Summary
Background
Genetic disorders and congenital anomalies are the leading causes of infant mortality. Diagnosis of most genetic diseases in neonatal and paediatric intensive care units (NICU and PICU) is not sufficiently timely to guide acute clinical management. We used rapid whole-genome sequencing (STATseq) in a level 4 NICU and PICU to assess the rate and types of molecular diagnoses, and the prevalence, types, and effect of diagnoses that are likely to change medical management in critically ill infants.
Methods
We did a retrospective comparison of STATseq and standard genetic testing in a case series from the NICU and PICU of a large children's hospital between Nov 11, 2011, and Oct 1, 2014. The participants were families with an infant younger than 4 months with an acute illness of suspected genetic cause. The intervention was STATseq of trios (both parents and their affected infant). The main measures were the diagnostic rate, time to diagnosis, and rate of change in management after standard genetic testing and STATseq.
Findings
20 (57%) of 35 infants were diagnosed with a genetic disease by use of STATseq and three (9%) of 32 by use of standard genetic testing (p=0·0002). Median time to genome analysis was 5 days (range 3–153) and median time to STATseq report was 23 days (5–912). 13 (65%) of 20 STATseq diagnoses were associated with de-novo mutations. Acute clinical usefulness was noted in 13 (65%) of 20 infants with a STATseq diagnosis, four (20%) had diagnoses with strongly favourable effects on management, and six (30%) were started on palliative care. 120-day mortality was 57% (12 of 21) in infants with a genetic diagnosis.
Interpretation
In selected acutely ill infants, STATseq had a high rate of diagnosis of genetic disorders. Most diagnoses altered the management of infants in the NICU or PICU. The very high infant mortality rate indicates a substantial need for rapid genomic diagnoses to be allied with a novel framework for precision medicine for infants in NICU and PICU who are diagnosed with genetic diseases to improve outcomes.
Funding
Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Human Genome Research Institute, and National Center for Advancing Translational Sciences.
Introduction
Since the progression of genetic diseases can be rapid in infants, diagnosis must be swift to permit timely consideration of interventions that lessen morbidity and mortality rates.1–7 There are more than 4300 genetic diseases of known causes. Collectively, they are the leading causes of infant mortality, particularly in neonatal intensive care units (NICUs),4 and in paediatric intensive care units (PICUs).8–18 The premise of genomic or precision medicine is that genetic diagnosis might allow supplementation of empirical, phenotype-driven management with genotype-differentiated treatment and genetic counselling.19–26 Timely molecular diagnoses of suspected genetic disorders had been largely precluded in acutely ill infants because of substantial clinical and genetic heterogeneity and tardiness of getting results from standard genetic tests, such as gene sequencing.5,19,20,23,24,27–32 Although appropriate NICU treat ment is one of the most cost-effective methods of high-cost health care, patients’ long-term outcomes are diverse.1,8,33,34 In genetic diseases with poor prognosis, rapid diagnosis might enable early discussions with parents about palliative care to minimise suffering.8,34
We previously reported methods for diagnosing genetic disorders with rapid whole-genome sequencing (STATseq) in 50 h.5 This method was used to simultaneously test almost all Mendelian illnesses and was postulated to give a diagnosis in time to guide clinical management of acutely ill infants and children in the NICU or PICU.5 In our study, we report the rate and types of genetic diagnoses with STATseq and standard genetic tests in the first 35 infants in a regional (level 4) NICU and PICU at a quaternary children's hospital and the prevalence, types, and results of medical findings.
Methods
Study design and patients
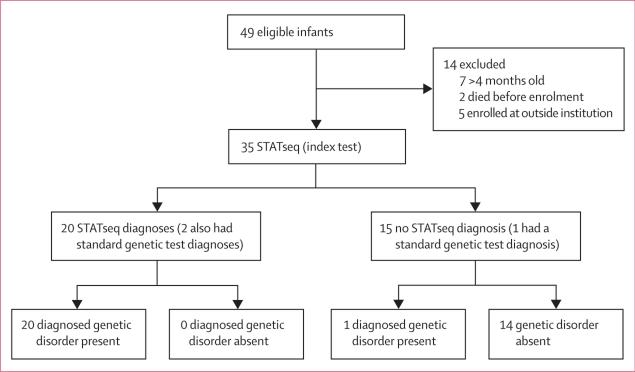
This study was undertaken at Children's Mercy–Kansas City, MO, USA. It was a retrospective comparison of the diagnostic rate, time to diagnosis, and types of molecular diagnoses of standard clinical genetic testing (reference test), as clinically indicated, with STATseq (index test) in a case series. Participants were parent–child trios, enrolled in a research biorepository who had genomic sequencing and standard diagnostic tests to diagnose monogenic disorders of unknown cause in the affected children.5,29 Affected infants and children with suspected genetic disorders were nominated for STATseq by the treating physician, typically a neonatologist (figure 1). A standard form of the primary signs and symptoms, past diagnostic test results, differential diagnosis or candidate genes, pertinent family history, availability of biological parents for enrolment, and whether the STATseq results might alter treatment was submitted for immediate assessment by a team of experts at the Center for Pediatric Genomic Medicine at the Children's Mercy–Kansas City. Infants had STATseq if the likely diagnosis was detectable with next-generation sequencing and had any potential to alter management or genetic counselling. Patients were not required to have standardised clinical examinations or diagnostic testing before referral; standard genetic testing for the cause was done as clinically indicated. Infants likely to have disorders associated with cytogenetic abnormalities were not accepted unless standard testing for those disorders was negative. About two-thirds of nominees were accepted for STATseq. About a half of accepted families were enrolled. The main reasons nominees were not enrolled were unavailability of one or more biological parents, preventing consent for the proband if before 28 days of life (DOL), parents wereyounger than 18 years of age and unable to consent, or parents refused to participate. 49 families with infants and children who were acutely ill or had died were enrolled and had STATseq of the parent–child trios. 35 of these families met inclusion criteria for this study: affected infant was younger than 4 months, was enrolled from a level 4 NICU or PICU at Children's Mercy–Kansas City between Nov 11, 2011, and Oct 1, 2014, had an acute illness of suspected monogenic cause, and did not have a genetic diagnosis. Of the 35 probands, 32 had standard clinical genetic testing. About 2400 infants younger than 4 months were admitted to the NICU or PICU during the study.
Figure 1. Summary of steps from patient nomination for STATseq to confirmatory result.
STATseq=rapid whole-genome sequencing.
This study was approved by the institutional review board at Children's Mercy–Kansas City. Parents provided written informed consent.
Procedures
The clinical features of affected infants were ascertained comprehensively through physician and family interviews and review of the medical records. Baseline demographics including age, sex, gestational age, birthweight, APGAR (Appearance, Pulse, Grimace, Activity, and Respiration) scores, and family history were obtained. Phenotypic features were translated into human phenotype ontology (HPO) terms and mapped to about 4300 monogenic diseases with the clinicopathological correlation software Phenomizer (appendix).3,35–37 Briefly, the Phenomizer uses term-similarity measures to calculate a similarity score for HPO terms entered by the user and terms used to label diseases in HPO. It then assigns a p value through statistical modelling to compare the similarity score obtained for the specific set of phenotypic terms entered to the distribution of similarity scores obtained with randomly chosen HPO term combinations. The p value is then used to rank the diseases.
