Abstract
Hydroxy FAs, one of the gut microbial metabolites of PUFAs, have attracted much attention because of their various bioactivities. The purpose of this study was to identify lactic acid bacteria with the ability to convert linoleic acid (LA) to hydroxy FAs. A screening process revealed that a gut bacterium, Lactobacillus acidophilus NTV001, converts LA mainly into 13-hydroxy-cis-9-octadecenoic acid and resulted in the identification of the hydratase responsible, fatty acid hydratase 1 (FA-HY1). Recombinant FA-HY1 was purified, and its enzymatic characteristics were investigated. FA-HY1 could convert not only C18 PUFAs but also C20 and C22 PUFAs. C18 PUFAs with a cis carbon-carbon double bond at the Δ12 position were converted into the corresponding 13-hydroxy FAs. Arachidonic acid and DHA were converted into the corresponding 15-hydroxy FA and 14-hydroxy FA, respectively. To the best of our knowledge, this is the first report of a bacterial FA hydratase that can convert C20 and C22 PUFAs into the corresponding hydroxy FAs. These novel hydroxy FAs produced by using FA-HY1 should contribute to elucidating the bioactivities of hydroxy FAs.
Keywords: arachidonic acid, docosahexaenoic acid, enzymology, fatty acid/metabolism, lipids, lipids/chemistry, lactic acid bacteria, hydroxy fatty acid, hydration, dehydration
Gut microbial metabolites of PUFAs have gained much attention because of their various physiological activities (1–5). Recently, we reported that a gut bacterium, Lactobacillus plantarum, generates hydroxy FAs, oxo FAs, and conjugated FAs from linoleic acid (LA) (6). The investigation of the presence of hydroxy FAs in the intestines of mice revealed that the metabolites of LA, 10-hydroxy-cis-12-octadecenoic acid and 13-hydroxy-cis-9-octadecenoic acid, were detected at higher levels in the intestines of specific pathogen-free (SPF) mice than in those of germ-free mice, indicating that gastrointestinal microbes play a role in modifying the FA profile of their host mice.
The physiological functions of 10-hydroxy FAs and their derivatives begin to become clear. 10-Hydroxy-cis-12-octadecenoic acid ameliorates intestinal epithelial barrier impairment via the G protein-coupled receptor 40 (GPR40)-MAPK/ERK kinase (MEK)-ERK pathway and may be useful in the treatment of tight junction-related disorders such as inflammatory bowel disease (3). 10-Oxo-cis-12-octadecenoic acid potently activates PPARγ, a master regulator of adipocyte differentiation, and may be involved in the regulation of host energy metabolism (5).
The production of 10-hydroxy-cis-12-octadecenoic acid from LA has been reported in many bacteria, including Lactobacillus acidophilus (7), L. plantarum (8, 9), Streptococcus bovis (10), and Stenotrophomonas nitritireducens (11). In our previous study, we reported that the enzyme linoleic acid hydratase (CLA-HY) from L. plantarum AKU 1009a catalyzes the hydration of the cis-9 double bond in C16 and C18 FAs, forming the corresponding 10-hydroxy FAs (12). Volkov et al. (13) reported that SPH, a hydratase from Streptococcus pyogenes M49, is a myosin-cross-reactive antigen (MCRA) family protein that catalyzes the hydration of the cis-9 and cis-12 double bonds in C16 and C18 FAs, forming the corresponding 10-hydroxy and 10,13-dihydroxy FAs.
The production of 13-hydroxy-cis-9-octadecenoic acid from LA has also been reported in some anaerobic bacteria. Hudson et al. (10) reported that a ruminant bacterium, S. bovis JB1, converts LA into 13-hydroxy-cis-9-octadecenoic acid, and Kishimoto et al. (14) reported that L. acidophilus IFO 13951 converts LA into the (S)-form of 13-hydroxy-cis-9-octadecenoic acid. We reported that Pediococcus sp. AKU 1080 converts LA into three products, 10-hydroxy-cis-12-octadecenoic acid, 13-hydroxy-cis-9-octadecenoic acid, and 10,13-dihydroxy-octadecanoic acid (15). However, the hydratase responsible for the conversion of LA to 13-hydroxy-cis-9-octadecenoic acid has not yet been identified, and the physiological activities of 13-hydroxy-cis-9-octadecenoic acid are unclear. As for the known bacterial hydratases, the length of the carbon chain in the substrate FA is limited to C16 and C18 (1, 12, 13, 16–19).
In this paper, we report the identification of a novel hydratase from L. acidophilus that converts LA into 13-hydroxy-cis-9-octadecenoic acid and produces various hydroxy FAs. Through the screening of lactic acid bacteria for the ability to convert LA into hydroxy FAs, we found that L. acidophilus NTV001 has a high ability to convert LA into 13-hydroxy-cis-9-octadecenoic acid. We subsequently identified the hydratase responsible for this conversion and named it fatty acid hydratase 1 (FA-HY1). Recombinant FA-HY1 was purified, and its enzymatic characteristics were investigated. FA-HY1 shows broad substrate specificity and catalyzes the hydration of C16, C18, C20, and C22 FAs. Thus, we succeeded in the production of 13-hydroxy-cis-9-octadecenoic acid and a variety of hydroxy FAs using FA-HY1, which is leading to the development of novel, potentially bioactive hydroxy FAs.
MATERIALS AND METHODS
Chemicals
FAs used as substrates were purchased from Nu-Chek Prep, Inc. (Elysian, MN), Larodan Fine Chemicals (Malmo, Sweden), and Cayman Chemical (MI). Eicosatetraenoic acid used as substrate was prepared from Mortierella alpina oil in our laboratory (20). FA-free (<0.02%) BSA was purchased from Sigma (St. Louis, MO). GC standard samples of 13-hydroxy-cis-9-octadecenoic acid, 10-hydroxy-cis-12-octadecenoic acid, and 10,13-dihydroxy-octadecanoic acid were prepared as previously described (15). All other chemicals used were of analytical grade and are commercially available.
Microorganism, cultivation, and reaction conditions for screening
Lactic acid bacteria used for this study were preserved in our laboratory (AKU Culture Collection, Division of Applied Life Science, Faculty of Agriculture, Kyoto University, Kyoto, Japan) and obtained from other culture collections (Japan Collection of Microorganisms, Saitama, Japan; ATCC, VA; and National Institute of Technology, Chiba, Japan). The bacteria were cultivated in Lactobacilli MRS broth (Difco, Detroit, MI). Screw-capped tubes (16.5 × 125 mm) containing 15 ml of Lactobacilli MRS medium were inoculated with each strain and then incubated, with shaking (120 strokes/min), under O2-limited conditions [<0.1% by O2 adsorbent (21)] in the sealed condition at 28°C or 37°C for 2–3 days. After cultivation, the cells were harvested by centrifugation (8,000 g, 10 min) and washed twice with 0.85% NaCl. The reactions were carried out in 1 ml of reaction mixture [100 mM potassium phosphate buffer (KPB), pH 6.5], containing 10 mM LA, 0.3 mg BSA, and washed cells of each strain at 37°C for 16 h with shaking (130 strokes/min) under anaerobic conditions maintained using Anaeropack Kenki (Mitsubishi Chemical, Tokyo, Japan). Lipid analyses were then carried out on the reaction mixtures.
