Abstract
Aim
To determine the prevalence and genotypic characterisation of extended spectrum beta-lactamases produced by gram negative bacilli isolated at Mbarara Regional Referral Hospital (MRRH).
Samples
Gram negative clinical isolates.
Study Design
Laboratory-based descriptive cross-sectional study.
Place and Duration of the Study
MRRH, June and August 2012.
Methods
Gram negative clinical isolates were sub cultured, and identified using biochemical tests. They were screened for ESBL by using oxyimino-cephalosporins and confirmed by double disc synergy Genotyping was performed using the PCR for TEM, SHV and CTX-M. Susceptibility pattern for the extended spectrum beta-lactamases, (ESBL) - positive isolates to other antibiotic classes was performed by the Kirby Bauer Technique.
Results
A total of 484 isolates were included in the study. The commonest ESBL producers were Escherichia coli (34%), followed by unidentified coliforms (19.3%) and Klebsiella spp. (12.7%). Phenotypically, 88/484 were ESBL producers while genotypically 213/ 484 possessed ESBL genes. The ESBL genes were blaCTX-M (146; 70%), blaSHV (72; 34%) and blaTEM (100; 47%). 87of 213 isolates expressed more than one ESBL gene. Of these 36 (7.4%) produced blaCTX-M/blaSHV, 28 (5.8%) blaCTX-M /blaTEM, 4 (0.8%) blaSHV/ blaTEM and 19 (3.9%) blaCTX-M/blaSHV/blaTEM. Sixty two (16%) were phenotypically and genotypically positive, 12 (3%) of the isolates were phenotypically positive but genotypically negative and 140 (37%) isolates were phenotypically negative but genotypically positive. The ESBL producers were highly susceptible to imipenem (95%), nitrofurantoin (66%) but less susceptible to ampicillin (4%) and ticarcillin (7%).
Conclusion
ESBL production among the Gram-negative clinical isolates at MRRH is very high with several isolates possessing multiple genes. The ESBL producers are highly susceptible to imipenem, but very resistant to ciprofloxacin.
Keywords: Antibiotics, resistance patterns, sensitivity patterns, ESBL, Uganda
1. INTRODUCTION
Resistance to β-lactam antimicrobial agents is on the rise worldwide. Extended-spectrum β-lactamases (ESBLs) are clinically important because they destroy cephalosporins, workhorse hospital antibiotics that are given as first-line agents to many severely-ill patients, including those with intra-abdominal infections, community acquired pneumonias and bacteraemias [1]. Delayed recognition of severe infections caused by ESBL producers, and inappropriate treatment with cephalosporins has been associated with increased mortality and morbidity. [2–4]. ESBLs are capable of hydrolyzing broad spectrum cephalosporins and monobactams.
In addition, ESBL-producing organisms exhibit co-resistance to many other classes of antibiotics resulting in limitation of therapeutic options. These enzymes can be chromosomal or plasmid-mediated and are encoded by genes such as TEM, SHV, CTX-M, VEB, PER, and OX [5].
Many ESBL producers are multi-resistant to non β-lactam antibiotics such as quinolones, aminoglycosides, chloromphenical and sulfamethoxazole-trimethoprim, narrowing treatment options [2,6,7]. Some producers achieve outbreak status, spreading among patients and locales, perhaps owing to particular pathogenicity traits.
The study was carried out to determine the prevalence and molecular characterization of Extended Spectrum Beta-Lactamases (ESBLs) produced by Gram-negative bacilli isolated at Mbarara Regional Referral Hospital (MRRH).
2. MATERIALS AND METHODS
This was a laboratory-based descriptive cross-sectional study that was conducted in the Microbiology Department of Mbarara University of Science and Technology. The study included all Gram-negative bacilli isolated from clinical samples (blood culture, urine, stool, peritoneal fluid, pus swab, high vaginal swab, cerebrospinal fluid, pleural fluid/aspirate, sputum, urethral and nasal swabs) collected from in patient and out patients of Mbarara Regional Referral Hospital between June and August 2012.
