Phospholipid Hydrolases
Hydrolases constitute an enormous proportion of our enzymes, being responsible for the initiation of most digestive processes and numerous physiological processes. They are defined as enzymes that use a molecule of water to degrade substrates including all four kinds of biological molecules; namely, nucleic acids (nucleases), proteins (proteases), lipids or fats (lipases), and carbohydrates or sugars (glycosidases). They include large families of acyl ester hydrolases, phosphate, pyrophosphate ester and diester hydrolases, and amide ester hydrolases.
Phospholipases constitute a class of hydrolases that catalyze the hydrolysis of acyl esters (deacylase activity) and phosphate esters (phosphodiesterase or phosphomonoesterase, also known as phosphohydrolase or phosphatase activity or sometimes pyrophosphatase activity) on phospholipids (diacylglycerophosphate esters and related compounds) (1). Phospholipases are defined by the position they hydrolize on the phospholipid backbone as shown in Fig. 1. Of course, many enzymes are named based on the initial assay used to discover or define their activity and then are later discovered to exhibit additional activities, and often their predominant physiological activity is quite different than their name implies. Also, there are certainly many enzymes not named as phospholipases that exhibit phospholipase activity, sometimes as a side or minor activity when presented with the appropriate phospholipid substrate.
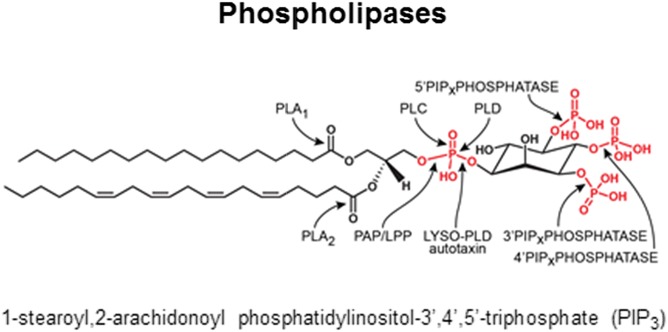
Fig. 1.
Site of hydrolase activity by phospholipase A1 (PLA1), phospholipase A2 (PLA2), phospholipase C (PLC), phosphatidic acid phosphatase (PAP)/lipid phosphate phosphatase (LPP), phospholipase D (PLD), lysophospholipid phospholipase D (LYSO-PLD, also known as autotaxin), and phosphatdiylinsositol polyphosphate phosphatase (PIPx phosphatase) which can be specific for the 3′, 4’ or 5′ position shown on a typical phospholipid, 1-steroyl,2-arachidonoyl-phosphatidylinositol-3′,4’,5′-triphosphate (PIP3).
Sphingolipids constitute a separate category of lipids from phospholipids (2, 3), yet they reside similarly in membranes and functionally often play similar roles to phospholipids with some overlapping biosynthetic pathways. Some of the enzymes that hydrolyze sphingolipids carry out similar reactions to the phospholipases, such as sphingomyelinase, which exhibits a phospholipase C activity toward sphingomyelin, but additionally there are deamidases that hydrolyze the acyl amide on the ceramide backbone of sphingolipids and glycosidases that hydrolyze carbohydrates of glycosphingolipids. Such enzymes are not included in this Thematic Review Series but are reviewed elsewhere (4).
Phospholipases Act on Membrane/Micellar Phospholipid Substrates
Phospholipases are major digestive enzymes and play a critical role in most physiological processes including the generation of numerous signaling lipids, and in aggregate, seem to affect all diseases in some manner. The phospholipid substrates generally exist in membranes or micelles due to their amphipathic character or low critical micelle concentration (CMC), yet the phospholipase enzymes can be either water-soluble and can associate with membranes/micelles to find their substrates or they may be membrane-bound proteins with an intimate supply of substrate. Thus, because phospholipases differ dramatically from “normal” water-soluble enzymes that bind water-soluble substrates in solution, special interfacial kinetics must be taken into account in studying phospholipases (5).