Cloning and expression of recombinant proteins
We conducted a homology search, using the BLAST program, of the genomes of L. acidophilus strains and found that there were two proteins in L. acidophilus ATCC4796 that share substantial homology with the CLA-HY protein sequence. We designed primers based on the genome sequence of the L. acidophilus ATCC4796 and amplified DNA regions containing each candidate gene (fa-hy1 and fa-hy2) using L. acidophilus NTV001 genomic DNA as the template. Genomic DNA was extracted from L. acidophilus NTV001 using a DNeasy Blood and Tissue kit (QIAGEN, Tokyo, Japan) according to the manufacturer’s instructions. Region 1-forward primer (5′-GATGCTTTTTCATGGCACTGGGT-3′) and region 1-reverse primer (5′-CACCAGTCGTCAAAGCTTCA-3′) were used to amplify the DNA region containing the fa-hy1 gene. Region 2-forward primer (5′-CGCTTGTTAACTGGTAGAACACATCA-3′) and region 2-reverse primer (5′-CGTGACTTTAACTGGGATGA-3′) were used to amplify the DNA region containing the fa-hy2 gene. The open reading frames of the two candidate genes in L. acidophilus NTV001 were identified by sequencing, using a Beckman-Coulter CEQ8000 (Beckman-Coulter, Fullerton, CA). Based on the sequence results, we designed primers to clone each candidate gene. The fa-hy1-forward primer (5′-ACACATATGCATTATAGTAGTGGTAAT-3′) and fa-hy1-reverse primer (5′-AAACTCGAGCTAAACCAACTTATACTTCT-3′) were used to amplify the fa-hy1 gene, including its stop codon (1,773 bp). The fa-hy2-forward primer (5′-CACCATATGTATTATTCCAATGGTAAT-3′) and fa-hy2-reverse primer (5′-CGCCTCGAGTTAGACTAAATTTGCTTCT-3′) were used to amplify the fa-hy2 gene, including its stop codon (1,776 bp). The underlined bases in these sequences point out the restriction enzyme recognition sites engineered into each primer. Each of the amplified products was treated with NdeΙ and XhoΙ and then ligated into the expression vector pET-21b (Novagen, WI), yielding the plasmids pET21b-fa-hy1 and pET21b-fa-hy2.
Escherichia coli Rosetta2 (DE3) cells were transformed with either pET21b-fa-hy1 or pET21b-fa-hy2, yielding E. coli Rosetta2 (DE3)/pET21b-fa-hy1 or E. coli Rosetta2 (DE3)/pE21b-fa-hy2. The cells were grown in Luria-Bertani medium containing 50 μg/ml ampicillin and 34 μg/ml chloramphenicol at 37°C until the optical density at 600 nm reached 0.5. Expression was induced by the addition of 1.0 mM isopropyl-β-d-thiogalactopyranoside, and the cells were grown at 16°C for an additional 16 h. The cells were harvested by centrifugation (12,000 g, 10 min), washed twice with 0.85% NaCl, and kept at −20°C until use.
Product analysis of reaction mixtures containing E. coli transformants with LA as the substrate
The reactions were carried out in 1 ml reaction mixtures (100 mM KPB, pH 6.5), containing 10 mM LA as the substrate, 0.5 mg BSA, and 60 mg of the washed cells of the appropriate E. coli transformant [E. coli Rosetta2 (DE3)/pET21b-fa-hy1 or E. coli Rosetta2 (DE3)/pET21b-fa-hy2]. Reaction mixtures were incubated at 37°C for 16 h with shaking (130 strokes/min) under anaerobic conditions maintained using Anaeropack Kenki (Mitsubishi Chemical). After incubation, total lipids were extracted, and the products were analyzed using GC. The peaks representing the reaction products were identified by comparing their retention times with those of reference standards, and the chemical structures of products were confirmed by GC/MS analysis of TMS derivatives.
Lipid analyses
Before lipid extraction, n-heptadecanoic acid was added to the reaction mixture as an internal standard. Lipids were extracted from the reaction mixture (1 ml) using 5 ml of chloroform-methanol-1.5% KCl (2:2:1, by volume) according to the Bligh-Dyer method, then the solvent was removed using a rotary evaporator. Methylation of FAs was carried out by incubation with 4% methanolic HCl at 50°C for 20 min. After methylation, the FA methyl esters were extracted with n-hexane, and the solvent was removed using a rotary evaporator. The resulting FA methyl esters were analyzed by GC using a Shimadzu GC-1700 gas chromatograph equipped with a flame-ionization detector and a split-injection system, fitted with a capillary column [SPB-1; 30 m length × 0.25 mm inner diameter (ID); Supelco, PA]. The initial column temperature of 180°C was maintained for 30 min, then increased to 220°C at a rate of 40°C/min and finally maintained at that temperature for 14 min. Helium was used as the carrier gas at a flow rate of 2.51 ml/min. FA methyl esters obtained from dehydration reactions were analyzed by GC using the Shimadzu GC-1700 gas chromatograph fitted with a capillary column (TC-70; 60 m length × 0.25 mm ID; GL Sciences, Tokyo, Japan). The initial column temperature of 180°C was maintained for 10 min, then increased to 260°C at a rate of 5°C/min, and finally maintained at that temperature for 9 min. Helium was used as the carrier gas at a flow rate of 0.97 ml/min.
Protein purification
All of the steps in the FA-HY1 purification described below were performed at 4°C. Washed cells of the E. coli transformant expressing FA-HY1 [E. coli Rosetta2 (DE3)/pET21b-fa-hy1] were suspended in 50 mM KPB (pH 6.0) and disrupted using an ultrasonic oscillator (Kubota, Tokyo, Japan). After ultracentrifugation at 100,000 g for 60 min, the supernatants were loaded on a Mono Q 10/100 GL column (GE Healthcare, Tokyo, Japan) equilibrated with 50 mM KPB (pH 6.0). The protein was eluted using a linear NaCl gradient from 0 to 1 M in 50 mM KPB (pH 6.0), at a flow rate of 2 ml/min, using an ÄKTA FPCL system (GE Healthcare). Fractions containing hydration activity were collected and concentrated by ultrafiltration using Vivaspin® Turbo 15 (10,000 molecular weight cut-off) (Sartorius, Tokyo, Japan) centrifugal concentrators. The concentrated solutions were loaded on a Superdex 200 10/300 GL column (GE Healthcare) equilibrated with 50 mM KPB (pH 6.0). Elution was performed at a flow rate of 0.5 ml/min. Fractions containing hydration activity were collected, concentrated as above, and kept at 4°C until use.