2.1 Culture and Identification of the Isolates
The specimens were cultured on Blood agar, Chocolate agar, Cystiene Lactose Electrolyte Deficient agar, (CLED), XLD and MacConkey agar depending on the sample type and incubated at 37°C over night. Identification of the isolates was carried out using conventional biochemical tests [8,9].
2.2 Phenotypic Screening for ESBLs
Screening for reduced susceptibility to third generation cephalosporins was carried out using cefotaxime and ceftazidime discs and double-disk synergy (DDS) method. The antibiotics used were; ceftazidime (30μg) and ceftazidime-clavulanic acid (20+10μg), cefotaxime (30μg) and cefotaxime-clavulanic acid (20+10μg). These antibiotics were placed at a distance of 30mm from each other. The plates were incubated overnight at 37°C. An increase of at east 5mm in the zone diameter for ceftazidime-clavulanic acid versus the zone diameter with ceftazidime tested alone was used to confirm the presence of ESBLs. A similar interpretation criterion was used when cefotaxime-clavulanic acid versus cefotaxime alone was uses, as recommended by the Clinical and Laboratory Standards Institute [10].
2.3 Molecular assays for detection of the β-Lactamase Genes
All the polymerase chain reaction (PCR) assays were performed at MBN Clinical Laboratories, Kampala Uganda where all the isolates were tested for the ESBL-resistance genes TEM, SHV and CTX-M, using primers as published earlier [11–13].
Frozen isolates were thawed and subcultured in Luria-Bertani and then incubated at 35 (+/−2) for 16–18 hrs.
2.3.1 DNA extractiion
2ml of the overnight culture was centrifuged at maximum speed of 13,000xg for 2 minutes, the supernatant fluid removed and the deposit re-suspended in 500 μl of TE buffer. This was spun again as before and the supernatant removed by gentle aspiration. To the deposit was added 100 μl of PCR water, vortexed, boiled for 15 minutes, cooled and centrifuged to collect the supernatant. This was then used as a DNA template for amplification of SHV, TEM, and CTX-M beta-lactamase genes. The extracted DNA from bacterial isolates was used as a template to detect SHV, TEM, and CTX-M beta-lactamase genes.
2.3.2 Amplification
PCR was carried out in a solution containing 200 μM concentration of dNTPs, 10 pM of each primer, 0.8mM/μl MgCl2, 0.5 U Taq polymerase and 50 ng DNA template in a final volume of 25 μl. The following thermocycler program was carried out for PCR experiments: 4 minutes denaturation at 94°C followed by 32 cycles of 1 minute at 94°C, 1 minute at the annealing temperature (55°C for blaSHV and blaCTX-M and 58°C for blaTEM) and 1 minute at 72°C with a final extension period of 10 minute at 72°C. K. pneumoniae 7881 and E. coli ATCC 35218 containing blaSHV, blaCTX-M and blaTEM gene were used as controls. 10 X master mix from AB gene was prepared (stable for one year at −20°C).