STATseq was done in accordance with a 50-h or 7-day protocol, depending on severity of the illness.5,29 The laboratory at Children's Mercy–Kansas City was licensed by the Clinical Laboratory Improvement Amendments and accredited by the College of American Pathologists. STATseq was done on specimens from both parents and affected infants simultaneously. Genomic DNA extraction from whole blood, library preparation, sequencing, and data analysis were done in accordance with established protocols.29 Genomic DNA was prepared with TruSeq PCR Free sample preparation (Illumina, San Diego CA); quantitation was with real-time PCR. Libraries were sequenced with Illumina HiSeq 2500 instruments (2 × 100 nucleotides) in rapid run (50-h protocol) or high-output mode (2 × 125 nucleotides, version 4 Illumina sequencing-by-synthesis chemistry, 7-day protocol). STATseq was done to a minimum of 90 Gb per sample (appendix), to provide an average 40 times coverage of the genome. Each sample met established quality metrics.
Genomic sequence data are available at the database of Genotypes and Phenotypes (accession phs000564). Sequences were aligned to the human reference NCBI 37 with the Genomic Short Read Nucleotide Alignment Program.38 Nucleotide variants were detected and genotyped with the Genome Analysis Toolkit (version 1.4, 1.6, or 3.2)39,40 and gave a mean of 4·9 million nucleotide variants per sample (appendix). Variants were annotated with RUNES, a non-commercial software from Children's Mercy–Kansas City.5,29 STATseq interpretations included different sources of evidence, including variant attributes, the gene involved, inheritance pattern, and clinical case history. Causative variants were identified primarily with VIKING software (version 0.9–1.6),5,29 a non-commercial software from Children's Mercy–Kansas City, by limitation to the American College of Medical Genetics categories 1–3 and allele frequency of less than 1% from an internal database.5,29,41–43 VIKING was used to display variants characterised by use of RUNES and, thereby, to interpret STATseq findings. VIKING allows input of patients’ clinical features to sort variants by candidate gene and has additional dynamic filters, including those for minor allele frequency, American College of Medical Genetics’ variant pathogenicity category, compound heterozygosity, and custom gene lists. VIKING enables custom classification of variants, visualisation of read alignments with the Integrated Genome Viewer (version 2.03–2.3.9), and export of analysis findings. Genomes contained about 825 potentially pathogenic variants (allele frequency <1%, American College of Medical Genetics categories 1–3). All inheritance patterns were assessed. When a single likely causative variant for a recessive disorder was identified, the locus was manually inspected with the Integrated Genome Viewer in the trio for uncalled variants.44 Expert interpretation and literature curation were done for likely causative variants with regard to evidence for pathogenicity.43
Although STATseq can give a provisional diagnosis of genetic disorders in 50 h,5,29 it is a research test, and Sanger sequencing was used to confirm all likely causative genotypes. During the study, the US Food and Drug Administration (FDA) granted non-significant risk status for reporting a provisional STATseq diagnosis to the treating physician in exceptional cases, when the results were likely to change medical management and the infant was likely to die imminently (FDA, Center for Devices and Radiological Health, Off ce of In Vitro Diagnostics and Radiological Health submission Q140271, May 8, 2014). Familial relationships were confirmed through segregation analysis of private variants in STATseq diagnoses associated with de-novo mutations. An infant was classified as having a definitive diagnosis if a pathogenic or likely pathogenic genotype, by use of the American College of Medical Genetics criteria, in a disease gene that overlapped with a reported phenotype was reported in the medical record.43 Expert consultation and functional confirmation were done when the participant's phenotype differed from the expected phenotype for that disease gene. No incidental findings were reported.
Of the 35 infants who had STATseq, 32 had standard genetic testing based on the physician's clinical judgment.5,29 Standard testing for the cause of genetic diseases included array comparative genomic hybridisation, fluorescence in-situ hybridisation, high-resolution analysis of chromosomes, sequencing of genes and gene panels, methylation studies, and gene deletion or duplication assays (appendix).
We assessed the diagnostic rate of standard genetic testing and STATseq and compared the clinical outcomes between participants who had a STATseq diagnosis and those who did not. Diagnostic measurements included the types of genetic diagnoses obtained and the time to diagnosis. Clinical assessments included acute clinical usefulness (yes or no), initiation of palliative care (yes or no), and 120-day mortality (yes or no). Additional descriptive details of acute clinical usefulness were also recorded. Palliative care initiation and mortality were ascertained from the medical records. Acute clinical usefulness was ascertained through reviews of medical records and surveys with the referring physicians.
Statistical analysis
The median or prevalence of baseline participant characteristics was calculated for individuals who had a STATseq diagnosis versus those who did not. An individual-level analysis was done to compare the number of diagnoses with standard genetic testing with those made with STATseq. Diagnosis with standard genetic testing was treated as a dichotomous variable (yes or no) for this comparison irrespective of whether a patient had different clinical tests or not. No individual had more than one diagnosis within this group. STATseq diagnosis was also treated as a dichotomous variable (yes or no). Three patients who had STATseq did not have standard genetic testing and were not included in the analysis. The diagnostic rates with standard genetic testing and STATseq were compared by use of a two-tailed McNemar's χ2 test for paired nominal data, giving two-sided p values for the 32 infants. 120-day mortality and initiation of palliative care was compared between patients with a genetic diagnosis and those without by use of a two-sided Fisher's exact test (n=35).
Role of the funding source
The funder had no role in the study design, the gathering, analysis, or interpretation of the data, or the writing of the report. LKW, JEP, LDS, CJS, IT, NAM, SES, JAC, GT, AN, LZ, EGF, and SFK had access to the raw data. The corresponding author had full access to all the data and the final responsibility to submit for publication.
Results
49 families with infants or children who were acutely ill or had died were enrolled and had rapid whole-genome sequencing (STATseq) of parent–child trios. 35 of these families met the inclusion criteria for this study (figure 2). The phenotypes for which infants had been nominated to have STATseq were diverse and were typically present at birth (table 1). The most common phenotypes were congenital (26%) and neurological anomalies (20%; table 1). However, infants usually had complex clinical features, and the primary reason for nomination for STATseq was one of several co-occurring phenotypes (appendix). For example, patient CMH487 was admitted to the NICU at birth with bronchopulmonary dysplasia and a ruptured omphalocele but was nominated for STATseq for acute liver failure on day of life (DOL) 71.
Figure 2. Flow diagram of the comparison of the diagnostic sensitivity and accuracy of STATseq with standard clinical genetic testing for infants in NICU.
STATseq=rapid whole-genome sequencing. NICU=neonatal intensive care unit.
Table 1.