UV-visible spectra and cofactor identification
UV-visible spectra of purified FA-HY1 (10 mg/ml), in the 300–500 nm range, were acquired using a UV-1700 UV-visible spectrophotometer (Shimadzu, Kyoto, Japan). To identify the flavin cofactor, samples of FA-HY1 (10 mg/ml) were heated at 95°C for 10 min. The precipitated protein was removed by centrifugation (20,000 g, 10 min), and the supernatants were used for HPLC analysis. Samples containing flavin adenine dinucleotide (FAD; 40 μM) or flavin mononucleotide (FMN; 40 μM) were prepared as reference standards. Reversed-phase HPLC separation was performed on a Cosmosil 5C18-MS-II packed column (3.0 mm ID × 150 mm length; Nacalai Tesque, Kyoto, Japan) with a methanol-5 mM ammonium acetate (20:80, by volume) solvent and a flow rate of 0.8 ml/min. The effluent was monitored at 350 nm using a UV detector.
Enzyme reaction conditions
The standard reaction conditions were as follows. Each reaction was carried out in 1 ml of reaction mixture (50 mM KPB, pH 6.0), containing 10 mM LA as a substrate, 0.3 mg BSA, 0.1 mM FAD, and 50 μg FA-HY1 at 37°C for 15 min with shaking (130 strokes/min). All reactions were performed in triplicate. Data presented in the figures and table represent the averages of three separate experiments that were reproducible within ± 10%.
The effects of cofactors were examined by adding cofactors in various combinations: 5 mM NAD+, 5 mM NADH, 5 mM NADP+, 5 mM NADPH, 0.1 mM FAD, and 0.1 mM FMN. The effect of FAD concentrations on enzyme activity was examined by varying its concentrations from 0 to 0.3 mM. The optimum reaction pH was determined using sodium citrate buffer (50 mM; pH 3.0–4.0), sodium succinate buffer (50 mM; pH 4.0–6.0), KPB (50 mM; pH 5.0–8.0), and Tris-HCl (50 mM; pH 7.0–9.0). The pH stability was determined by measuring the enzyme activity after incubating the enzyme at 4°C for 24 h in the following buffers: sodium citrate buffer (50 mM; pH 3.0–4.0), sodium succinate buffer (50 mM; pH 4.0–6.0), KPB (50 mM; pH 5.0–8.0), and Tris-HCl (50 mM; pH 7.0–9.0). The optimum reaction temperature was examined by varying the temperature from 20°C to 60°C in 50 mM KPB (pH 6.0). Thermal stability was determined by measuring the enzyme activity in 50 mM KPB (pH 6.0) after incubating the enzyme at various temperatures (4°C–60°C) for 30 min.
Kinetic analysis
Reactions were carried out under standard reaction conditions, with some modification of the enzyme and LA substrate concentrations. The kinetics of LA hydration were studied using purified enzyme (1 μg/ml) and 10–100 μM LA substrate; the reaction time was 5 min. The Km value was evaluated with the amounts of the product. The kinetic parameters were calculated by fitting the experimental data to either the Hill equation or the Michaelis-Menten equation, using KaleidaGraph 4.0 (Synergy Software Inc., PA) software.
Substrate specificity of FA-HY1
To investigate the substrate specificity of FA-HY1, the purified FA-HY1 was incubated with various FAs. The reactions were carried out in 1 ml reaction mixtures (50 mM KPB, pH 6.0), containing 10 mM substrate, 0.3 mg BSA, 0.1 mM FAD, and 0.1 mg of the purified enzyme. All reaction mixtures were incubated at 37°C for 16 h with shaking (130 strokes/min) under anaerobic conditions maintained using Anaeropack Kenki (Mitsubishi Chemical).
Isolation and identification of newly generated FAs
Reaction products were separated using an Isolera One automated flash purification system equipped with a SNAP Ultra 10 g cartridge (Biotage, Stockholm, Sweden). The mobile phase, n-hexane-diethyl ether (8:2–6:4; by volume), was used to elute the column at a flow rate of 12 ml/min, and the effluent was monitored at 200 and 233 nm using a UV detector. Solvents were removed from the isolated FAs using a rotary evaporator. After methylation, the methyl esters of the isolated FAs were transformed into their pyrrolidide and TMS derivatives. Pyrrolidide derivatives were prepared by treating the FA methyl esters with pyrrolidine-acetic acid (10:1, by volume) in screw-cap tubes for 1 h at 100°C, followed by extraction with n-hexane. The organic extract was washed with water and dried over anhydrous Na2SO4, and then the solvent was removed under vacuum in a rotary evaporator. TMS derivatives were prepared by treating the FA methyl esters with a mixture of TMS agent (pyridine-hexamethyldisilazane-trimethylchlorosilane, 9:3:1, by volume) in screw-cap tubes for 30 min at 60°C, followed by extraction with chloroform. The chemical structures of isolated FAs were determined by GC/MS, proton NMR (1H-NMR), 1H-1H double-quantum-filtered chemical-shift correlation spectroscopy (DQF-COSY), and two-dimensional nuclear Overhauser effect spectroscopy (NOESY).
Determination of absolute configuration and enantiomeric excess
The absolute configurations of 13-hydroxy-cis-9-octadecenoic acid and 13-hydroxy-cis-9,cis-15-octadecadienoic acid were determined using Mosher’s method (22). The enantiomeric excess (ee) values of the 13-hydroxy-cis-9-octadecenoic acid and 13-hydroxy-cis-9,cis-15-octadecadienoci acid were determined using HPLC as described below. First, 13-oxo-cis-9-octadecenoic acid and 13-oxo-cis-9,cis-15-octadecadienoic acid were synthesized from each hydroxy FA using the Jones oxidation method (23). Then, (RS)-13-hydroxy-cis-9-octadecenoic acid and (RS)-13-hydroxy-cis-9,cis-15-octadecadienoic acid were synthesized by reduction of each keto FA to the hydroxy FA using NaBH4 (24). HPLC separation was performed on a Chiralpak IA column (4.6 mm ID × 250 mm length; Daicel, Tokyo, Japan), using acetonitrile-water (65:35, by volume) containing 0.2% formic acid as the solvent, at a flow rate of 1.0 ml/min. The effluent was monitored at 205 nm using a UV detector.