2.3.1.1 Gel electrophoresis
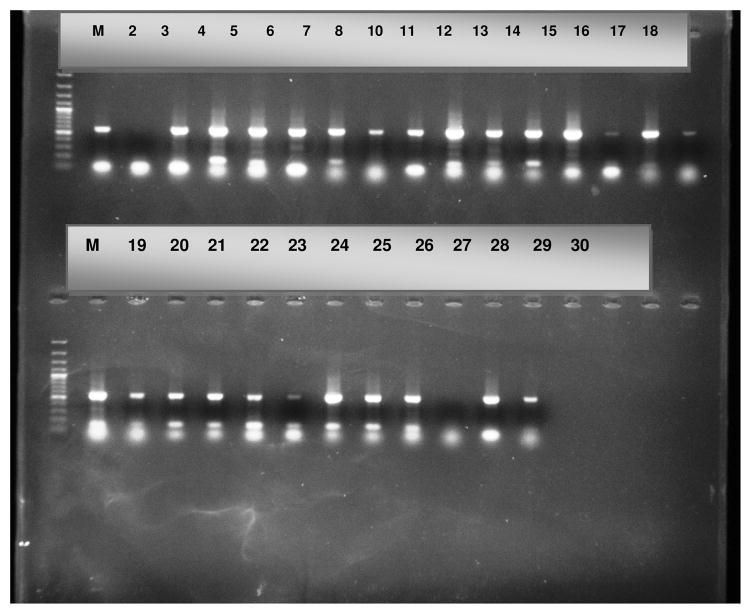
All PCR amplicons were verified by gel electrophoresis for amplicons of the following sizes (blaTEM 859bp, blaSHV 865bp, and blaCTX-M 544bp) which was performed at a voltage of 120V for 1 hour (Fig. 2).
Fig. 2.
Amplified PCR products
2.4 Resistance Patterns of ESBL-Positive Isolates
The susceptibility pattern of the confirmed ESBL-positive isolates was performed for the following antibiotics: gentamicin (10μg), ampicillin (10μγ), nitrofurantoin (300μg), ciprofloxacin (5μg), chloromphenicol (30μg), septrin (1.25/23.75μg), ticarcillin (75μg), imipenem (10μg) and nalidixic acid (30μg). The results were expressed as susceptible or resistant according to the criteria recommended by the Clinical Laboratory Standards Institute (CLSI) [10].
2.5 Quality Control
Klebsiella pneumoniae ATCC 700603 and Escherichia coli ATCC 25922 were used for the quality control of testing methods.
2.6 Data Analysis
Data was entered using EPI info version 7, exported to Excel and Stata version 11 (Stata Corporation, College Station, TX, USA) statistical packages for analysis.
3. RESULTS
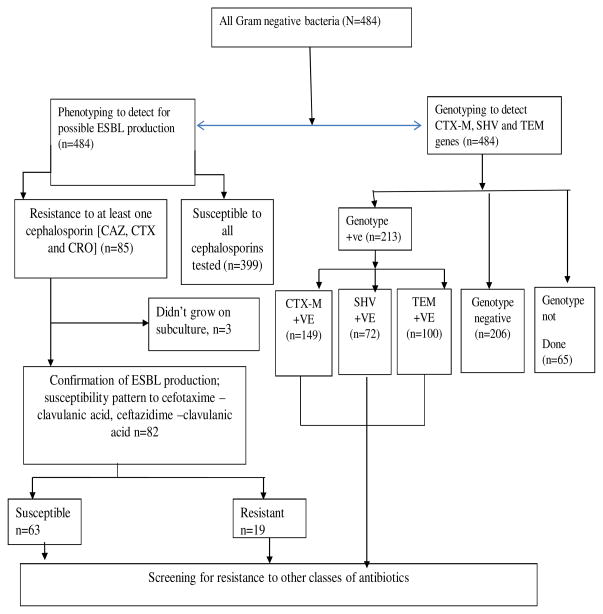
Of the 484 isolates phenotypically screened, 85 (29.9 %) isolates were resistant to at least one cephalosporin (ceftazidime, cefotaxime or ceftriaxone). Three of these isolates did not grow, and on phenotypic confirmation, 19 (23.2%) of the 82 isolates were confirmed to be ESBL (Fig. 1). The most common ESBL producing bacterial isolate was Escherichia coli (34.0%), Klebsiella spp. (12.7%) and Salmonella spp (9.9 %) (Fig. 3). Of the 419 isolates that were genotyped, 321 (50.8 %) isolates carried an ESBL coding gene. Majority of the isolates carried the blaCTX-M (146/213), followed by the blaTEM (100/213) and blaSHV (72/213).
Fig. 1.
Experimental design
Fig. 3.