Phospholipases by definition must find their substrates that reside in a membrane or micelle, so the enzymes generally must first associate with the membrane or micelle and often are membrane-bound. The most detailed description of how a phospholipase acts to associate with a membrane and then to sequester its substrate phospholipid molecule in its active site has been developed for phospholipase A2 (PLA2) utilizing deuterium exchange mass spectrometry (6) to elucidate the interactions for docking and molecular dynamics to refine the structure, as illustrated in Fig. 2 (7).
Fig. 2.
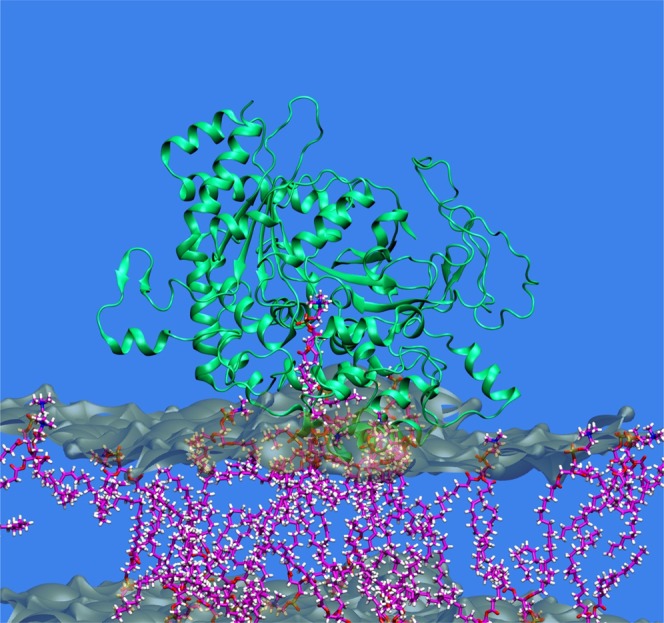
A phospholipase typically associates with a membrane and then sequesters a phospholipid substrate molecule in its catalytic site to carry out hydrolysis. The catalytic domain of human cPLA2 based on its crystal structure is shown optimized after extracting a typical substrate 1-palmitoyl,2-arachidonoyl phosphatidylcholine (PAPC) molecule into its catalytic site from the membrane containing 1-palmitoyl,2-oleoyl phosphatidylcholine (POPC) bilayers based on extensive molecular dynamics stimulations. Adapted from (7).
Phospholipase Focus
This series will begin with a consideration of an acyl hydrolase specific for the sn-2 position (stereospecific nomenclature) of the glycerolphosphate backbone of phospholipids, phospholipase A2, because that is the most well studied phospholipase and actually consists of a superfamily of enzymes including some 16 groups and many subgroups representing six main types of PLA2 (8). The secreted PLA2s are among the most well studied as they have been known for over a century from studies on snake venom and the pancreatic digestive enzyme, and many mechanistic and inhibitory studies have been carried out on their numerous isoforms.
In the first review in this series, which appears in this issue of the Journal of Lipid Research, Makoto Murakami and colleagues from the Tokyo Metropolitan Institute of Science describe “A new era of secreted phospholipases A2 (sPLA2)” (9). This will be followed with a review on “Cytosolic phospholipase A2: physiological function and role in disease” by Christina Leslie from the National Jewish Health Sciences in Denver (10), which will appear in the August issue. In the September issue will be a review by Sasanka Ramanadham and collaborators from the University of Alabama at Birmingham on “Calcium-independent phospholipases A2 (iPLA2s) and their roles in biological processes and diseases” (11).
