Abstract
NADPH/NADP+ homeostasis is critical for countering oxidative stress in cells. Nicotinamide nucleotide transhydrogenase (TH), a membrane enzyme present in both bacteria and mitochondria, couples the proton motive force to the generation of NADPH. We present the 2.8 Å crystal structure of the transmembrane proton channel domain of TH from Thermus thermophilus and the 6.9 Å crystal structure of the entire enzyme (holo-TH). The membrane domain crystallized as a symmetric dimer, with each protomer containing a putative proton channel. The holo-TH is a highly asymmetric dimer with the NADP(H)-binding domain (dIII) in two different orientations. This unusual arrangement suggests a catalytic mechanism in which the two copies of dIII alternatively function in proton translocation and hydride transfer.
Nicotinamide nucleotide transhydrogenase (TH) is an integral membrane protein that can utilize the proton motive force for hydride transfer from NADH to NADP+ forming NADPH (1): NADH + NADP+ + H+out ↔ NAD+ + NADPH + H+in, where “out” and “in” respectively denotes the space outside the prokaryotic plasma membrane and cytosol (or mitochondrial intermembrane space and matrix in eukaryotes). The resultant NADPH is utilized for amino acid biosynthesis and by glutathione reductase and glutathione peroxidase to remove reactive oxygen species (2). Mutations in Nnt genes, which encode for TH, are correlated with familial glucocorticoid deficiency of the adrenal cortex (3). Mice with mutations in the Nnt genes exhibit glucose intolerance and impaired secretion of insulin, as in type-2 diabetes (4, 5).
Functional TH exists as a dimer (6), where each protomer contains three domains: a hydrophilic NAD(H)-binding domain (dI), a transmembrane domain (dII), and a hydrophilic NADP(H)-binding domain (dIII) (Fig. 1A). Despite topological variations of TH subunits across species (Fig. S1), dIII is always a C-terminal extension of dII. Hydride transfer between NAD(H) and NADP(H) occurs across the dI-dIII interface; proton translocation is across dII. One proton is translocated across the membrane per hydride transferred between nucleotides (7).
Fig. 1. TH function and structure.
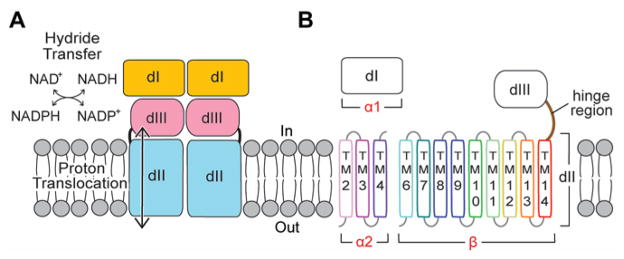
(A) Schematic representation of TH function. The NAD(H)-binding domain (dI) and the NADP(H)-binding domain (dIII) catalyze hydride transfer. Proton translocation occurs through the transmembrane domain (dII). “In” represents prokaryotic cytosol or mitochondrial matrix, and “out” is the space outside the prokaryotic plasma membrane or mitochondrial intermembrane space. (B) A Tt TH protomer is encoded by three genes (α1, α2, β): α1 encodes dI; α2 encodes three transmembrane (TM) helices of dII; β gene encodes the remaining nine helices of dII, the hinge region, and dIII. TM1 and TM5 of dII are missing in Tt TH.
Structures of hydrophilic dI and dIII were previously solved for TH isolated from bovine (8) and human (9) mitochondria as well as from Rhodospirillum rubrum (10) and Escherichia coli (11). Co-crystallization of dI with dIII results in an asymmetric heterotrimer (dI)2:dIII (12, 13). Here, we determined the structures of the dimeric (dI)2 and the heterotrimeric (dI)2:dIII from Thermus thermophilus (Tt) TH (Fig. S2A; Table S1). In each of the heterotrimer structures, the NADP(H)-binding site of dIII is near the NAD(H)-binding site of one copy of dI, poised for direct hydride transfer. If a second copy of dIII were present and obeyed local 2-fold symmetry, there would be a steric clash between the two copies of dIII (12, 13) (Fig. S2B). The structures, therefore, present a fundamental puzzle: how the second copy of dIII is oriented in the intact enzyme (14, 15), and how hydride transfer at dI-dIII interface is coupled to proton translocation across dII, approximately 40 Å away (6, 16). Solving the structures of dII and the intact enzyme from Tt TH addresses these questions.