Characteristics of infants by STATseq diagnosis
| Total | Infants with a STATseq diagnosis | Infants without a STATseq diagnosis | |
|---|---|---|---|
| Infants | 35 (100%) | 20 (100%) | 15 (100%) |
| Consanguinity or isolated population | 1 (3%) | 1 (5%) | 0 |
| Sex, male | 18 (51%) | 9 (45%) | 9 (60%) |
| Family history of similar disorder | 5 (14%) | 4 (20%) | 1 (7%) |
| Gestational age (weeks; mean, range) | 37 (29-41) | 37 (29-40) | 37 (30-41) |
| Premature (<37 weeks' gestation) | 13 (37%) | 8 (40%) | 5 (33%) |
| Birthweight (kg; mean, range) | 2·7 (0·7-4·5) | 2·8 (0·7-4·5) | 2·6 (1·1-3·6) |
| Low birthweight (<2500 g) | 7 (20%) | 3 (15%) | 4 (27%) |
| Very low birthweight (1000-1500 g) | 4 (11%) | 2 (10%) | 2 (13%) |
| Extremely low birthweight (<1000 g) | 1 (3%) | 1 (5%) | 0 |
| APGAR (mean, range) | |||
| 1 min | 4·9 (0-9·0) | 5·3 (0-9·0) | 4·5 (0-8·0) |
| 5 min | 6·6 (0-9·0) | 7·1 (0-9·0) | 5·9 (0-9·0) |
| Deaths | 15 (43%) | 11 (55%) | 4 (27%) |
| Age at death (days; mean, range) | 81 (2-595) | 46 (16-100) | 218 (2-595) |
| Principal phenotypic feature | |||
| Symptom onset (days; mean, range) | 0·3 (0-7·0) | 0·5 (0-7·0) | 0 (0) |
| Multisystem congenital anomalies | 9 (26%) | 5 (25%) | 4 (27%) |
| Neurological | 7 (20%) | 4 (20%) | 3 (20%) |
| Cardiac findings or heterotaxy | 5 (14%) | 3 (15%) | 2 (13%) |
| Hydrops or pleural effusion | 4 (11%) | 2 (10%) | 2 (13%) |
| Metabolic findings (including hypoglycaemia) | 4 (11%) | 2 (10%) | 2 (13%) |
| Renal | 1 (3%) | 0 | 1 (7%) |
| Arthrogryposis | 2 (6%) | 2 (10%) | 0 |
| Respiratory | 1 (3%) | 0 | 1 (7%) |
| Hepatic | 1 (3%) | 1 (5%) | 0 |
| Dermatological | 1 (3%) | 1 (5%) | 0 |
| Testing (days; median, range) | |||
| Age at enrolment | 26 (1-71) | 32 (2-71) | 17 (1-63) |
| Interval from enrolment to analysis | 5 (3-153) | 6 (3-153) | 5 (3-46) |
| Interval from analysis to report | 9 (1-878) | 9 (1-878) | .. |
| Interval from enrolment to report | 23 (5-912) | 23 (5-912) | .. |
Data are number (%), unless otherwise indicated. STATseq=rapid whole-genome sequencing. APGAR=Appearance, Pulse, Grimace, Activity, and Respiration. 32 patients had standard genetic testing and the percentage diagnosed is based on this denominator.
32 infants had standard genetic screening (comprised of 94 tests; appendix) and 12 of these infants also had genetic screening with next-generation sequencing-based targeted gene panels. The mean age when standard genetic testing was ordered was DOL 20 (SD 33), and the median time from then to reporting of the genetic diagnosis was 16 days (range 1–162). Standard testing led to genetic diagnoses in three (9%) of 32 infants (by use of microarray comparative genomic hybridisation in one infant, and single gene sequencing in two infants figure 2; appendix). STATseq replicated two of these diagnoses; but the third genetic diagnosis was of a structural variation, which was only retrospectively detected with STATseq data and considered non-diagnostic by STATseq.
STATseq provided genetic diagnoses for 20 (57%) of 35 infants, and 18 of these diagnoses were not identified with standard genetic testing. The number of diagnoses with STATseq was significantly higher than that with the standard genetic testing (p=0·0002; figure 2; table 2; appendix). No standard or STATseq diagnoses were false positives.
Table 2.
Presentations, genetic diagnoses, and inheritance patterns in infants
| Clinical indication for STATseq | Causal gene |
Disease | Atypical presentation or incomplete diagnosis |
Inheritance pattern |
Chromosome coordinates of causal variants (hg19) |
Causal variants | |
|---|---|---|---|---|---|---|---|
| CMH064 | Desquamating skin rash | GJB2 | Keratitis-ichthyosis-deafness syndrome | Yes | Autosomal dominant, de novo | 13:20763634-20763636 | c.85_87del (p.Phe29del) |
| CMH172 | Status epilepticus | BRAT1 | Lethal neonatal rigidity and multifocal seizure syndrome | .. | Autosomal recessive, homozygous | 7:2583573-2583574 | c.453_454insATCTTCTC (p.Leu152IlefsTer70) |
| CMH184 | Heterotaxy | MMP21 | Heterotaxy | .. | Autosomal recessive, compound heterozygous | 10:127462732-127462732; 10:127460915-127466819 | c.365del (p.Met122SerfsTer55); exon 1-3 deletion |
| CMH487 | Acute liver failure | PRF1 | Familial haemophagocytic lymphohistiocytosis type 2 | Yes | Autosomal recessive, compound heterozygous | 10:72358167-72358167; 10:72360387-72360387 | c.1310C>T (p.Ala437Val); c.272C>T (p.Ala91Val) |
| CMH545 | Bilateral chylous effusions | PTPN11 | Noonan syndrome | .. | Autosomal dominant, de novo | 12:112915523-112915523 | c.922A>G (p.Asn308Asp) |
| CMH569 | Hyperinsulinaemic hypoglycaemia | ABCC8 | Familial hyperinsulinism type 1 | .. | Autosomal dominant | 11:17424218-17424218 | c.3640C>T (p.Arg1214Trp) |
| CMH578 | Hypertrophic cardiomyopathy, increased neck folds, low set ears, and hypotonia | PTPN11 | Noonan syndrome | .. | Autosomal dominant, de novo | 12:112926258-112926258 | c.1391G>C (p.Gly464Ala) |
| CMH586 | Failure to thrive, lactic acidosis, and hypoglycaemia | MT:TE | Reversible cytochrome C oxidase deficiency | .. | Mitochondrial | Mitochondria: 14674-14674 | tRNA-Glu; nucleotide 73 T>C |
| CMH629 | Seizures, arthrogryposis, and pulmonary hypoplasia | SCN2A | Epileptic encephalopathy | Yes | Autosomal dominant, de novo | 2:166245193-166245193 | c.4877G>A (p.Arg1626Gln) |
| CMH659 | Arthrogryposis, vesicoureteric reflux, ventricular and atrial septal defects, lissenencephaly, and absent corpus collusum | KAT6B | Say-Barber-Biesecker-Young-Simpson syndrome | .. | Autosomal dominant, de novo | 10:76784946-76784949 | c.3603_3606del (p.Thr1203ArgfsTer21) |
| CMH672 | Seizures | KCNQ2 | Epileptic encephalopathy | .. | Autosomal dominant, de novo | 20:62070965-62070965 | c.913T>C (p.Phe305Leu) |
| CMH678 | Intrauterine growth retardation, cardiomegaly, atrioventricular canal defect, osteopenia, microcolon, and large optic nerves | GNPTAB | Mucolipidosis III α/β | .. | Autosomal recessive, compound heterozygous | 12:102164296-102164296; 12:102164276-102164277 | c.1001G>A (p.Arg334Gln); c.1017_1020dupTGCA (p.Pro341CysfsTer22) |