Dehydration reaction catalyzed by FA-HY1
Dehydration reactions, catalyzed by FA-HY1, were examined using 13-hydroxy-cis-9-octadecenoic acid as the substrate. To produce 13-hydroxy-cis-9-octadecenoic acid, reactions were carried out in 1 ml of reaction mixture (100 mM KPB, pH 6.5), containing 100 mg LA, 10 mg BSA, 0.1 mM FAD, and 300 mg of washed cells of the E. coli transformant expressing FA-HY1. Reaction mixtures were incubated at 37°C for 16 h with shaking (130 strokes/min) under anaerobic conditions. After incubation, total lipids were extracted, and the 13-hydroxy-cis-9-octadecenoic acid was purified using an Isolera One automated flash purification system equipped with a SNAP Ultra cartridge, as described above.
Dehydration reactions were carried out in 1 ml of reaction mixture (50 mM KPB, pH 6.0), containing 10 mM purified 13-hydroxy-cis-9-octadecenoic acid, 0.3 mg BSA, 0.1 mM FAD, and 0.1 mg FA-HY1. Reaction mixtures were incubated at 37°C for 16 h with shaking (130 strokes/min) under anaerobic conditions. After incubation, total lipids were extracted and analyzed using GC, as described above. Reaction products were separated by reverse-phase HPLC, using a Shimadzu SLC-10A system equipped with a Cosmosil Cholester column (10 mm ID × 250 mm length; Nacalai Tesque). The column was eluted at a flow rate of 5 ml/min using a mobile phase consisting of acetonitrile-water (8:2, by volume) containing 0.2% acetic acid. The effluent was monitored at 205 nm using a UV detector. The chemical structures of the purified FAs were determined using data obtained from GC/MS analysis of the pyrrolidide derivatives, as well as 1H-NMR, DQF-COSY, and NOESY analyses.
RESULTS
Screening of lactic acid bacteria that convert LA into hydroxy FAs
The ability of lactic acid bacteria to convert LA into hydroxy FAs was investigated. We tested more than 300 strains belonging to genera such as Lactobacillus, Leuconostoc, Enterococcus, Streptococcus, Pediococcus, and so forth. Most of the lactic acid bacteria converted LA into 10-hydroxy-cis-12-octadecenoic acid, and some of them produced CLA (cis-9,trans-11-octadecadienoic acid and trans-9,trans-11-octadecadienoic acid). We found that L. acidophilus AKU1222 (NTV001) converted substantial quantities of LA into 13-hydroxy-cis-9-octadecenoic acid. Further analysis revealed that 96% of the product was 13-hydroxy-cis-9-octadecenoic acid, and the remainder consisted of 10-hydroxy-cis-12-octadecenoic acid and 10,13-dihydroxy-octadecanoic acid. We attempted to identify and clone the gene in L. acidophilus NTV001 that encodes the hydratase responsible for this conversion.
Identification of the hydratase gene in L. acidophilus NTV001
To identify the gene encoding the hydratase that converts LA into 13-hydroxy-cis-9-octadecenoic acid, the protein sequence of CLA-HY (569 amino acids) was used as the query in a homology search for the genomes of L. acidophilus strains. We found two candidate proteins in the genome of L. acidophilus ATCC4796. Each of these proteins has been annotated as an MCRA and shares 32% amino acid identity with CLA-HY. Based on the sequence of the L. acidophilus ATCC4796 genome, we succeeded in cloning two candidate genes (fa-hy1 and fa-hy2) from L. acidophilus NTV001 (see Materials and Methods). The fa-hy1 gene (accession number LC030242) of L. acidophilus NTV001 consists of a 1,773 bp open reading frame that encodes a polypeptide of 590 amino acids. This polypeptide shares 100% amino acid identity with the product of the corresponding gene from L. acidophilus ATCC4796. The fa-hy2 gene (accession number LC030243) of L. acidophilus NTV001 consists of a 1,776 bp open reading frame that encodes a polypeptide of 591 amino acids. This polypeptide shares 99% amino acid identity with the product of the corresponding gene from L. acidophilus ATCC4796. FA-HY1 shares 57% amino acid identity with FA-HY2.
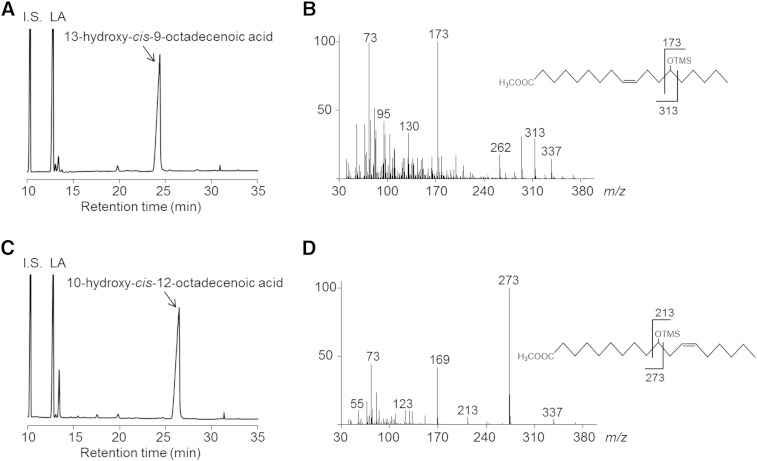
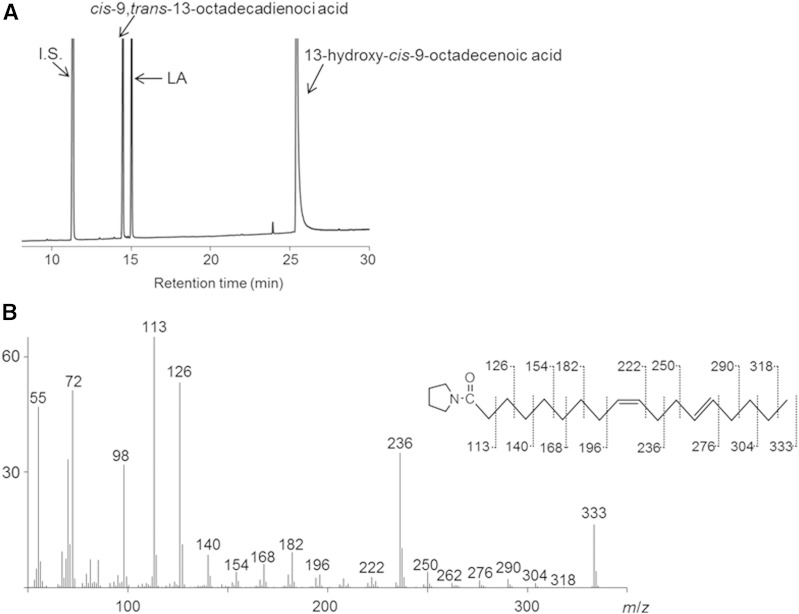
Next, the products obtained from reaction mixtures, containing each E. coli transformant [E. coli Rosetta2 (DE3)/pET21b-fa-hy1 or E. coli Rosetta2 (DE3)/pET21b-fa-hy2] as the catalyst and LA as the substrate, were analyzed. A representative GC chromatogram from the analysis of products obtained with FA-HY1 is shown in Fig. 1A. The retention time of the newly generated product was 24.4 min, which was the same as that of the standard 13-hydroxy-cis-9-octadecenoic acid. This chemical structure was confirmed by GC/MS analysis of its TMS derivative (Fig. 1B). Thus, FA-HY1 was responsible for the production of 13-hydroxy-cis-9-octadecenoic acid from LA. The detailed characterization of FA-HY1 is described in the following sections. A representative GC chromatogram from the analysis of the products obtained with FA-HY2 is shown in Fig. 1C. The retention time of the newly generated product is 26.4 min, the same as that of the standard 10-hydroxy-cis-12-octadecenoic acid. This chemical structure was confirmed by GC/MS analysis of its TMS derivative (Fig. 1D). Thus, FA-HY2 was responsible for the production of 10-hydroxy-cis-12-octadecenoic acid from LA. FA-HY2 converted oleic acid, γ-linolenic acid, and α-linolenic acid into the corresponding 10-hydroxy FAs. FA-HY2 most efficiently converted γ-linolenic acid. When the activity of LA hydration was defined as 100%, FA-HY2 converted oleic acid, γ-linolenic acid, and α-linolenic acid into the corresponding 10-hydroxy FAs with relative activities of 160%, 166%, and 137%, respectively.