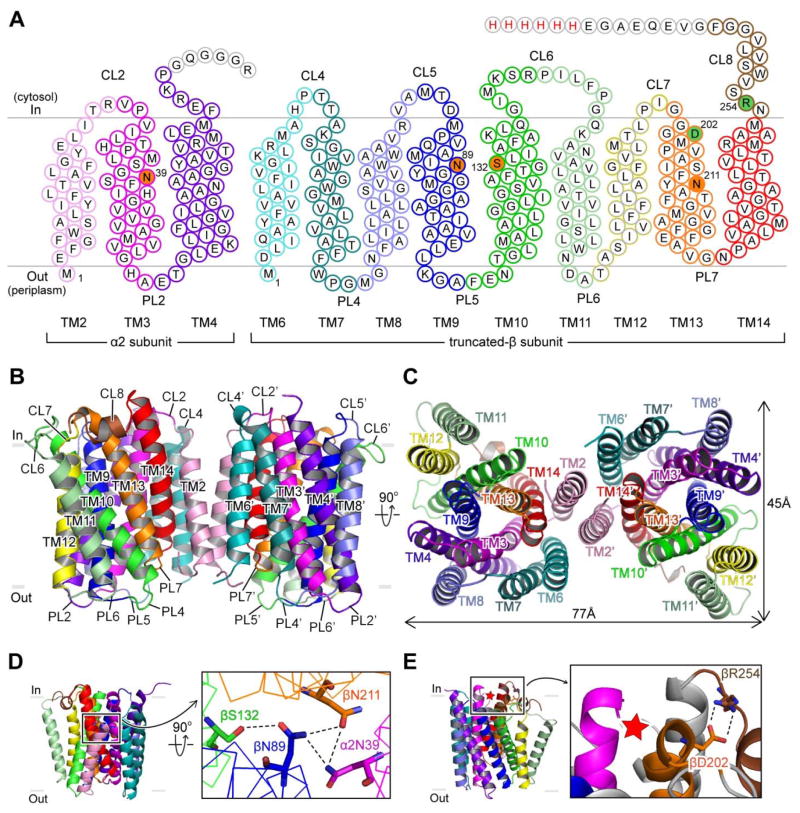
Tt TH is encoded by three genes (α1, α2, β): α1 encodes dI; α2 encodes the first three transmembrane (TM) helices of dII; and β encodes nine TM helices of dII linked with dIII (Fig. 1B). Based on sequence alignments, two helices (TM1, TM5) present in TH from some species are missing in Tt TH dII (Fig. S3). To determine the crystal structure of dII, we made a construct that expresses the α2 gene and a truncated version of β gene in which dIII is replaced by a histidine tag (Fig. 2A). The resulting dII contains 12 TM spans and crystalizes as a symmetric dimer with a 2-fold axis normal to the membrane plane (Fig. 2B; Table S2). The topology is consistent with previous studies of TH from different species, so the fold of dII observed in Tt TH is probably universal (17) (Fig. S3).
Fig. 2. Structural details of proton channel.
(A) Schematic diagram of the Tt dII construct used for crystallization, labeling transmembrane (TM) helices, cytoplasmic loops (CL) and periplasmic loops (PL). The Tt β gene was truncated at CL8 by a histidine tag (red font). Residues filled in orange form hydrogen bonds within the proton pathway. Residues filled in green form a conserved salt-bridge. Residues in grey outlines are disordered in the crystal structure. (B, C) Atomic model of the dII dimer viewed from (B) within the membrane and (C) from the periplasm. TM3, TM9-10 and TM13 form the proton channel. TM2 is at the dimer interface. (D) Residues α2N39 (TM3), βN89 (TM9), βS132 (TM10) and βN211 (TM13) form a hydrogen-bond network (black dotted lines). (E) Asp-Arg salt bridge formed by βD202 (TM13) and βR254 (CL8) is located on the membrane surface adjacent to the proton channel opening (red asterisk).