| CMH680 | Seizures | SCN2A | Epileptic encephalopathy | .. | Autosomal dominant, de novo | 2:166201137-166201137 | c.2635G>A (p.Gly879Arg) |
| CMH725 | Several congenital anomalies | CHD7 | CHARGE syndrome: coloboma of the eye, heart defects, atresia of the choanae, retardation of growth or development, genital or urinary abnormalities, and ear abnormalities and deafness | .. | Autosomal dominant, de novo | 8:61655225-61655225 | c.1234C>T (p.Gln412Ter) |
| CMH809 | Hypertrophic cardiomyopathy, hepatomegaly, and thrombocytopenia | PTPN11 | LEOPARD syndrome: Noonan syndrome with multiple lentigines | .. | Autosomal dominant, de novo | 12:112926897-112926897 | c.1517A>C (p.Gln506Pro) |
| CMH846 | Seizure, polyhydramnios, respiratory failure, flat face, facial nerve palsy | PHOX2B | Central hypoventilation syndrome | .. | Autosomal dominant, de novo | 4:41747937-41747938 | c.831dupC (p.Gly278ArgfsTer82) |
| CMH855 | Hypoplastic right heart, tricuspid stenosis, diabetes, biliary atresia, and absent gallbladder | GATA6 | Pancreatic agenesis and congenital heart defects | .. | Autosomal dominant, de novo | 18:19752065-19752065 | c.960del (p.Asn320LysfsTer26) |
| CMH873 | Acute renal failure with nephrotic syndrome, and cataracts | LAMB2 | Pierson syndrome | .. | Autosomal recessive, compound heterozygous | 3:49159603-49159604; 3:49158878-49158878 | c.4773dupG (p.Arg1592AlafsTer7); c.5248C>T (p.Gln1750Ter) |
| CMH890 | Craniosynostosis, bilateral choanal atresia, micrognathia, and ventriculomegaly | FGFR2 | Pfeiffer syndrome | .. | Autosomal dominant, de novo | 10:123274794-123274794 | c.1124A>G (p.Tyr375Cys) |
| CMH902 | Pulmonary hypertension, abnormal ears, multicystic kidney, labial hypoplasia, and brain cyst | CHD7 | CHARGE syndrome: coloboma of the eye, heart defects, atresia of the choanae, retardation of growth or development, genital or urinary abnormalities, and ear abnormalities and deafness | .. | Autosomal dominant, de novo | 8:61757922-61757929 | c.5164_5171del (p.Phe1722GlyfsTer12) |
STATseq=rapid whole-genome sequencing.
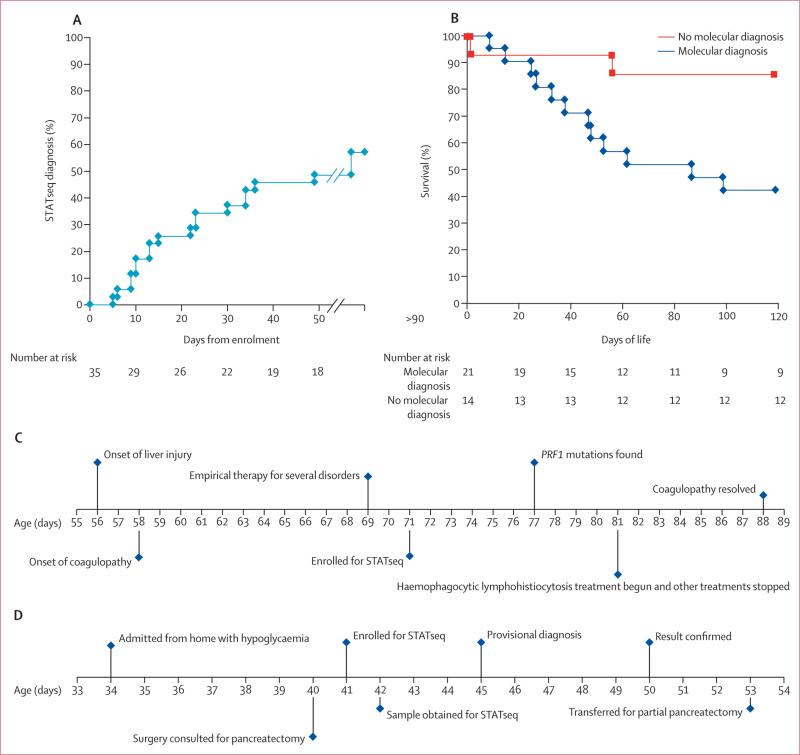
The mean age at enrolment for STATseq was DOL 26 (SD 21), and the median time to confirmed, reported diagnosis was 23 days (range 5–912; table 1). The median interval from enrolment to STATseq completion and start of variant analysis was 5 days (3–153; table 1). The outlier CMH064 (153 days from enrolment to STATseq completion and start of variant analysis) was the first enrollee, when STATseq methods were still in development. Sanger sequencing was done only for variants thought to be disease causing (n=24), and there was a 100% confirmation (table 2; appendix). 13 (65%) of 20 STATseq diagnoses were reported before discharge or death. Four infants died within 4 days of enrolment, and STATseq was incomplete at the time of their deaths (table 3). Reasons for longer time to diagnosis with STATseq than with standard genetic testing were development of software for detection of structural variants during the study, publication of novel disease-gene associations during the study, or infants whose phenotypes differed sufficiently from previous reports so as to require extensive analysis and external expert consultation.
Table 3.
Effect of genetic diagnoses on infant management in the NICU or PICU
| Any clinical usefulness of STATseq |
Results returned before discharge or death |
Genetic or reproductive counselling change |
Subspecialty consult (non- genetic) initiated |
Medication change |
Procedure change |
Diet change |
Palliative care initiated |
Imaging change |
Patient transferred to different facility |
Time from enrolment to diagnosis (days) |
Age at diagnosis (days) |
Age at death (days) |
Age at discharge from NICU or PICU (days) |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CMH064 | No | No | .. | .. | .. | .. | .. | .. | .. | .. | 415 | * | 54 | 54 |
| CMH172 | Yes | No | Yes | .. | .. | .. | .. | .. | .. | .. | 49 | * | 39 | 39 |
| CMH184 | No | No | .. | .. | .. | .. | .. | .. | .. | .. | 912 | 956 | .. | 75 |
| CMH487 | Yes | Yes | .. | .. | Yes | .. | .. | .. | .. | .. | 36 | 107 | .. | 386 |
| CMH545 | Yes | Yes | Yes | .. | .. | .. | .. | Yes | Yes | .. | 13 | 69 | 88 | 88 |
| CMH569 | Yes | Yes | .. | Yes | Yes | Yes | .. | .. | .. | Yes | 9 | 50 | .. | 53 |
| CMH578 | No | Yes | .. | .. | .. | .. | .. | .. | .. | .. | 6 | 8 | 48 | 21 |
| CMH586 | Yes | No | Yes | .. | Yes | .. | Yes | .. | .. | .. | 34 | 98 | .. | 70 |
| CMH629 | No | No | .. | .. | .. | .. | .. | .. | .. | .. | 167 | * | 63 | 63 |
| CMH659 | Yes | Yes | .. | .. | .. | .. | .. | Yes | .. | .. | 23 | 61 | .. | 115 |
| CMH672 | Yes | Yes | .. | .. | Yes | .. | .. | .. | .. | .. | 22 | 26 | .. | 33 |
| CMH678 | Yes | Yes | .. | .. | .. | .. | .. | Yes | .. | .. | 10 | 28 | 34 | 34 |
| CMH680 | Yes | Yes | .. | .. | .. | .. | Yes | .. | .. | .. | 10 | 24 | .. | 143 |
| CMH725 | No | No | .. | .. | .. | .. | .. | .. | .. | .. | 23 | 65 | .. | 42 |
| CMH809 | Yes | Yes | .. | .. | .. | .. | .. | .. | Yes | .. | 5 | 7 | 16 | 16 |
| CMH846 | Yes | Yes | .. | .. | .. | .. | .. | Yes | Yes | .. | 9 | 16 | 28 | 28 |
| CMH855 | Yes | Yes | Yes | .. | .. | Yes | .. | Yes | .. | .. | 13 | 62 | .. | 101 |
| CMH873 | No | No | .. | .. | .. | .. | .. | .. | .. | .. | 30 | * | 26 | 25 |
| CMH890 | Yes | Yes | .. | .. | .. | Yes | .. | Yes | .. | .. | 15 | 35 | 49 | 49 |
| CMH902 | No | Yes | .. | .. | .. | .. | .. | .. | .. | .. | 34 | 53 | 100 | 100 |
| Total (yes) or mean | 13 | 13 | 4 | 1 | 4 | 3 | 2 | 6 | 3 | 1 | 92 | 104 | 11 | 77 |
| Percentage of diagnoses | 65% | 65% | 20% | 5% | 20% | 15% | 10% | 30% | 15% | 5% | .. | .. | 55% | .. |
STATseq=rapid whole-genome sequencing. NICU=neonatal intensive care unit. PICU=paediatric intensive care unit.
