as a prognostic factor in patients with stage IIIB/IV nonsmall cell lung cancer. Cancer. 2010;116:670–5. doi: 10.1002/cncr.24813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paik PK, Johnson ML, D'Angelo SP, et al. Driver mutations determine survival in smokers and never-smokers with stage IIIB/IV lung adenocarcinomas. Cancer. 2012;118:5840–7. doi: 10.1002/cncr.27637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riely GJ, Kris MG, Rosenbaum D, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clinical cancer research. 2008;14:5731–4. doi: 10.1158/1078-0432.CCR-08-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dogan S, Shen R, Ang DC, et al. Molecular Epidemiology of EGFR and KRAS Mutations in 3,026 Lung Adenocarcinomas: Higher Susceptibility of Women to Smoking-Related KRAS-Mutant Cancers. Clin Cancer Res. 2012;18(22):6169–77. doi: 10.1158/1078-0432.CCR-11-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackman DM, Miller VA, Cioffredi LA, et al. Impact of epidermal growth factor receptor and KRAS mutations on clinical outcomes in previously untreated non-small cell lung cancer patients: results of an online tumor registry of clinical trials. Clin Cancer Res. 2009;15:5267–73. doi: 10.1158/1078-0432.CCR-09-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andreyev HJ, Norman AR, Cunningham D, et al. Kirsten ras mutations in patients with colorectal cancer: the 'RASCAL II' study. British journal of cancer. 2001;85:692–6. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winder T, Mundlein A, Rhomberg S, et al. Different types of K-Ras mutations are conversely associated with overall survival in patients with colorectal cancer. Oncology reports. 2009;21:1283–7. doi: 10.3892/or_00000352. [DOI] [PubMed] [Google Scholar]
- 19.Imamura Y, Morikawa T, Liao X, et al. Specific mutations in KRAS codons 12 and 13, and patient prognosis in 1075 BRAF wild-type colorectal cancers. Clinical cancer research. 2012;18:4753–63. doi: 10.1158/1078-0432.CCR-11-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Mulla F, MacKenzie EM. Differences in in vitro invasive capacity induced by differences in Ki- Ras protein mutations. The Journal of pathology. 2001;195:549–56. doi: 10.1002/path.995. [DOI] [PubMed] [Google Scholar]
- 21.Guerrero S, Casanova I, Farre L, Mazo A, Capella G, Mangues R. K-ras codon 12 mutation induces higher level of resistance to apoptosis and predisposition to anchorage-independent growth than codon 13 mutation or proto-oncogene overexpression. Cancer research. 2000;60:6750–6. [PubMed] [Google Scholar]
- 22.Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. The New England journal of medicine. 2008;359:1757–65. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 23.De Roock W, Jonker DJ, Di Nicolantonio F, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. Jama. 2010;304:1812–20. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 24.Mao C, Qiu LX, Liao RY, et al. KRAS mutations and resistance to EGFR-TKIs treatment in patients with non-small cell lung cancer: a meta-analysis of 22 studies. Lung Cancer. 2010;69:272–8. doi: 10.1016/j.lungcan.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 25.Linardou H, Dahabreh IJ, Kanaloupiti D, et al. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. The lancet oncology. 2008;9:962–72. doi: 10.1016/S1470-2045(08)70206-7. [DOI] [PubMed] [Google Scholar]
- 26.Pirker R, Pereira JR, Szczesna A, et al. Cetuximab plus chemotherapy in patients with advanced non-small-cell lung cancer (FLEX): an open-label randomised phase III trial. Lancet. 2009;373:1525–31. doi: 10.1016/S0140-6736(09)60569-9. [DOI] [PubMed] [Google Scholar]
- 27.Khambata-Ford S, Harbison CT, Hart LL, et al. Analysis of potential predictive markers of cetuximab benefit in BMS099, a phase III study of cetuximab and first-line taxane/carboplatin in advanced non-small-cell lung cancer. Journal of clinical oncology. 2010;28:918–27. doi: 10.1200/JCO.2009.25.2890. [DOI] [PubMed] [Google Scholar]
- 28.Ihle NT, Byers LA, Kim ES, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. Journal of the National Cancer Institute. 2012;104:228–39. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shepherd FA, Domerg C, Hainaut P, et al. Pooled Analysis of the Prognostic and Predictive Effects of KRAS Mutation Status and KRAS Mutation Subtype in Early-Stage Resected Non-Small-Cell Lung Cancer in Four Trials of Adjuvant Chemotherapy. Journal of clinical oncology. 2013;31(17):2173–81. doi: 10.1200/JCO.2012.48.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodenhuis S, Boerrigter L, Top B, et al. Mutational activation of the K-ras oncogene and the effect of chemotherapy in advanced adenocarcinoma of the lung: a prospective study. Journal of clinical oncology. 1997;15:285–91. doi: 10.1200/JCO.1997.15.1.285. [DOI] [PubMed] [Google Scholar]