Each dII protomer constitutes a proton pathway where helices TM3-4, TM9-10, and TM13-14 arrange as a hexagram (Fig. 2C). Conserved motifs located at a level corresponding to the middle of the membrane are found within the interior of the hexagram on TM3 (NXXH/S), TM9 (NXXG or HXXV) and TM13 (NXXT/S) (Fig. S4). In the Tt dII structure, α2N39 (TM3), βN89 (TM9), βS132 (TM10) and βN211 (TM13) form a hydrogen-bond network (Fig. 2D). Of specific interest is βN89 (TM9), this residue is at the equivalent position as E. coli βH91, which has been identified as important for proton translocation by mutagenesis studies (2, 18, 19). Two other residues that have been implicated in proton translocation in E. coli TH are βS139 (20) and βN222 (18), which are equivalent to βS132 (TM10) and βN211 (TM13) in Tt TH. No ordered water molecules are observed in the structure; hence, this may correspond to an occluded conformation of the proton channel.
Based on previous cysteine crosslinking studies, a salt bridge between E. coli βD213 and βR265 has been postulated and implicated as important for both proton translocation in dII and NADP(H) binding in dIII (18, 20, 21). The structure shows that this Asp-Arg salt bridge is formed between the equivalent residues in Tt TH, βD202 (TM13) and βR254 (“hinge region” connecting dII and dIII), and is positioned on the membrane surface adjacent to the entrance of the proton channel (Fig. 2E).
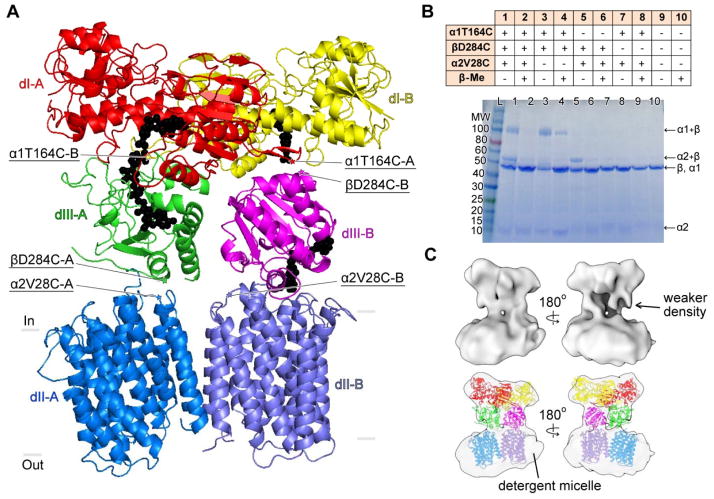
Using the structures of the dI dimer, the dII dimer and the dIII subunits, the crystal structure of Tt holo-TH was solved at 6.9 Å resolution by molecular replacement in Phaser (22) (Fig. 3A; Table S3). In the 200 kDa holo-TH dimer, two copies of dIII are positioned between the dI and dII dimers in different orientations (Fig. S5A). One copy of dIII (dIII-A) has its NADP(H)-binding site “face-up” to interact with the NAD(H)-binding site of dI-A as in the (dI)2:dIII heterotrimer structures (Fig. S2A). The second copy of dIII (dIII-B) occupies the space between dI-B and dII-B, but remarkably, it is flipped over 180° with its NADP(H)-binding site “face-down” toward dII-B. Though the structures of the “hinge region” between dII and dIII are unknown, both orientations of dIII can be accommodated by the full-length linker in all species (Fig. S5B).
Fig. 3. Structural asymmetry of holo-TH.
(A) Crystal structure shows two orientations of dIII: “face-up” dIII-A (green) and “face-down” dIII-B (magenta) with respect to their NAD(P)H-binding sites. Nucleotides, NAD(H) in dI and NADP(H) in dIII, are modeled as black spheres based on the heterotrimer structure (11, 12). Subunits sandwiching dIII-A and dIII-B are respectively labeled as A and B. Underlined labels mark the mutations used for crosslinking experiments. (B) SDS-PAGE shows the crosslinking of dIII in two orientations. The table on top describes samples in lane 1–10. For the wild-type enzyme (lane 10), subunits α1 and β each run with the same apparent molecular weight (MW) of about 45 kDa; the α2 subunit shows a faint band at 10kDa. βD284C on dIII can either crosslink with α1T164C in dI (lanes 1 and 3) forming α1+β, or with α2V28C on dII (lanes 1 and 5) forming α2+β. The presence of β-Me largely eliminates the crosslinking (lanes 2, 4 and 6). (C) Cryo-EM structure reveals an asymmetric dimer: one copy of dIII exhibits weaker density than the other.