STATseq was incomplete at the time of death.
Nine (45%) of 20 STATseq diagnoses were diseases that were not part of the differential diagnosis at time of enrolment. In one acutely ill infant (CMH487), a provisional molecular diagnosis, which was likely to change medical management, was reported verbally on day 3, before confirmatory testing.
No phenotypic feature was associated with an increase in genetic diagnosis with STATseq. Recurrent genes with causative variants were PTPN11 (three variants), CHD7 (two), and SCN2A (two); all of which occurred de novo (table 2; appendix). Dominant de-novo mutations were the most common mechanism of genetic disease (13 [65%] of 20 patients). One infant had a dominantly inherited disease, with a paternally inherited variant and somatic loss of the maternal allele. Genome sequencing provided good coverage of the mitochondrial genome, giving one diagnosis of a maternally inherited disease. Four of five patients with autosomal recessive inheritance were compound heterozygous, and one, from a genetically isolated population, was homozygous (table 2).
The median stay in the NICU or PICU was 42 days (range 3–387). 14 (40%) of 35 infants died within 120 days. The 120-day mortality was higher in infants who had a genetic diagnosis with either STATseq or standard testing than in those who did not (12 [57%] of 21 [including one infant diagnosed with standard testing who died at 10 days] vs two [14%] of 14 infants, respectively; p=0·46; table 3; figure 3B; appendix). Palliative care was initiated in a higher number of infants with genetic diagnoses than in those without (six [29%] of 21 with genetic diagnosis vs none of 14 with no diagnosis; p=0·06; table 3).
Figure 3. Characteristics of patients with STATseq diagnoses.
(A) Time from enrolment to confirmed diagnosis with STATseq in 18 infants. Two infants who were diagnosed at days 167 and 912 are not shown. (B) Kaplan-Meier survival curves of infants in NICU and PICU who did (n=21) or did not (n=14) receive a diagnosis of a genetic disease. 12 infants diagnosed with genetic diseases died before day of life 120, whereas three infants without a genetic disease died. (C) Timecourse for development of acute liver failure, enrolment, STATseq diagnosis, and treatment in infant CMH487. (D) Time course for admission, enrolment, diagnosis, and treatment of infant CMH569 with hyperinsulinaemic hypoglycaemia. STATseq=rapid whole-genome sequencing. NICU=neonatal intensive care unit. PICU=paediatric intensive care unit.
The short-term clinical effect of STATseq diagnoses was assessed by chart reviews and surveys with referring physicians (table 3). 13 (65%) of 20 STATseq diagnoses were useful in the acute clinical management of the infants (table 3). Reasons for clinical usefulness were diverse and included starting palliative care, medication changes, and change in genetic counselling. Of 13 diagnoses made before discharge or death, 11 (85%) were useful in the acute clinical management of the infants. In four (31%) of 13 timely diagnoses (four [20%] of 20 STATseq diagnoses and four [11%] of 35 infants), the change in acute management or outcome was both substantial and favourable. Two examples of substantial favourable outcomes are shown in panels 1 and 2. Other examples are shown in the appendix.
In several cases, review of reports identified potential treatments that were novel or for which evidence of effectiveness was only anecdotal. For example, in CMH809, with PTPN11-associated hypertrophic cardiomyopathy (LEOPARD syndrome), an N-of-1 trial of everolimus, an inhibitor of mTOR-dependent MEK/ERK activation, was internally discussed as a potential therapy, but not implemented.45–48 The infant died on DOL 17.
Discussion
Rapid, clinical genome sequencing (STATseq) was feasible in a NICU or PICU and provided genetic diagnoses for most of the enrolled infants with a wide range of clinical presentations. Since genetic diseases are the leading cause of death in the NICU and PICU, and overall infant mortality,2,4,5,8–11,13,15,16,21,26,33,34 these results might have broad implications for the NICU or PICU practice.
57% of the cases had a definitive diagnosis with STATseq, significantly higher than that with standard genetic tests (9%). Nine genetic diagnoses were not suspected before STATseq, and thus patients were not given standard genetic testing for the specific genes. Additionally, the rapidity of STATseq diagnosis and absence of clinician masking might have reduced the extent of standard genetic testing in some cases, contributing to the large difference in diagnostic yield. The rate of diagnosis with STATseq was higher than that reported for whole-exome sequencing,6,7,19,25,29,49–53 especially in view of the absence of consanguinity in our study. Several factors might have contributed to this difference. A priori, genome sequencing is both faster and more complete than is whole-exome sequencing.29,54 Use of parent–infant trios in our study allowed identification of de-novo mutations for the most common mechanism of disease. Several recent reports of diagnostic yield with whole-exome sequencing did not include parent–infant trios.6,7,53 The phenotypes of infants in this study were usually incomplete of classical genetic disease descriptions, as shown by nine STATseq diagnoses being for diseases that were not initially considered by the clinician, and the average STATseq-based diagnosis ranked 806 most likely on a software-derived list of differential diagnoses. By contrast, the mean rank among 32 older children diagnosed with whole-exome sequencing was 279.29 The manifestation of classical genetic disease phenotypes seems to take time in affected infants. Access to the medical record to establish a detailed phenotypic description for clinicopathological correlation helped overcome this obstacle. This level of phenotypic detail—or iterative genotype-driven ascertainment of clinical features—is not available to commercial laboratories, and could also contribute to their lower reported diagnostic rates. Additionally, the cases reported in our study were a small subset of the total admissions to the NICU and PICU during the study and had a strong pretest probability of genetic disease. By contrast, recent case series of the diagnostic yield of whole-exome sequencing included predominantly older children who had non-diagnostic standard genetic testing for longer than in this study, potentially reducing the pretest probability of a positive test. However, despite these sources of bias, the higher percentage of diagnosis with STATseq might be the result of higher prevalence of genetic disease in level 4 NICU and PICU populations, as opposed to older children reported in previous whole-exome sequencing studies. Although STATseq can be used to provide a provisional diagnosis of genetic disorders in 50 h,5,29 the fastest time to reported diagnosis in our study was 5 days and the median was 23 days (table 1). There were several reasons for this difference in speed. First, some diagnoses were made after improvements in methods or publication of novel disease-gene associations during the study. Second, extensive analysis and expert consultation were needed in cases for which diagnoses differed from expected presentations. Third, STATseq is a research test, and confirmation with a clinical test is mandatory before the results are reported. Confirmatory Sanger sequencing typically took 1 week. During the study, however, the FDA granted non-significant risk status to our return of a provisional STATseq-based diagnosis to the treating physician in exceptional cases, for which the results were likely to change management and death was likely to be imminent. The fastest provisional diagnosis was 3 days. In the future, Sanger sequencing might not be mandatory, because of the reproducibility of high-quality STATseq results, which would improve the turnaround time and provide increased opportunity for clinical intervention.