Fig. 1.
Analysis of LA hydration by E. coli transformants expressing FA-HY1 and FA-HY2. A: Representative GC chromatogram of a reaction mixture containing E. coli Rosetta2 (DE3)/pET21b-fa-hy1 and LA. The internal standard (I.S.) is n-heptadecanoic acid. B: GC/MS spectrum of the TMS derivative of the methyl ester of the newly generated product from the hydration of LA by E. coli Rosetta2 (DE3)/pET21b-fa-hy1. C: Representative GC chromatogram of the reaction mixture containing E. coli Rosetta2 (DE3)/pET21b-fa-hy2 and LA. D: GC/MS spectrum of the TMS derivative of the methyl ester of the newly generated product from the hydration of LA by E. coli Rosetta2 (DE3)/pET21b-fa-hy2.
Purification of FA-HY1
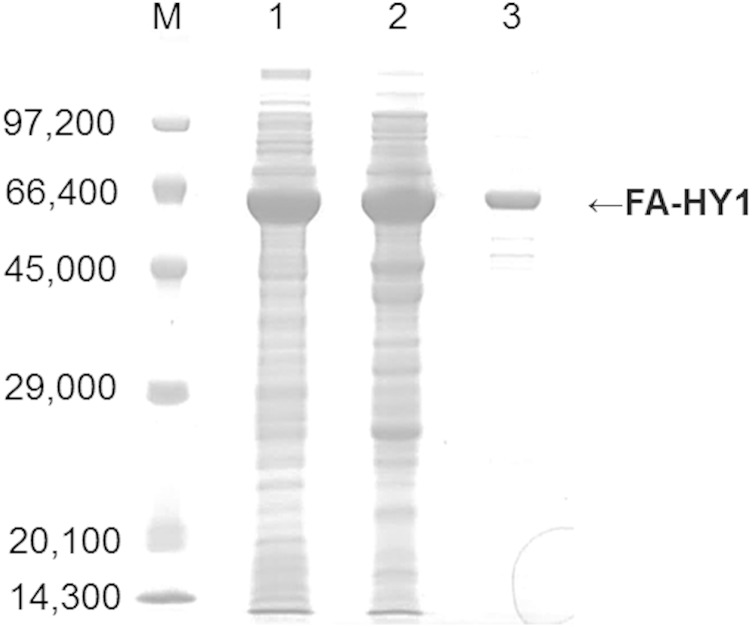
In order to characterize FA-HY1, recombinant FA-HY1 was purified from the E. coli transformant, E. coli Rosetta2 (DE3)/pET21b-fa-hy1. The purified FA-HY1 migrated as a single band on an SDS-PAGE gel (Fig. 2). The apparent molecular mass of the band, ∼66 kDa, corresponds to the calculated molecular mass (67 kDa). The molecular mass of the native enzyme, determined by gel filtration using a Superdex 200 10/300 GE column, was estimated to be 132 kDa (data not shown). Taken together, the SDS-PAGE and gel filtration results suggest that native FA-HY1 is a homodimer.
Fig. 2.
SDS-PAGE analysis of purified FA-HY1. M, protein molecular mass (Da) marker; lane 1, cell-free extracts of E. coli expressing FA-HY1; lane 2, Mono Q 10/100 GL column eluate; lane 3, Superdex 200 10/300 GL column eluate. The 12.5% SDS-PAGE gels were stained with Coomassie blue and destained by subsequent washing in distillated water.
Spectral properties and cofactor identification
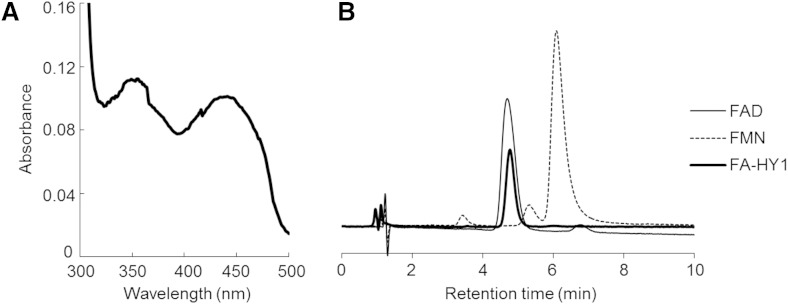
Concentrated FA-HY1 (10 mg/ml) has a light yellow color. The UV spectrum of FA-HY1 shows two peaks at ∼350 and 450 nm (Fig. 3A), indicating that FA-HY1 is a flavoprotein. To identify the flavin cofactor in FA-HY1, the protein was heat-denatured, and the protein-free supernatant was assayed using HPLC (Fig. 3B). The retention time of the flavin in the FA-HY1 supernatant was 4.7 min, which was the same as the retention time of an FAD standard. These results demonstrate that FA-HY1 is an FAD-containing enzyme.
Fig. 3.
Spectral properties of FA-HY1 and cofactor identification. A: FA-HY1 spectra were acquired at an enzyme concentration of 10 mg/ml. B: The HPLC chromatograms of protein-free FA-HY1 supernatant, 40 μM FAD, and 40 μM FMN are shown. The retention times of the peaks from the protein-free FA-HY1 supernatant and the FAD standard were both 4.7 min; the retention time of FMN was 6.0 min.