The fourth type of PLA2 is PAP-acetyl hydrolase (PAFAH), also known as lipoprotein-association phospholipase A2 (Lp-PLA2), and this enzyme has been considered recently in the Journal by Diana Stafforini and Guy Zimmerman from the University of Utah (12). The lysosomal phospholipase A2 (L-PLA2) and the adipose phospholipase A (A-PLA2) have been less well studied than the other four types and are included in a recent comprehensive review on phospholipases A2 (8). As hydrolytic enzymes, the PLA2s are not just degradative and signaling enzymes, as some of them play an important functional role in producing the lysophospholipid substrate for acyl transferases to produce remodeled phospholipids containing a polyunsaturated fatty acid in their sn-2 position. Describing a more physiological setting, Nicolas Bazan from the Louisiana State University in New Orleans will describe recent work on the remodeling of phospholipids to become enriched in the omega-3 fatty acid docosahexenoic acid (DHA) and very long chain polyunsaturated fatty acids, as well as the role of these phospholipids in photoreceptor cell function and retinal degeneration.
Phospholipase A1 (PLA1) hydrolyzes the acyl chain on the sn-1 position of phospholipids. Hiroyuki Arai from the University of Tokyo and Junken Aoki from Tohoku University will review our current knowledge about phospholipase A1, including its biology and pathology.
Phosphohydrolases can cleave phospholipids on the glycerol side of the phosphodiester bond defined as phospholipase C (PLC), as reviewed by Lucio Cocco and collaborators from the University of Bologna on those PLCs acting on phosphatidylinositol polyphosphates in their article titled “Phosphoinositide-specific phospholipase C (PI-PLC) in health and disease” (13). Relatedly, David Brindley and colleagues from the University of Alberta in Edmonton review lipid phosphate phosphatase (LPP), also referred to as phosphatidic acid phosphatase (PAP2), in their paper, “Lipid phosphate phosphatases and their roles in mammalian physiology and pathology” (14). Importantly, members of this family hydrolyze phosphatidic acid (PA), lysophosphatidic acid (LPA), sphingosine-1-phosphate (S1P), ceramide-1-phosphate (C1P), and diacylglycerol pyrophosphate (DGPP), all of which are important signaling lipids. Phosphatidic acid phosphatase 1 (PAP1), which is also known as “lipin”, has been reviewed previously by Karen Reue and Jennifer R. Dwyer from the University of California, Los Angeles in the Journal (15).
Phosphohydrolases that cleave phospholipids on the polar or distal side of the phosphodiester bond are called phospholipase D (PLD), and are reviewed by Michael Frohman and his colleague at the State University of New York in Stony Brook in their article “Physiological and pathophysiological roles for phospholipase D” (16). Gordon Mills and colleagues at the MD Anderson Cancer Center at the University of Texas in Houston have reviewed those PLD enzymes acting on lysophospholipids in their paper titled “Autotaxin, a lysophospholipase D with pleomorphic effects in oncogenesis and cancer progression” (17).
Finally, phospholipases generally referred to as phosphatases can hydrolyze the various phosphate esters on specific positions on the inositol sugar attached to phosphatidylinositol polyphosphates, and Michael Wakelem and colleagues from the Babraham Institute in Cambridge will describe the various phosphatidylinositolphosphate phosphatases and cancer.
In this Thematic Review Series, we include for each type of phospholipase the reaction it primarily carries out, noting secondary reactions, the names (Group numbering or other system, systematic name, and common names), primary sequence differences between the various forms and homologies, three-dimensional structure(s) when crystal structures are solved, the biological functions, and the disease implications. All of the identified enzymes of the type covered by the review are summarized, though the reviews generally focus on the major form(s) that have been most well studied and their signaling function and disease implications.
Acknowledgments
I wish to thank the National Institutes of Health for grant GM 20,501-39 that has supported work in my laboratory on phospholipases for some forty years. I wish to thank Dr. Varnavas Mouchlis and Dr. Eoin Fahy for aid in the figure preparations.