A prerequisite for broad adoption of STATseq in NICU or PICU populations is demonstration of improved outcomes. There were three outcomes, or their proxies, in this study, namely short-term mortality, morbidity, and suffering in infants with a dire prognosis. The mortality rate in infants with a diagnosis of a genetic disease was very high (57% at 120 days). However, 120-day mortality was 14% in the infants who were enrolled but did not have a genetic diagnosis, and overall mortality in the Children's Mercy–Kansas City NICU was 4% (23 of 563 neonates of gestational age >36 weeks admitted in 2013 died). Reported rates of neonatal 28-day mortality in NICUs vary widely (from 0·8% to 6·2%).7–9 If the finding that most of the infants in a NICU with diagnosable genetic diseases die within 120 days is substantiated in future studies, then vigorous effort will be warranted to address this apparent, unmet medical need.
Among infants who died, the average age was 0·5 days at symptom onset, 26 days at enrolment, and 46 days at death (table 1). 65% of STATseq diagnoses were reported before discharge or death. Thus, the interval for diagnosis and institution of genotype-directed interventions that could lessen morbidity and mortality was very short. Nevertheless, treating physicians adjudged STATseq diagnoses to have been helpful in acute clinical care in 65% of infants (table 3). The main types of change in care associated with diagnoses were in medications, genetic counselling, and medical procedures. In four cases, described in the panels and appendix, acute management or outcome, or both, were substantively and favourably changed, or had the potential to have been changed. A major goal in future studies of rapid genome sequencing in NICUs will be patient ascertainment and enrolment at symptom onset to maximise the interval for implementation of precision medicine.
A genetic diagnosis that confers a dire prognosis can empower early discussions with the infant's parents about palliative care to minimise suffering.33 End-of-life decisions are common in neonatal genetic diseases, with the primary cause of most deaths being withdrawal or withholding of care.10 Less than 10% of neonatal deaths occur despite maximal intensive care.7–9 We noted that STATseq-based diagnosis enabled such prognostic determination and discussion of initiation of palliative care when the prognosis was dismal.8–12,16,18 Indeed, palliative care was given to 30% of infants with STATseq diagnoses.
In families desiring the full complement of intensive care, optimum management of each infant could be regarded as an N-of-1-genome case study, as exemplified by CMH809. This management could be accomplished, for example, with a specific precision neonatology consultant team in large level 4 NICUs and PICUs, ascertainment of candidate infants at admission, facilitation of genetic diagnosis by use of STATseq, immediate provision of prognostic and therapeutic guidance and counselling in very rare disorders, and provision of treatment protocols for rapid implementation of specialised treatments, services, and studies for infants diagnosed with genetic diseases.46
This study was limited by its size (35 families), retrospective analysis, and absence of a randomised masked control group. It was restricted to infants younger than 4 months in one level 4 NICU or PICU, for whom the presentation suggested a genetic diagnosis that had some potential to affect management or counselling. Sufficient time has not elapsed since study inception to ascertain long-term outcomes. Full assessment of the usefulness of STATseq to alter infant morbidity and mortality will need prospective study, with enrolment at or close to birth, more timely STATseq results than achieved in our study, and rapid initiation of individualised treatment. In addition to usefulness, the cost-effectiveness of such testing and treatment needs to be ascertained to shape medical practice. The psychosocial effect of diagnoses for parents or health-care providers was not measured. Broader ethical concerns such as identification of carrier status and incidental findings unrelated to the neonatal presentation of the infants also warrant further investigation. Some of these limitations, and the generalisability of the results reported here to broader newborn populations will be assessed in a randomised masked study (ClinicalTrials. gov, number NCT02225522).
The experience so far suggests a novel framework for implementation of precision medicine in a level 4 NICU or PICU.48 STATseq has the potential to alter clinical management or genetic counselling and refine treatment plans for infants in accordance with their diagnoses. However, additional study is needed before STATseq is used routinely in level 4 NICUs or PICUs.
Supplementary Material
Research in context.
Evidence before this study
We searched PubMed with the terms “diagnosis”, “infant”, and “genome sequencing” or “exome sequencing”. There were no date or language restrictions. We found several case reports and five retrospective clinical series related to the usefulness of genomic sequencing for molecular diagnosis in infants in whom the differential diagnosis included monogenic diseases. The exome is the 1·5% of the genome that encodes proteins, and exome sequencing has until now been favoured over genome sequencing because of costs. The evidence before this study was that whole-genome or exome sequencing results in a molecular diagnosis in 25–73% of children with possible monogenic diseases. No prospective studies of the usefulness of genome or exome sequencing for diagnosis of monogenic diseases have yet been published. The usefulness of genome or exome sequencing for molecular diagnosis in neonatal and paediatric intensive care units (NICUs or PICUs), where genetic diseases are the leading cause of death and daily costs of care are commensurate with those of whole-genome sequencing, have not been investigated. The temporal dynamics of monogenic disease progression in the NICU or PICU, or the implications for calibration of time-to-diagnosis and time-to-intervention have not been investigated.
Added value of this study
We report that 57% of 35 infants were diagnosed with monogenic disease in NICU or PICU with rapid whole-genome sequencing (STATseq). Our study is the first of clinical usefulness of genomic diagnoses in acute illnesses in infants in NICU or PICU. In agreement with the results of two previous reports of the clinical effect of genomic diagnoses in children with non-acute neurodevelopmental disabilities, we noted that 65% of STATseq diagnoses had immediate clinical usefulness, including a strongly favourable effect on management in 20% and institution of palliative care in 30% of infants.
Implications of all the available evidence
The clinical implication of the available evidence is that clinicians should consider genome or exome sequencing for diagnosis in NICU or PICU for infants in whom the differential diagnosis includes monogenic diseases. However, further studies are needed to identify the characteristics of infants in whom the likelihood of a diagnosis is sufficiently likely to warrant the cost of genome sequencing, and how rapid molecular diagnoses can be integrated into clinical workflows to improve outcomes in newborns with acute genetic diseases.
Panel 1: Clinical effect of STATseq diagnosis in liver failure.