Effects of cofactors on the activity of FA-HY1
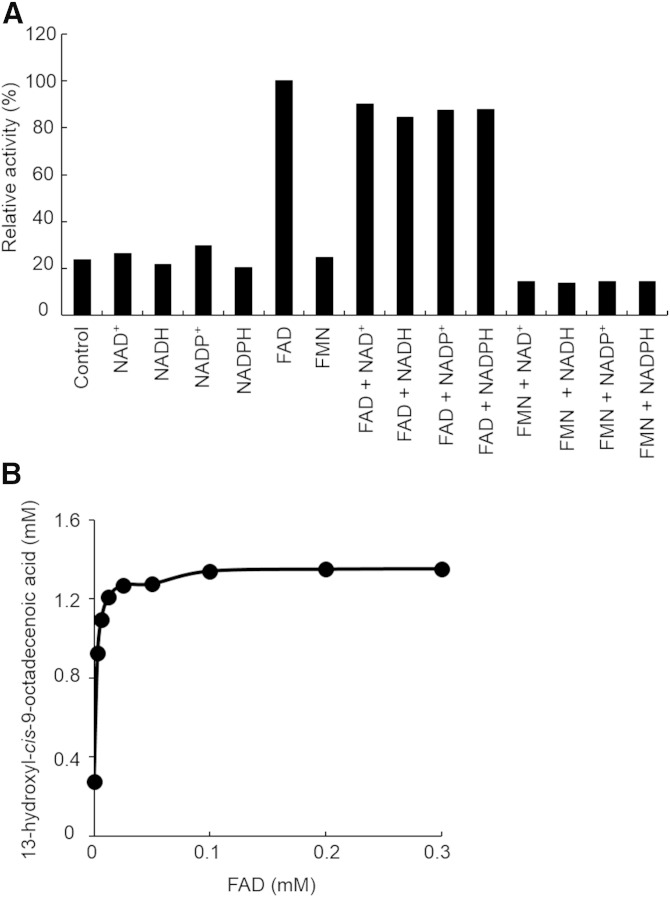
Addition of NAD+, NADH, NADP+, NADPH, or FMN to the reaction mixture did not affect the activity of FA-HY1. However, addition of FAD increased the activity (Fig. 4A). The activity of FA-HY1 increased with increasing FAD concentration, reaching a plateau above 0.1 mM FAD (Fig. 4B).
Fig. 4.
Effects of added cofactors on the activity of FA-HY1. A: Each cofactor was added to a reaction mixture containing 50 mM KPB (pH 6.0), 10 mM LA, 0.3 mg/ml BSA, and 50 µg/ml enzyme. Reactions were conducted at 37°C for 15 min with shaking (130 strokes/min). Control reaction was carried out without the addition of cofactors. B: Reactions were carried out by varying the concentration of FAD from 0 to 0.3 mM.
Enzymatic properties of FA-HY1
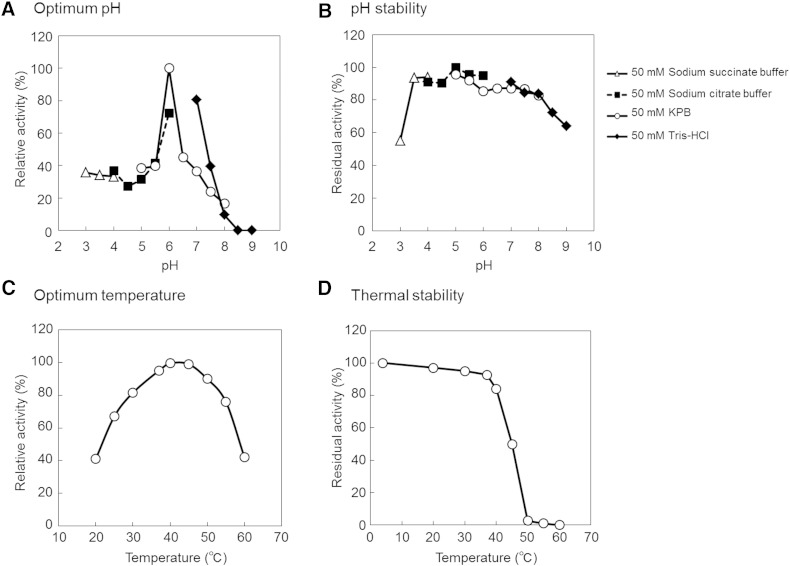
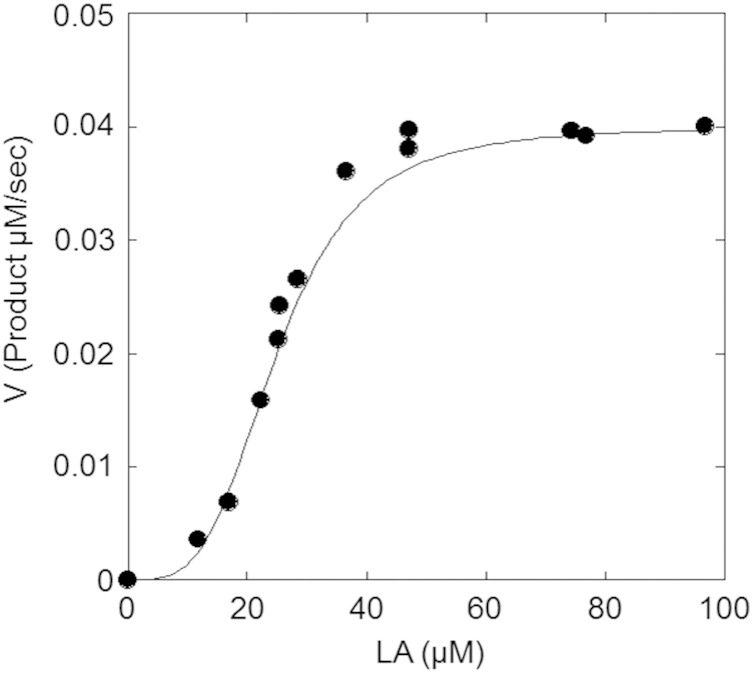
FA-HY1 exhibited maximum activity at 50 mM KPB, pH 6.0 (Fig. 5A), and was stable between pH 3.5 and 8.0 (Fig. 5B). The optimum reaction temperature was found to be 40°C (Fig. 5C), and the enzyme retained 84% activity after incubation at 40°C for 30 min (Fig. 5D). The substrate concentration-velocity curve for LA hydration showed a sigmoid shape (Fig. 6). Fitting the data to the Hill equation resulted in an apparent Km value for LA of 24 μM, a kcat value of 2.4 s−1, and a Hill coefficient of 3.6.
Fig. 5.