REFERENCES
- 1.Dennis E. A. 1983. Phospholipases. In The Enzymes. 3rd edition. Vol. 16. P. Boyer, editor. Academic Press, New York. 307–53. [Google Scholar]
- 2.Fahy E., Subramaniam S., Brown H. A., Glass C. K., Merrill A. H., Jr, Murphy R. C., Raetz C. R., Russell D. W., Seyama Y., Shaw W., et al. 2005. A comprehensive classification system for lipids. J. Lipid Res. 46: 839–861. [DOI] [PubMed] [Google Scholar]
- 3.Fahy E., Subramaniam S., Murphy R. C., Nishijima M., Raetz C. R. H., Shimizu T., Spener F., van Meer G., Wakelam M. J. O., Dennis E. A. 2009. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 50: S9–S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merrill A. H. 2011. Sphingolipid and glycosphingolipid metabolic pathways in the era of sphingolipidomics. Chem. Rev. 111: 6387–6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carman G. M., Deems R. A., Dennis E. A. 1995. Lipid signaling enzymes and surface dilution kinetics. J. Biol. Chem. 270: 18711–18714. [DOI] [PubMed] [Google Scholar]
- 6.Cao J., Burke J. E., Dennis E. A. 2013. Using hydrogen-deuterium exchange mass spectrometry to define the specific interactions of the phospholipase A2 superfamily with lipid substrates, inhibitors and membranes. J. Biol. Chem. 288: 1806–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mouchlis V. D., Bucher D., McCammon J. A., Dennis E. A. 2015. Membranes serve as allosteric activators of phospholipase A2 enabling it to extract, bind, and hydrolyze phospholipid substrates. Proc. Natl. Acad. Sci. USA. 112: E516–E525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dennis E. A., Cao J., Hsu Y. H., Magrioti V., Kokotos G. 2011. Phospholipase A2 enzymes: physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem. Rev. 111: 6130–6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murakami M., Sato H., Miki Y., Yamamoto K., Taketomi Y. 2015. A new era of secreted phospholipases A2 (sPLA2). J. Lipid Res 56: 1248–1261. [DOI] [PMC free article] [PubMed]
- 10.Leslie C. C. Cytosolic phospholipase A2: physiological function and role in disease. J. Lipid Res. Epub ahead of print. April 2, 2015; 10.1194/jlr.R057588. [DOI] [PMC free article] [PubMed]
- 11.Ramanadham S., Ali T., Ashley J. W., Bone R. N., Hancock W. D., Lei X. Calcium-independent phospholipases A2 (iPLA2s) and their roles in biological processes and diseases. J. Lipid Res. Epub ahead of print. May 28, 2015; 10.1194/jlr.R058701. [DOI] [PMC free article] [PubMed]
- 12.Stafforini D. M., Zimmerman G. A. 2014. Unraveling the PAF-AH/Lp-PLA2 controversy. J. Lipid Res. 55: 1811–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cocco L., Follo M. Y., Manzoli L., Suh P. G. Phosphoinositide-specific phospholipase C (PI-PLC) in health and disease. J. Lipid Res. Epub ahead of print. March 27, 2015; 10.1194/jlr.R057984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang X., Benesch M. G. K., Brindley D. N. Lipid phosphate phosphatases and their roles in mammalian physiology and pathology. J. Lipid Res. Epub ahead of print. March 26, 2015; 10.1194/jlr.R058362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reue K., Dwyer J. R. 2009. Lipin proteins and metabolic homeostasis. J. Lipid Res. 50: S109–S114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson R. K., Frohman M. A. Physiological and pathophysiological roles for phospholipase D. J. Lipid Res. Epub ahead of print. April 29, 2015; 10.1194/jlr.R059220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Federico L., Kang J. J., Vellano C. P., Mills G. B. Autotaxin, a lysophospholipase D with pleomorphic effects in oncogenesis and cancer progression. J. Lipid Res. Epub ahead of print. May 14, 2015; 10.1194/jlr.R060020. [DOI] [PMC free article] [PubMed] [Google Scholar]