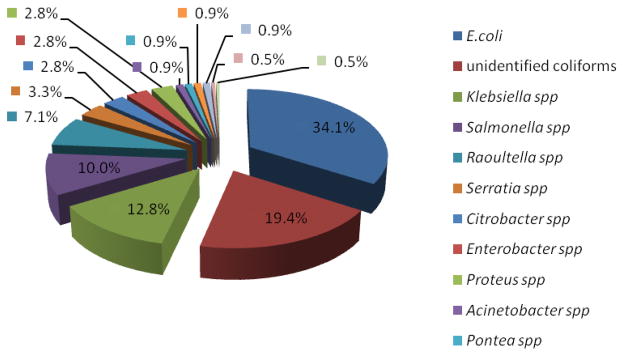
ESBL-producing bacteria
As shown in Table 1, eighty seven, 87 (40.8 %) out of the 213 isolates produced more than one ESBL gene genes. Majority of these isolates produced both CTX-M and SHV.
Table 1.
Combinations of ESBL genes produced by the isolates
| Genotype | Frequency | Percentage | Cum. freq |
|---|---|---|---|
| CTX-M and SHV (n=87) | 36 | 41.4 | 41.4 |
| CTX-M and TEM (n=87) | 28 | 32.2 | 73.6 |
| SHV and TEM (n=87) | 04 | 4.6 | 78.2 |
| CTX-M, SHV and TEM (n=87) | 19 | 21.8 | 100 |
Gel electrophoresis displaying the amplified blaSHV (862 bp) genes. Lane 2 positive control and Lane 3 negative control. Lanes 4–27, 29 and 30 are positive, Lane 28 is the only negative one. Lanes M represent the molecular weight marker (50 bp DNA ladder, Promega, Madison, USA).
As shown in Fig. 4, the ESBL producers were highly resistant to ticarcillin, ampicilin, tetracycline, and chloramphenicol but were highly susceptibility to imipenem.
Fig. 4.
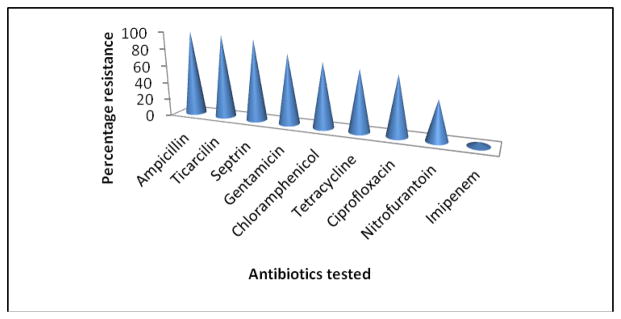
Resistance pattern of ESBL producing organisms
4. DISCUSSION
Using genotypic methods, the prevalence of ESBL in this study, was about 51 %. This correlates to what has been reported in other studies such as that from Israel (47 %) [14], and Nigeria (36.6 %)[15], but is much higher than that reported from Italy (6.3 %), Greece (27.4 %), Netherlands (2.0 %) and Germany (2.6%) [16]. The high prevalence has been majorly reported in low and middle income countries. This implies that the prevalence of ESBL may have a relationship with the economic status of a society. It may also have to do with the easy access to antibiotics in the study area. In our study, the commonest ESBL gene was CTX-M. This is different from studies performed elsewhere, where the commonest gene is SHV [17]. Furthermore, 87 out of the 213 isolates were able to express more than one resistance gene; blaCTX-M/ blaSHV, blaCTX-M /blaTEM, blaSHV/ blaTEM and blaCTX-M/ blaSHV/blaTEM. This is in agreement with studies carried out in other parts of the world that one bacterial isolate can express more than one resistance gene for example in Turkey, 68 had SHV and TEM, 61 (55.9 %) TEM and CTX-M, and 54 (49.5 %) SHV and CTX-M genes [17] and also in Iran, 7 isolates (21.87%) contained both genes TEM and SHV[18].