CMH487, a full-term infant admitted to the NICU at birth with several congenital anomalies, required tracheostomy, and was ventilator dependent (figure 3C). On day of life (DOL) 56, he developed acute hepatic failure. Extensive testing did not reveal the cause. Steroids were initiated empirically on DOL 67, with some improvement in hepatic failure. Intravenous immunoglobulin was given on DOL 69. The infant–parent trio was enrolled on DOL 71. The STATseq genotype was suggestive of type 2 haemophagocytic lymphohistiocytosis on DOL 74, which was confirmed and reported on DOL 77 with recommendations for functional studies. Despite a small overlap with the classic presentation, the diagnosis was confirmed by the absence of natural killer cell function. Disease-specific treatment (intravenous immunoglobulin and corticosteroids) was continued, and empirical therapies for other disorders were discontinued on DOL 81. Coagulopathy resolved on DOL 88. The patient is now 23 months old, at home, has normal liver function, and has undergone several surgical procedures for correction of congenital anomalies.
Panel 2: Clinical effect of STATseq diagnosis in hypoglycaemia.
CMH569 was admitted to the PICU on day of life (DOL) 34 with a blood glucose concentration of 1 mmol/L (figure 3D). Hypoglycaemia persisted despite glucose infusion of greater than 13 mg/kg per min and maximum dose of diazoxide. Testing showed hyperinsulinaemia (6·4 μIU/mL). The infant–parent trio was enrolled on DOL 41. The genotype obtained with STATseq was suggestive of ABCC8-associated familial hyperinsulinism type 1, which was reported provisionally on DOL 45. The presence of one, paternally derived mutation and clinical presentation suggested the focal form of the familial hyperinsulinism (pancreatic adenomatous hyperplasia that involved a portion of the pancreas), caused by biallelic mutations in ABCC8. Focal familial hyperinsulinism is inherited autosomal dominantly, but only manifests when the mutation is on the paternally derived allele and there is somatic loss of the maternal allele in a β-cell precursor. The confirmed diagnosis was reported on DOL 50. 18F-fluorodopa PET was used to confirm and localise the focal pancreatic lesions, which changed the surgical approach and clinical outcome: targeted resection of focal pancreatic lesions was done, avoiding insulin-requiring diabetes mellitus. STATseq shortened the PICU stay by about 3 weeks and the morbidity (and potential mortality) associated with breakthrough hypoglycaemia. The patient is now 19 months old and euglycaemic. The patient maintained normal blood glucose during a fasting challenge, indicating no persistent hyperinsulinism.
Acknowledgments
This work was supported by gifts from the Marion Merrell Dow Foundation, Children's Mercy–Kansas City, Patton Trust, W T Kemper Foundation, Pat & Gil Clements Foundation, Claire Giannini Foundation, Black & Veatch, and grants from Eunice Kennedy Shriver National Institute of Child Health and Human Development and National Human Genome Research Institute (U19HD077693), and National Center for Advancing Translational Sciences (CTSA grant TL1TR000120). We thank Hongying Daisy Dai, the divisions of Clinical Genetics and Neonatology at Children's Mercy–Kansas City, Keijan Zhang (Cincinnati Children's Hospital, OH, USA), and the families for their participation in this research study.
Footnotes
Contributors
SFK and LKW were responsible for the idea and design of the study. LKW, JEP, LDS, CJS, IT, NAM, SES, JAC, EGF, SMH, and SFK were involved in the acquisition, analysis, or interpretation of data. SFK, LKW, JEP, LDS, IT, and JAC drafted the manuscript. CJS, EGF, SES, PMA, SLC, MAC, RTF, JAH, HK, RJM, JLR, and SLT were involved in the revision of the manuscript for important intellectual content. SFK, EGF, CJS, NAM, SES, SMH, GT, AN, MC, LZ, and IT obtained funding, administrative, technical, or material support. LKW, JEP, SFK, and JAC supervised the study.
Declaration of interests
We declare no competing interests.
References
- 1.Couce ML, Bana A, Boveda MD, Perez-Munuzuri A, Fernandez-Lorenzo JR, Fraga JM. Inborn errors of metabolism in a neonatology unit: impact and long-term results. Pediatr Int. 2011;53:13–17. doi: 10.1111/j.1442-200X.2010.03177.x. [DOI] [PubMed] [Google Scholar]
- 2.Dixon-Salazar TJ, Silhavy JL, Udpa N, et al. Exome sequencing can improve diagnosis and alter patient management. Sci Transl Med. 2012;4:138ra78. doi: 10.1126/scitranslmed.3003544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohler S, Schulz MH, Krawitz P, et al. Clinical diagnostics in human genetics with semantic similarity searches in ontologies. Am J Hum Genet. 2009;85:457–64. doi: 10.1016/j.ajhg.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin JA, Hamilton BE, Osterman MJK, Curtin SC, Mathews TJ. Births: final data for 2012. US Department of Health and Human Services; Hyattsville, MD: 2013. [Google Scholar]
- 5.Saunders CJ, Miller NA, Soden SE, et al. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med. 2012;4:154ra35. doi: 10.1126/scitranslmed.3004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of Mendelian disorders. N Engl J Med. 2013;369:1502–11. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–79. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiner J, Sharma J, Lantos J, Kilbride H. How infants die in the neonatal intensive care unit: trends from 1999 through 2008. Arch Pediatr Adolesc Med. 2011;165:630–34. doi: 10.1001/archpediatrics.2011.102. [DOI] [PubMed] [Google Scholar]
- 9.Wilkinson DJ, Fitzsimons JJ, Dargaville PA, et al. Death in the neonatal intensive care unit: changing patterns of end of life care over two decades. Arch Dis Child Fetal Neonatal Ed. 2006;91:F268–71. doi: 10.1136/adc.2005.074971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagen CM, Hansen TW. Deaths in a neonatal intensive care unit: a 10-year perspective. Pediatr Crit Care Med. 2004;5:463–68. doi: 10.1097/01.pcc.0000128893.23327.c1. [DOI] [PubMed] [Google Scholar]
- 11.Berger TM, Hofer A. Causes and circumstances of neonatal deaths in 108 consecutive cases over a 10-year period at the Children's Hospital of Lucerne, Switzerland. Neonatology. 2009;95:157–63. doi: 10.1159/000153100. [DOI] [PubMed] [Google Scholar]
- 12.Ray JG, Urquia ML, Berger H, Vermeulen MJ. Maternal and neonatal separation and mortality associated with concurrent admissions to intensive care units. CMAJ. 2012;184:E956–62. doi: 10.1503/cmaj.121283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.March of Dimes . 2012 natality and 2010 infant mortality data updated. March of Dimes; White Plains, NY: 2012. [Google Scholar]
- 14.Yoon PW, Olney RS, Khoury MJ, Sappenfield WM, Chavez GF, Taylor D. Contribution of birth defects and genetic diseases to pediatric hospitalizations. A population-based study. Arch Pediatr Adolesc Med. 1997;151:1096–103. doi: 10.1001/archpedi.1997.02170480026004. [DOI] [PubMed] [Google Scholar]
- 15.O'Malley M, Hutcheon RG. Genetic disorders and congenital malformations in pediatric long-term care. J Am Med Dir Assoc. 2007;8:332–34. doi: 10.1016/j.jamda.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 16.Pinar H. Postmortem findings in term neonates. Semin Neonatol. 2004;9:289–302. doi: 10.1016/j.siny.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Stevenson DA, Carey JC. Contribution of malformations and genetic disorders to mortality in a children's hospital. Am J Med Genet A. 2004;126A:393–97. doi: 10.1002/ajmg.a.20409. [DOI] [PubMed] [Google Scholar]