To confirm the existence of the two dIII orientations in holo-TH, pairs of cysteine residues were inserted into holo-TH for crosslinking experiments using the purified enzyme solubilized in detergent. The structure predicts that residue βD284 (dIII) should be at the interface with dII in the face-up conformation, but at the interface with dI in the face-down conformation. Residue βD284 was replaced by a cysteine (βD284C) in combination with a cysteine placed in dI (α1T164C) and/or dII (α2V28C). SDS-PAGE analysis (Fig. 3B) showed that dIII can crosslink either to dII, forming the α2+β product, or to dI, forming the α1+β product. The crosslinks were largely reversed by β-mercaptoethanol (β-Me). Activity measurements showed that hydride transfer from NADH to NADP+ and then back to an NAD+ analogue (so-called cyclic activity), which is not coupled to proton translocation (23), is minimally influenced by the crosslinking (Table S4). In contrast, hydride transfer from NADPH directly to the NAD+ analogue, which is coupled to proton translocation (24), is decreased by crosslinking and is increased after disrupting the crosslinks. Although the data are not quantitative, it is clear that the face-down conformation of dIII is present in the active form of holo-TH, and is consistent with the importance of domain dynamics for catalytic function (21).
In addition to crystallography, we performed single-particle cryo-electron microscopy (cryo-EM) imaging of detergent-solubilized Tt holo-TH, yielding a structure at 18 Å resolution (Figs. 3C, S6). The structure shows that the enzyme is an asymmetric dimer: one copy of dIII is more disordered than the other.
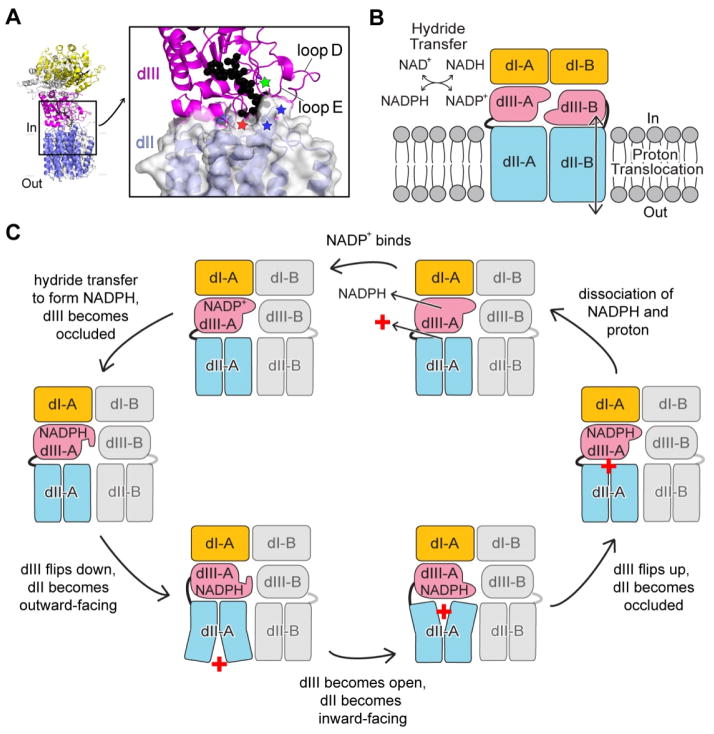
Previous biochemical data have shown that mutations in a conserved Asp residue in the NADP(H)-binding site of dIII, E. coli βD392, abolish both hydride transfer and proton translocation (16); and the rate-limiting step in forward catalysis is the release of the product NADP(H) from dIII (23). The holo-TH structure provides mechanistic implications of how dIII can be involved in proton translocation: the face-down conformation of dIII brings Tt βD378 (equivalent to E. coli βD392) (16) and regions of dIII that regulate NADP(H) binding, including loops D and E (14, 25), close to the surface of the proton channel and the Asp-Arg salt-bridge (Fig. 4A). Hence, it is reasonable that the face-down conformation of dIII is competent for proton translocation, whereas its face-up conformation allows hydride transfer (Fig. 4B). The division-of-labor assignation works into the binding-change model of the catalytic cycle proposed by Jackson and colleagues (26). The model is consistent with previous biochemical determinations: 1) TH exhibits half-of-the-sites reactivity such that inhibition of one protomer within the dimeric enzyme inhibits the second protomer (23, 24, 27); 2) There are “open” and “occluded” conformational states of dIII via its loops D and E, summarized in (26). The model assumes that dIII is mobile, which is a reasonable hypothesis because previous cysteine mutations of the Asp-Arg salt-bridge (Fig. 2E) lead to crosslinking and inhibition of both proton translocation in dII and NADP(H) binding in dIII (21). The membrane domain is proposed to have a single buried protonatable site (in most cases, a histidine in TM3 or TM9), and this residue can either be a) deprotonated and occluded, b) accessible to protonation by “outward-facing” or, c) accessible to protonation by “inward-facing”. We speculate that changes in dIII, which include the protonation state of Tt βD378 (E. coli βD392) (28), the conformation of loops D and E (14, 25), and the redox state of NADP(H), are coupled to conformational/protonation changes in the membrane domain coupled to proton translocation (Fig. 4C).