- 31.Guan JL, Zhong WZ, An SJ, et al. KRAS Mutation in Patients with Lung Cancer: A Predictor for Poor Prognosis but Not for EGFR-TKIs or Chemotherapy. Ann Surg Oncol. 2013;20(4):1381–8. doi: 10.1245/s10434-012-2754-z. [DOI] [PubMed] [Google Scholar]
- 32.Camps C, Jantus-Lewintre E, Cabrera A, et al. The identification of KRAS mutations at codon 12 in plasma DNA is not a prognostic factor in advanced non-small cell lung cancer patients. Lung Cancer. 2011;72:365–9. doi: 10.1016/j.lungcan.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Kalikaki A, Koutsopoulos A, Hatzidaki D, et al. Clinical outcome of patients with non-small cell lung cancer receiving front-line chemotherapy according to EGFR and K-RAS mutation status. Lung Cancer. 2010;69:110–5. doi: 10.1016/j.lungcan.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 34.Pao W, Wang TY, Riely GJ, et al. KRAS Mutations and Primary Resistance of Lung Adenocarcinomas to Gefitinib or Erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brevet M, Johnson ML, Azzoli CG, Ladanyi M. Detection of EGFR mutations in plasma DNA from lung cancer patients by mass spectrometry genotyping is predictive of tumor EGFR status and response to EGFR inhibitors. Lung Cancer. 2011;73:96–102. doi: 10.1016/j.lungcan.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumoto S, Iwakawa R, Takahashi K, et al. Prevalence and specificity of LKB1 genetic alterations in lung cancers. Oncogene. 2007;26:5911–8. doi: 10.1038/sj.onc.1210418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji H, Ramsey MR, Hayes DN, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–10. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 38.Huang CL, Taki T, Adachi M, et al. Mutations of p53 and K-ras genes as prognostic factors for non-small cell lung cancer. Int J Oncol. 1998;12:553–63. doi: 10.3892/ijo.12.3.553. [DOI] [PubMed] [Google Scholar]
- 39.Fukuyama Y, Mitsudomi T, Sugio K, Ishida T, Akazawa K, Sugimachi K. K-ras and p53 mutations are an independent unfavourable prognostic indicator in patients with non-small-cell lung cancer. Br J Cancer. 1997;75:1125–30. doi: 10.1038/bjc.1997.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koivunen JP, Kim J, Lee J, et al. Mutations in the LKB1 tumour suppressor are frequently detected in tumours from Caucasian but not Asian lung cancer patients. British journal of cancer. 2008;99:245–52. doi: 10.1038/sj.bjc.6604469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Z, Cheng K, Walton Z, et al. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature. 2012;483:613–7. doi: 10.1038/nature10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carretero J, Shimamura T, Rikova K, et al. Integrative genomic and proteomic analyses identify targets for Lkb1-deficient metastatic lung tumors. Cancer cell. 2010;17:547–59. doi: 10.1016/j.ccr.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahoney CL, Choudhury B, Davies H, et al. LKB1/KRAS mutant lung cancers constitute a genetic subset of NSCLC with increased sensitivity to MAPK and mTOR signalling inhibition. British journal of cancer. 2009;100:370–5. doi: 10.1038/sj.bjc.6604886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. Journal of clinical oncology. 2005;23:5900–9. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 45.Massarelli E, Varella-Garcia M, Tang X, et al. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clinical cancer research. 2007;13:2890–6. doi: 10.1158/1078-0432.CCR-06-3043. [DOI] [PubMed] [Google Scholar]
- 46.Zhu CQ, da Cunha Santos G, Ding K, et al. Role of KRAS and EGFR as biomarkers of response to erlotinib in National Cancer Institute of Canada Clinical Trials Group Study BR.21. Journal of clinical oncology. 2008;26:4268–75. doi: 10.1200/JCO.2007.14.8924. [DOI] [PubMed] [Google Scholar]
- 47.Douillard JY, Shepherd FA, Hirsh V, et al. Molecular predictors of outcome with gefitinib and docetaxel in previously treated non-small-cell lung cancer: data from the randomized phase III INTEREST trial. Journal of clinical oncology. 2010;28:744–52. doi: 10.1200/JCO.2009.24.3030. [DOI] [PubMed] [Google Scholar]