In this study, the predominant ESBL producers were Escherichia coli, unidentified coliform and Klebsiella spp. These findings are similar to previous studies reported elsewhere [19–23]. The susceptibility pattern of the ESBL-producing isolates in this study indicates a cross resistance of these ESBLs to many other common antibiotics. This has been recorded in other studies [6,7,24–27]. The mechanism behind this multiresistance phenomenon is genetic; it may be chromosomal or plasmid mediated. The gene encoding for resistance to ESBL and other antibiotic classes (e.g. quinolones) are often located on the same mobile DNA element (plasmid) [28], thus propagation of this plasmid during conjugation leads to development of multidrug resistance in previously sensitive organisms.
There was a discrepancy between genotypic and phenotypic methods for the detection of ESBL. This is due to the fact that genotypic assays are basically used to show the presence or absence of a defined resistance determinant gene. This equally is supported by the fact that the low prevalence phenotypically could possibly be due to non-expression of the genes. However low-level resistance mechanisms can be detected genotypically which otherwise can be difficult to detect using phenotypic methods because this is dependent on gene expression. Thus, if the genes are not expressed, then detection becomes difficult to achieve.
5. CONCLUSION
The prevalence of ESBL in our study is high, with the commonest ESBL gene being CTX-M, and a high proportion of isolates containing multiple genes. These ESBL producing isolates are highly resistant to ampicilin, ticarcilin, tetracycline and chloramphenicol are highly susceptible to imipenem.
Acknowledgments
The project described was supported by the MESAU-MEPI programmatic Award through award no 1R24TW00886 from the Fogarty International Centre.
Footnotes
ETHICAL APPROVAL
Ethical approval was obtained from various bodies as permission was sought from Department of Microbiology, Mbarara University of Science and Technology, Faculty of Medicine Research and Ethics committee, Institutional review committee and National Council for Sciences and Technology Institutional Review Committee.
COMPETING INTERESTS
Authors have declared that no competing interests exist.
Authors’ contributions
This work was carried out in collaboration between all authors. Author AM participated in the planning of the study, data entry and drafting of manuscript author FB supervised the laboratory work, author YB participated in the planning of the study and data collection. Author JB participated in the planning of the study and drafting and critical review of the manuscript. All authors read and approved the final manuscript.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the Fogarty International Center or the National Institutes of Health
References
- 1.Sobhan G, Nourkhoda S, Zamberi BS, Vasante KN, Mariana NS, Iraj PE, et al. Phenotypic and genotypic assay for detection of extended spectrum Beta-lactamases production by Klebsiella pneumonia isolates in Emam Reza Hospital in Tabriz, Iran. Journal of Pure and Applied Microbiology. 2011;5(1):01–10. [Google Scholar]
- 2.Schwaber MJ, Carmeli Y. Mortality and delay in effective therapy associated with extended-spectrum β-lactamase production in Enterobacteriaceae bacteraemia: A systematic review and meta-analysis. Journal of Antimicrobial Chemotherapy. 2007;60(5):913–920. doi: 10.1093/jac/dkm318. [DOI] [PubMed] [Google Scholar]