Fig. 4. Possible role of face-down dIII in proton translocation.
(A) Regions of dIII that regulate NADP(H) binding, namely loop D, loop E and Tt βD378 (green asterisk), are close to the proton channel opening (red asterisk) and the Asp-Arg salt-bridge (blue asterisks). dII is shown as surface. NADP(H) is modeled as black spheres. (B) “Division of labor” of enzymatic activities within a TH dimer: one half of the dimer carries out hydride transfer and the other translocates proton. (C) Coordination between dII and dIII in one half of the TH dimer for the forward reaction. A proton is represented by a red cross. Two halves of the TH dimer cycle out of phase; for simplicity of representation, the other half is shown in grey and remains unchanged.
In general terms, we postulate that the two halves of the TH dimer cycle out of phase between occluded and open states of dIII that are coordinated to inward-facing and outward-facing conformations of the proton channel. After hydride transfer reaction, dIII becomes occluded in the face-up orientation, whereupon it flips to the face-down orientation without NADP(H) exchange with the solvent pool of NADP(H). For the forward reaction (NADPH product with a proton translocated inward), the interaction between the occluded dIII and the membrane domain converts dII from the deprotonated occluded state to become outward-facing, followed by the conversion of dIII to the open state, switching dII to inward-facing and eventually flipping up of dIII. Dissociations of both NADPH from dIII and the proton from dII to the solvent are required for the catalytic cycle to proceed. For hydride transfer in the reverse direction, the sequence of events is similar except that dII proceeds from proton-occluded→inward-facing→outward-facing (Fig. S7). The proton motive force is coupled to the dissociation of NADP(H) and, therefore, to the equilibrium for hydride transfer between NAD(H) and NADP(H).
Supplementary Material
Acknowledgments
We thank Y. Hatefi for commitment and inspiration on TH; M. Yeager, H. Heaslet, J. Chartron and R. Akhouri for their work on E. coli holo-TH; M. Saier and Z. Zhang for providing the bacterial strain; M. Soltis and D. Terrell for the Gryphon robot; V. Cherezov and M. Caffrey for helpful comments and suggestions; and W. Liu for the help of using the RockImager. We thank the staff of the Stanford Synchrotron Radiation Lightsource and the staff of the Northeastern Collaborative Access Team at the Advanced Photon Source for assistance with data collection. This research was funded by NIH 5R01GM061545 2004 – 2008, NIGMS 1R01GM103838 - 01A1, GM095600 (to R.B.G) and supported by PA-10-228, Structural Biology of Membrane Proteins. The work was performed at the National Resource for Automated Molecular Microscopy which is supported by a grant from the NIH NIGMS (P41GM103310). A. Moeller is supported by NIH GM073197.
Footnotes
Atomic coordinates and diffraction data for the structures reported here are deposited in the RCSB Protein Data Bank under accession codes 4O93 and 4O9U.
The authors declare no competing financial interests.
Author Contributions: For the crystallography research, J.H.L. and C.D.S. designed experiments; J.H.L., M.Y. and C.D.S. performed experiments; J.H.L. and C.D.S. analyzed the data. For the cryo-EM research, A.M. and J.A.S. designed and performed experiments, and analyzed data. L.A.S. and R.B.G. contributed the Tt TH operon for expression and initial purification. L.A.S., J.H.L. and R.B.G. created holo-TH constructs, expressed and assayed mutants. J.H.L., R.B.G. and C.D.S. wrote the manuscript.