Enzymatic properties of FA-HY1. A: Reactions were carried out in the indicated buffer containing 10 mM LA, 0.3 mg/ml BSA, 0.1 mM FAD, and 0.5 µg/ml enzyme at 37°C for 15 min with shaking. The value obtained with 50 mM KPB (pH 6.0) was defined as 100%. B: The enzyme was preincubated at 4°C for 24 h in the indicated buffer. The reactions were carried out at 37°C for 15 min, with shaking, in 50 mM KPB (pH 6.0) containing 10 mM LA, 0.3 mg/ml BSA, 0.1 mM FAD, and 0.5 µg/ml preincubated enzyme. The value obtained with 50 mM sodium citrate buffer (pH 5.0) was defined as 100%. C: Reactions were carried out in 50 mM KPB (pH 6.0) containing 10 mM LA, 0.3 mg/ml BSA, 0.1 mM FAD, and 0.5 µg/ml enzyme at the indicated temperature for 15 min. The value at 40°C was defined as 100%. D: The enzyme was preincubated for 30 min at indicated temperature in 50 mM KPB (pH 6.0). The reactions were carried out in 50 mM KPB (pH 6.0) containing 10 mM LA, 0.3 mg/ml BSA, 0.1 mM FAD, and 0.5 µg/ml preincubated enzyme at 37°C for 15 min, with shaking.
Fig. 6.
Kinetic parameters of FA-HY1 for LA. Reactions were carried out in 50 mM KPB (pH 6.0) containing the indicated concentrations of LA, 0.2 mg/ml BSA, 0.1 mM FAD, and 1 µg/ml enzyme at 37°C for 5 min with shaking (130 strokes/min).
Substrate specificity of FA-HY1
To examine the substrate specificity of FA-HY1, various FAs were incubated with purified FA-HY1. Hydroxy FA products generated by FA-HY1 are listed in Table 1. C18 FAs with a cis carbon-carbon double bond at the Δ12 position, such as LA, pinolenic acid (cis-5,cis-9,cis-12-octadecatrienoic acid), columbinic acid (trans-5,cis-9,cis-12-octadecatrienoic acid), γ-linolenic acid (cis-6,cis-9,cis-12-octadecatrienoic acid), α-linolenic acid (cis-9,cis-12,cis-15-octadecatrienoic acid), and stearidonic acid (cis-6,cis-9,cis-12,cis-15-octadecatetraenoic acid) were converted into the corresponding 13-hydroxy FAs. In contrast, FAs with a trans carbon-carbon double bond at the Δ12 position (linoelaidic acid, trans-9,trans-12-octadecadienoic acid) and FA esters (methyl linoleate and ethyl linoleate) were not hydrated. C20 FAs with a cis carbon-carbon double bond at the Δ14 position, such as cis-11,cis-14-eicosadienoic acid, sciadonic acid (cis-5,cis-11,cis-14-eicosatrienoic acid), dihomo-γ-linolenic acid (cis-8,cis-11,cis-14-eicosatrienoic acid), cis-11,cis-14,cis-17-eicosatrienoic acid, and arachidonic acid (cis-5,cis-8,cis-11,cis-14-eicosatetraenoic acid), were converted into the corresponding 15-hydroxy FAs. In particular, sciadonic acid and arachidonic acid were converted efficiently to the corresponding 15-hydroxy FAs, giving yields of 51.7% and 45.7% against the added PUFAs, respectively. However, eicosatetraenoic acid (cis-8,cis-11,cis-14,cis-17-eicosatetraenoic acid) was converted into the corresponding 12-hydroxy FA, and EPA was not hydrated. Mead acid (cis-5,cis-8,cis-11-eicosatrienoic acid) was converted into the corresponding 12-hydroxy FA. DHA was converted into the corresponding 14-hydroxy FA; however, the yield was low. FA-HY1 showed broad substrate specificity and could convert C16–22 FAs into the corresponding hydroxy FAs.
TABLE 1.
Substrate specificity of FA-HY1
| Substrate | Product(s) | Yield (%) |
| 16:1Δ9cis | 10-Hydroxy-16:0 | 2.2 |
| 18:1Δ9cis | 10-Hydroxy-18:0 | 0.6 |
| 18:1Δ11cis | 12-Hydroxy-18:0 | 58.8 |
| 18:1Δ11trans | No products detected | — |
| 18:2Δ9cis, Δ12cis | 13-Hydroxy-cis-9-18:1 | 48.1 |
| 18:2Δ9trans, Δ12trans | No products detected | — |
| 18:3Δ5cis, Δ9cis, Δ12cis | 13-Hydroxy-cis-5,cis-9-18:2 | 57.0 |
| 18:3Δ5trans, Δ9cis, Δ12cis | 13-Hydroxy-trans-5,cis-9-18:2 | 45.5 |
| 18:3Δ6cis, Δ9cis, Δ12cis | 13-Hydroxy-cis-6,cis-9-18:2 | 56.6 |
| 10-Hydroxy-cis-6,cis-12-18:2 | 0.1 | |
| 18:3Δ9cis, Δ12cis, Δ15cis | 13-Hydroxy-cis-9,cis-15-18:2 | 53.8 |
| 18:4Δ6cis, Δ9cis, Δ12cis, 15cis | 13-Hydroxy-cis-6,cis-9,cis-15-18:3 | 12.1 |
| 20:2Δ11cis, Δ14cis | 15-Hydroxy-cis-11-20:1 | 9.2 |
| 20:3Δ5cis, Δ8cis, Δ11cis | 12-Hydroxy-cis-5,cis-8-20:2 | 18.7 |
| 20:3Δ5cis, Δ11cis, Δ14cis | 15-Hydroxy-cis-5,cis-11-20:2 | 51.7 |
| 20:3Δ8cis, Δ11cis, Δ14cis | 15-Hydroxy-cis-8,cis-11-20:2 | 14.1 |
| 12-Hydroxy-cis-8,cis-14-20:2 | 4.7 | |
| 20:3Δ11cis, Δ14cis, Δ17cis | 12-Hydroxy-cis-14,cis-17-20:2 | 0.4 |
| 15-Hydroxy-cis-11,cis-17-20:2 | 0.2 | |
| 20:4Δ5cis, Δ8cis, Δ11cis, Δ14cis | 15-Hydroxy-cis-5,cis-8,cis-11-20:3 | 45.7 |
| 20:4Δ8cis, Δ11cis, Δ14cis, Δ17cis | 12-Hydroxy-cis-8,cis-14,cis-17-20:3 | 2.1 |
| EPA | No products detected | — |
| DHA | 14-Hydroxy-cis-4,cis-7,cis-10,cis-16,cis-19-22:5 | 0.6 |
| Methyl linoleate | No products detected | — |
| Ethyl linoleate | No products detected | — |
16:0, hexadecanoic acid; 16:1, hexadecenoic acid; 18:0, octadecanoic acid; 18:1, octadecenoic acid; 18:2, octadecadienoic acid; 18:3, octadecatrienoic acid; 18:4, octadecatetraenoic acid; 20:1, eicosadecenoic acid; 20:2, eicosadienoic acid; 20:3, eicosatrienoic acid; 20:4, eicosatetraenoic acid; 22:5, docosapentaenoic acid.