- 3.Tumbarello M, Sanguinetti M, Montuori E, et al. Predictors of mortality in patients with bloodstream infections caused by extended-spectrum-β-lactamase-producing Enterobacteriaceae: Importance of inadequate initial antimicrobial treatment. Antimicrobial Agents and Chemotherapy. 2007;51(6):987–1994. doi: 10.1128/AAC.01509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucher HW, Bradley S, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clinical Infectious Diseases. 2009;48(1):1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 5.Bush K, Jacoby GA. Updated functional classification of β-lactamases. Antimicrobial Agents and Chemotherapy. 2010;54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lautenbach E, Strom BL, Bilker WB, Patel JB, Edelstein PH, Fishman NO. Epidemiological investigation of fluoroquinolone resistance in infections due to extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumonia. Clinical Infectious Diseases. 2001;33(8):1288–1294. doi: 10.1086/322667. [DOI] [PubMed] [Google Scholar]
- 7.Pitout J, Nordmann K, Laupland B, Poirel L. Emergence of Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) in the community. Journal of Antimicrobial Chemotherapy. 2005;56(1):52–59. doi: 10.1093/jac/dki166. [DOI] [PubMed] [Google Scholar]
- 8.Galia Menashe, Borer Abraham, Yagupsky Pablo, Peled Nechama, Gilad Jacob, Fraser Drora, Riesenberg Klaris, Schlaeffer Francisc. Clinical significance on mortality of extended spectrum beta lactamase producing enterobacteriaceae isolates in nosocomial bacteraemia. Scandinavian Journal of infectious diseases. 2001;33(3):188–197. doi: 10.1080/00365540151060806. [DOI] [PubMed] [Google Scholar]
- 9.Biochemical tests for identification of medical bacteria. 3. London: Lippincott Williams and Wilkins; 2000. [Google Scholar]
- 10.Forbes BA, Sahm DF, Weissfeld AS. Bailey & Scott’s Diagnostic Microbiology. 11. USA: Mosby, Inc; 2002. [Google Scholar]; Clinical and Laboratory Standards Institute. Twentieth Informational Supplement, M100-S20. Vol. 30. UCLSI; Wayne, PA: 2010. Performance Standards for Antimicrobial Susceptibility Testing; p. 1. [Google Scholar]
- 11.Paterson DL, Hujer KM, Hujer AM, Yeiser B, Bonomo MD, Rice LB, Bonomo RA the International Klebsiella Study Group. Extended-spectrum β-lactamases in Klebsiella pneumoniae bloodstream isolates from seven countries: Dominance and widespread prevalence of SHV- and CTX-M-type β-lactamases. Antimicrobial Agents and Chemotherapy. 2003;47(11):3554–3560. doi: 10.1128/AAC.47.11.3554-3560.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hujer KM, Hujer AM, Hulten EA, Bajaksouzian S, Adams J, Donskey CJ, Ecker DJ, Massire C, Eshoo MW, Sampath R, Thomson JM, Rather PN, Craft DW, Fishbain JT, Ewell AJ, Jacobs MR, Paterson DL, Bonomo RA. Analysis of antibiotic resistance genes in multidrug resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrobial Agents and Chemotherapy. 2006;50(12):4114–4123. doi: 10.1128/AAC.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karami N, Hannoun C. Colonization dynamics of ampicillin resistant Escherichia coli in the infantile colonic microbiota. Journal of Antimicrobial Chemotherapy. 2008;62:703–708. doi: 10.1093/jac/dkn263. [DOI] [PubMed] [Google Scholar]
- 14.Galia Menashe, Johnson BM, Yagupsky Pablo, Peled Nechama, Gilad Jacob, Fraser Drora, Riesenberg Klaris, Schlaeffer Francisc. Clinical significance on mortality of extended spectrum beta lactamase producing enterobacteriaceae isolates in nosocomial bacteraemia. Scandinavian Journal of infectious diseases. 2001;33(3):188–197. doi: 10.1080/00365540151060806. [DOI] [PubMed] [Google Scholar]
- 15.Osazuwa F, Osazuwa EO. Detection of ESBL producing Klebsiella pneumonae and their antibiotic susceptibility pattern. Research Journal of Pharmaceutical, Biological and Chemical Sciences. 2011;2(1):603. [Google Scholar]