- 18.Soneda A, Teruya H, Furuya N, et al. Proportion of malformations and genetic disorders among cases encountered at a high-care unit in a children's hospital. Eur J Pediatr. 2012;171:301–05. doi: 10.1007/s00431-011-1534-2. [DOI] [PubMed] [Google Scholar]
- 19.Bainbridge MN, Wiszniewski W, Murdock DR, et al. Whole-genome sequencing for optimized patient management. Sci Transl Med. 2011;3:87re3. doi: 10.1126/scitranslmed.3002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzaga-Jauregui C, Lupski JR, Gibbs RA. Human genome sequencing in health and disease. Annu Rev Med. 2012;63:35–61. doi: 10.1146/annurev-med-051010-162644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green ED, Guyer MS. National Human Genome Research I. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470:204–13. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 22.Kingsmore SF, Dinwiddie DL, Miller NA, Soden SE, Saunders CJ. Adopting orphans: comprehensive genetic testing of Mendelian diseases of childhood by next-generation sequencing. Expert Rev Mol Diagn. 2011;11:855–68. doi: 10.1586/erm.11.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kingsmore SF, Saunders CJ. Deep sequencing of patient genomes for disease diagnosis: when will it become routine? Sci Transl Med. 2011;3:87ps23. doi: 10.1126/scitranslmed.3002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lupski JR, Belmont JW, Boerwinkle E, Gibbs RA. Clan genomics and the complex architecture of human disease. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lupski JR, Reid JG, Gonzaga-Jauregui C, et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med. 2010;362:1181–91. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawn JE, Kinney M. Preterm birth: now the leading cause of child death worldwide. Sci Transl Med. 2014;6:263ed21. doi: 10.1126/scitranslmed.aaa2563. [DOI] [PubMed] [Google Scholar]
- 27.Biesecker LG, Green RC. Diagnostic clinical genome and exome sequencing. N Engl J Med. 2014;370:2418–25. doi: 10.1056/NEJMra1312543. [DOI] [PubMed] [Google Scholar]
- 28.Shashi V, McConkie-Rosell A, Rosell B, et al. The utility of the traditional medical genetics diagnostic evaluation in the context of next-generation sequencing for undiagnosed genetic disorders. Genet Med. 2014;16:176–82. doi: 10.1038/gim.2013.99. [DOI] [PubMed] [Google Scholar]
- 29.Soden S, Saunders CJ, Willig LK, et al. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci Transl Med. 2014;6:265ra168. doi: 10.1126/scitranslmed.3010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soden SE, Saunders CJ, Dinwiddie DL, et al. A systematic approach to implementing monogenic genomic medicine. J Genomes Exomes. 2013;1:15–24. [Google Scholar]
- 31.ACMG . Points to consider in the clinical application of genomic sequencing. American College of Medical Genetics; Bethesda, MD: 2012. [DOI] [PubMed] [Google Scholar]
- 32.Worthey EA, Mayer AN, Syverson GD, et al. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011;13:255–62. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- 33.Downing GJ, Zuckerman AE, Coon C, Lloyd-Puryear MA. Enhancing the quality and efficiency of newborn screening programs through the use of health information technology. Semin Perinatol. 2010;34:156–62. doi: 10.1053/j.semperi.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 34.Lantos JD, Meadow WL. Costs and end-of-life care in the NICU: lessons for the MICU? J Law Med Ethics. 2011;39:194–200. doi: 10.1111/j.1748-720X.2011.00588.x. [DOI] [PubMed] [Google Scholar]
- 35.Kohler S, Doelken SC, Rath A, Ayme S, Robinson PN. Ontological phenotype standards for neurogenetics. Hum Mutat. 2012;33:1333–39. doi: 10.1002/humu.22112. [DOI] [PubMed] [Google Scholar]
- 36.Ullah MZ, Aono M, Seddiqui MH. Estimating a ranked list of human hereditary diseases for clinical phenotypes by using weighted bipartite network. Conf Proc IEEE Eng Med Biol Soc. 2013;2013:3475–78. doi: 10.1109/EMBC.2013.6610290. [DOI] [PubMed] [Google Scholar]
- 37.Zemojtel T, Kohler S, Mackenroth L, et al. Effective diagnosis of genetic disease by computational phenotype analysis of the disease-associated genome. Sci Transl Med. 2014;6:252ra123. doi: 10.1126/scitranslmed.3009262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–81. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;11:1–11. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richards CS, Bale S, Bellissimo DB, et al. ACMG recommendations for standards for interpretation and reporting of sequence variations: revisions 2007. Genet Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 42.Maddalena A, Bale S, Das S, Grody W, Richards S. Committee ALQA. Technical standards and guidelines: molecular genetic testing for ultra-rare disorders. Genet Med. 2005;7:571–83. doi: 10.1097/01.gim.0000182738.95726.ca. [DOI] [PubMed] [Google Scholar]
- 43.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015 doi: 10.1038/gim.2015.30. published online March 5. DOI:10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith LD, Saunders CJ, Dinwiddie DL, et al. Exome sequencing reveals de novo germline mutation of the mammalian target of rapamycin (MTOR) in a patient with megalencephaly and intractable seizures. J Genomes Exomes. 2013;2:63–72. [Google Scholar]
- 46.Schramm C, Edwards MA, Krenz M. New approaches to prevent LEOPARD syndrome-associated cardiac hypertrophy by specifically targeting Shp2-dependent signaling. J Biol Chem. 2013;288:18335–44. doi: 10.1074/jbc.M113.483800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marin TM, Keith K, Davies B, et al. Rapamycin reverses hypertrophic cardiomyopathy in a mouse model of LEOPARD syndrome-associated PTPN11 mutation. J Clin Invest. 2011;121:1026–43. doi: 10.1172/JCI44972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith LD, Kingsmore SF. N-of-1 genomic medicine for the rare pediatric genetic diseases. Expert Opin Orphan Drugs. 2014;2:1279–90. [Google Scholar]
- 49.Srivastava S, Cohen JS, Vernon H, et al. Clinical whole exome sequencing in child neurology practice. Ann Neurol. 2014;76:473–83. doi: 10.1002/ana.24251. [DOI] [PubMed] [Google Scholar]
- 50.Need AC, Shashi V, Hitomi Y, et al. Clinical application of exome sequencing in undiagnosed genetic conditions. J Med Genet. 2012;49:353–61. doi: 10.1136/jmedgenet-2012-100819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lupski JR, Gonzaga-Jauregui C, Yang Y, et al. Exome sequencing resolves apparent incidental findings and reveals further complexity of SH3TC2 variant alleles causing Charcot-Marie-Tooth neuropathy. Genome Med. 2013;5:57. doi: 10.1186/gm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Farwell KD, Shahmirzadi L, El-Khechen D, et al. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet Med. 2014 doi: 10.1038/gim.2014.154. published online Nov 6. http://dx.doi.org/10.1038/gim.2014.154. [DOI] [PubMed]
- 53.Lee H, Deignan JL, Dorrani N, et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312:1880–87. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jacob HJ. Genetic diagnosis through whole-exome sequencing. N Engl J Med. 2014;370:1069. doi: 10.1056/NEJMc1315908. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.