Materials and Methods
References and Notes
- 1.Hatefi Y, Yamaguchi M. FASEB journal. 1996;10:444–452. doi: 10.1096/fasebj.10.4.8647343. [DOI] [PubMed] [Google Scholar]
- 2.Arkblad EL, Tuck S, Pestov NB, Dmitriev RI, Kostina MB, Stenvall J, Tranberg M, Rydström J. Free Radic Biol Med. 2005;38:1518–1525. doi: 10.1016/j.freeradbiomed.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 3.Meimaridou E, et al. Nature genetics. 2012;44:740–742. doi: 10.1038/ng.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freeman HC, Hugill A, Dear NT, Ashcroft FM, Cox RD. Diabetes. 2006;55:2153–2156. doi: 10.2337/db06-0358. [DOI] [PubMed] [Google Scholar]
- 5.Toye AA, et al. Diabetologia. 2005;48:675–686. doi: 10.1007/s00125-005-1680-z. [DOI] [PubMed] [Google Scholar]
- 6.Pedersen A, Karlsson GB, Rydström J. Journal of Bioenergetics and Biomembranes. 2008;40:463–473. doi: 10.1007/s10863-008-9170-x. [DOI] [PubMed] [Google Scholar]
- 7.Bizouarn T, Sazanov LA, Aubourg S, Jackson JB. Biochimica et biophysica acta. 1996;1273:4–12. doi: 10.1016/0005-2728(95)00125-5. [DOI] [PubMed] [Google Scholar]
- 8.Prasad GS, Sridhar V, Yamaguchi M, Hatefi Y, Stout CD. Nat Struct Biol. 1999;6:1126–1131. doi: 10.1038/70067. [DOI] [PubMed] [Google Scholar]
- 9.White SA, Peake SJ, McSweeney S, Leonard G, Cotton NP, Jackson JB. Structure. 2000;8:1–12. doi: 10.1016/s0969-2126(00)00075-7. [DOI] [PubMed] [Google Scholar]
- 10.Prasad GS, et al. Biochemistry. 2002;41:12745–12754. doi: 10.1021/bi020251f. [DOI] [PubMed] [Google Scholar]
- 11.Johansson T, Oswald C, Pedersen A, Törnroth S, Okvist M, Karlsson BG, Rydström J, Krengel U. Journal of molecular biology. 2005;352:299–312. doi: 10.1016/j.jmb.2005.07.022. [DOI] [PubMed] [Google Scholar]
- 12.Cotton NP, White SA, Peake SJ, McSweeney S, Jackson JB. Structure. 2001;9:165–176. doi: 10.1016/s0969-2126(01)00571-8. [DOI] [PubMed] [Google Scholar]
- 13.Sundaresan V, Chartron J, Yamaguchi M, Stout CD. Journal of molecular biology. 2005;346:617–629. doi: 10.1016/j.jmb.2004.11.070. [DOI] [PubMed] [Google Scholar]
- 14.Sundaresan V, Yamaguchi M, Chartron J, Stout CD. Biochemistry. 2003;42:12143–12153. doi: 10.1021/bi035006q. [DOI] [PubMed] [Google Scholar]
- 15.Venning JD, et al. J Biol Chem. 2001;276:30678–30685. doi: 10.1074/jbc.M104429200. [DOI] [PubMed] [Google Scholar]
- 16.Meuller J, Hu X, Bunthof C, Olausson T, Rydström J. Bba-Bioenergetics. 1996;1273:191–194. doi: 10.1016/0005-2728(95)00154-9. [DOI] [PubMed] [Google Scholar]
- 17.Bizouarn T, Meuller J, Axelsson M, Rydström J. Bba-Bioenergetics. 2000;1459:284–290. doi: 10.1016/s0005-2728(00)00163-8. [DOI] [PubMed] [Google Scholar]
- 18.Bragg PD, Hou C. Arch Biochem Biophys. 2001;388:299–307. doi: 10.1006/abbi.2001.2298. [DOI] [PubMed] [Google Scholar]
- 19.Bizouarn T, et al. Biochimica et biophysica acta. 2000;1457:211–228. doi: 10.1016/s0005-2728(00)00103-1. [DOI] [PubMed] [Google Scholar]
- 20.Yamaguchi M, Stout CD, Hatefi Y. J Biol Chem. 2002;277:33670–33675. doi: 10.1074/jbc.M204170200. [DOI] [PubMed] [Google Scholar]
- 21.Althage M, Bizouarn T, Rydström J. Biochemistry. 2001;40:9968–9976. doi: 10.1021/bi0103157. [DOI] [PubMed] [Google Scholar]
- 22.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Journal of applied crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutton M, Day JM, Bizouarn T, Jackson JB. Eur J Biochem. 1994;219:1041–1051. doi: 10.1111/j.1432-1033.1994.tb18587.x. [DOI] [PubMed] [Google Scholar]
- 24.Yamaguchi M, Hatefi Y. J Biol Chem. 1995;270:16653–16659. doi: 10.1074/jbc.270.28.16653. [DOI] [PubMed] [Google Scholar]
- 25.Johansson C, Pedersen A, Karlsson BG, Rydström J. Eur J Biochem. 2002;269:4505–4515. doi: 10.1046/j.1432-1033.2002.03144.x. [DOI] [PubMed] [Google Scholar]
- 26.Jackson JB. Biochimica et biophysica acta. 2012;1817:1839–1846. doi: 10.1016/j.bbabio.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 27.Jackson JB, White SA, Quirk PG, Venning JD. Biochemistry. 2002;41:4173–4185. doi: 10.1021/bi012078d. [DOI] [PubMed] [Google Scholar]