- 16.Bouchillon SK, Johnson BM, Hoban DJ, Johnson JL, Dowzicky MJ, Wu DH, Visalli MA, Bradford PA. Determining incidence of extended spectrum β-lactamase producing Enterobacteriaceae, vancomycin-resistant Enterococcus faecium and methicillin-resistant Staphylococcus aureus in 38 centres from 17 countries: The pearls study 2001–2002, AUGUST. 2004;24(2):119–124. doi: 10.1016/j.ijantimicag.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Spanu T, Luzzaro F, Perilli M, Amicosante G, Toniolo A, Fadda G. Occurrence of extended-spectrum β-lactamases in members of the family enterobacteriaceae in italy: implications for resistance to β-lactams and other antimicrobial drugs. Antimicrobial Agents and Chemotherapy. 2002;46:196–202. doi: 10.1128/AAC.46.1.196-202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yazdi M, Nazemi A, Mirinargasi M, Jafarpour M, Sharifi SH. Genotypic versus Phenotypic methods to detect Extended-Spectrum Beta-Lactamases (ESBLs) in Uropathogenic Escherichia coli. Annals of Biological Research. 2012;3(5):2454–245. [Google Scholar]
- 19.Nasehi Leila, Shahcheraghi Fereshteh, Nikbin Vajihe Sadat, NematzadehIran Shoeib. PER, CTX-M, TEM and SHV Beta-lactamases in Clinical Isolates of Klebsiella pneumoniae Isolated from Tehran. Iran Journal of Basic Medical Sciences. 2010;13(3) [Google Scholar]
- 20.Bhattacharya S. ESBL-From petri dish to the patient. Indian Journal of Medical Microbiology. 2006;24:20–4. doi: 10.4103/0255-0857.19889. [DOI] [PubMed] [Google Scholar]
- 21.Tsering Dechen C, Das Shyamasree, Adhiakari Luna, Pal Ranabir, Singh Takhellambam SK. Extended Spectrum Beta-lactamase Detection in Gram-negative Bacilli of Nosocomial Origin. Journal of Global Infectious Disease. 2009;1(2):87–92. doi: 10.4103/0974-777X.56247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Astal Z, Sharif FA, Abdallah SA, Fahd MI. Extended-spectrum beta-lactamases in E. coli isolated from community-acquired urinary tract infections in the Gaza Strip, Palestine. Annual Saudi Medical Journal. 2004;24:55–57. doi: 10.5144/0256-4947.2004.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vandana KE, Honnavar P. AmpC Beta-Lactamase among ESBL producing E. coli and Klebsiella pneumoniae. Journal of Clinical Diagnostic Research. 2009;3:1635–1656. [Google Scholar]
- 24.Shah AA, Hasan F, Ahmed S, Hameed A. Characteristics, epidemiology and clinical importance of emerging strains of Gram-negative bacilli producing extended-spectrum beta-lactamases. Research in Microbiology. 2004;155:409–421. doi: 10.1016/j.resmic.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 25.Ali AM, Rafi S, Qureshi AH. Frequency of extended spectrum beta lactamase producing gram negative bacilli among clinical isolates. at clinical laboratories of Army Medical College, Rawalpindi. Journal of Ayub Medical College Abbottabad. 2004;16(1):35–7. [PubMed] [Google Scholar]
- 26.Ali Arif Maqsood, Abbasi Brig Shahid Ahmed, Ahmed Mubasher. Frequency of extended spectrum beta-lactamases (esbl) producing nosocomial isolates. Pakistan Armed Forces Medical Journal. 2009;3 [Google Scholar]
- 27.Schwaber J, Navon-Venezia S, Schwartz D, Carmeli Y. High levels of antimicrobial coresistance among extended-spectrum-β-lactamase-producing Enterobacteriaceae. Antimicrobial Agents and Chemotherapy. 2005;49 (5):2137–2139. doi: 10.1128/AAC.49.5.2137-2139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morosini MI, García-Castillo M, Coque TM, et al. Antibiotic coresistance in extended-spectrum-β-lactamase-producing Enterobacteriaceae and in vitro activity of tigecycline. Antimicrobial Agents and Chemotherapy. 2006;50(8):2695–2699. doi: 10.1128/AAC.00155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]