- 28.Pedersen A, Johansson T, Rydström J, Karlsson BG. Bba-Bioenergetics. 2005;1707:254–258. doi: 10.1016/j.bbabio.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 29.Bizouarn T, van Boxel GI, Bhakta T, Jackson JB. Nucleotide binding affinities of the intact proton-translocating transhydrogenase from Escherichia coli. Biochimica et biophysica acta. 2005;1708:404–410. doi: 10.1016/j.bbabio.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 30.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nature Protocols. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McPhillips TM, McPhillips SE, Chiu HJ, Cohen AE, Deacon AM, Ellis PJ, Garman E, Gonzalez A, Sauter NK, Phizackerley RP, Soltis SM, Kuhn P. Blu-Ice and the Distributed Control System: software for data acquisition and instrument control at macromolecular crystallography beamlines. J Synchrotron Radiat. 2002;9:401–406. doi: 10.1107/s0909049502015170. [DOI] [PubMed] [Google Scholar]
- 32.Tanley SW, Diederichs K, Kroon-Batenburg LM, Schreurs AM, Helliwell JR. Experiences with archived raw diffraction images data: capturing cisplatin after chemical conversion of carboplatin in high salt conditions for a protein crystal. J Synchrotron Radiat. 2013;20:880–883. doi: 10.1107/S0909049513020724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Powell HR, Johnson O, Leslie AG. Autoindexing diffraction images with iMosflm. Acta crystallographica. Section D, Biological crystallography. 2013;69:1195–1203. doi: 10.1107/S0907444912048524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta crystallographica. Section D, Biological crystallography. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta crystallographica. Section D, Biological crystallography. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta crystallographica. Section D, Biological crystallography. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goujon M, McWilliam H, Li WZ, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic acids research. 2010;38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.The PyMOL Molecular Graphics System, Version 1.5.0.4. Schrödinger, LLC; [Google Scholar]
- 43.Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. Journal of microscopy. 1987;146:113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 44.Moeller A, Kirchdoerfer RN, Potter CS, Carragher B, Wilson IA. Organization of the influenza virus replication machinery. Science. 2012;338:1631–1634. doi: 10.1126/science.1227270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. Journal of structural biology. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 46.Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau P-W, Lyumkis D, Potter CS, Carragher B. Appion: An integrated, database-driven pipeline to facilitate EM image processing. Journal of structural biology. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: Software tools to facilitate particle selection in single particle electron microscopy. Journal of structural biology. 2009;166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheres SHW, Núñez-Ramírez R, Sorzano COS, Carazo JM, Marabini R. Image processing for electron microscopy single-particle analysis using XMIPP. Nature Protocols. 2008;3:977–990. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. Journal of structural biology. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 50.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. Cryo-EM Structure of a Fully Glycosylated Soluble Cleaved HIV-1 Envelope Trimer. Science. 2013 doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. Journal of structural biology. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 53.Hohn M, Tang G, Goodyear G, Baldwin PR, Huang Z, Penczek PA, Yang C, Glaeser RM, Adams PD, Ludtke SJ. SPARX, a new environment for Cryo-EM image processing. Journal of structural biology. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 54.Lyumkis D, Brilot AF, Theobald DL, Grigorieff N. Likelihood-based classification of cryo-EM images using FREALIGN. Journal of structural biology. 2013;183:377–388. doi: 10.1016/j.jsb.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.