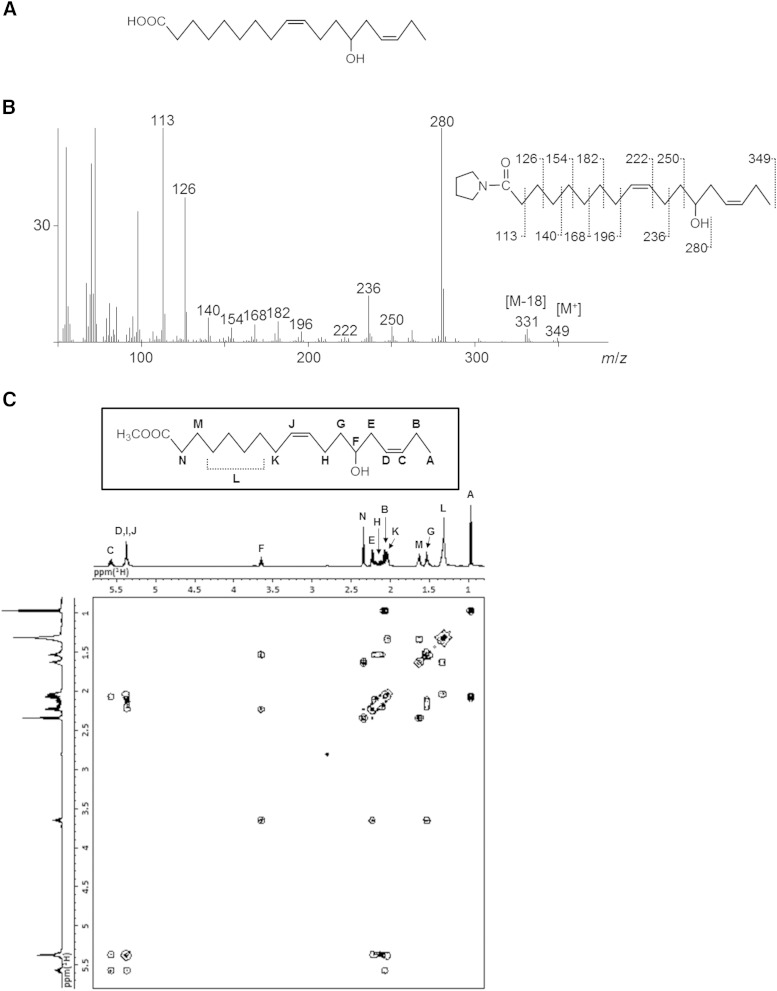
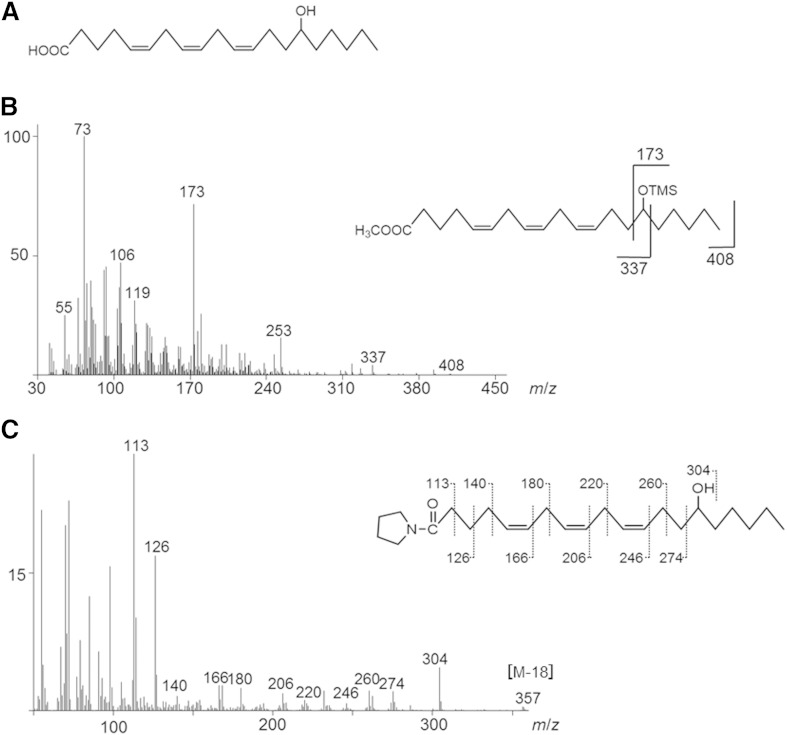
The chemical structure of the product from α-linolenic acid, 13-hydroxy-cis-9,cis-15-octadecadienoic acid, was determined by GC/MS analysis of its pyrrolidide derivative, as well as NMR analysis (Fig. 7). The chemical structure of the product from arachidonic acid, 15-hydroxy-cis-5,cis-8,cis-11-eicosatrienoic acid, was determined by GC/MS analysis of its TMS and pyrrolidide derivatives (Fig. 8). In the same way, the chemical structures of other products were also determined through a combination of GC/MS and NMR analyses.
Fig. 7.
Structural identification of 13-hydroxy-cis-9,cis-15-octadecadienoic acid. A: The structure of 13-hydroxy-cis-9,cis-15-octadecadienoic acid. B: GC/MS spectrum of the pyrrolidide derivative of 13-hydroxy-cis-9,cis-15-octadecadienoic acid. C: DQF-COSY spectrum of 13-hydroxy-cis-9,cis-15-octadecadienoic acid.
Fig. 8.
Structural identification of 15-hydroxy-cis-5,cis-8,cis-11-eicosatrienoic acid. A: The structure of 15-hydroxy-cis-5,cis-8,cis-11-eicosatrienoic acid. B: GC/MS spectrum of the TMS derivative of 15-hydroxy-cis-5,cis-8,cis-11-eicosatrienoic acid methyl ester. C: GC/MS spectrum of the pyrrolidide derivative of 15-hydroxy-cis-5,cis-8,cis-11-eicosatrienoic acid.
The absolute stereochemistry of 13-hydroxy-cis-9-octadecenoic acid and 13-hydroxy-cis-9,cis-15-octadecadienoic acid produced by FA-HY1 was determined to be (R) and (S), respectively, based on analyses of their NMR spectra. The enantiomeric purity (ee) of each of the products was >99% by HPCL analysis (data not shown).
Dehydration reaction catalyzed by FA-HY1
To examine the dehydration reaction catalyzed by FA-HY1, the substrate 13-hydroxy-cis-9-octadecenoic acid was incubated with purified FA-HY1. Fig. 9A shows a GC chromatogram of the methylated FAs produced by FA-HY1 from 13-hydroxy-cis-9-octadecenoic acid. The dehydration reaction catalyzed by FA-HY1 generated cis-9,trans-13-octadecadienoic acid and LA with the yields of 20.3% and 19.3%, respectively. The chemical structure of the cis-9,trans-13-octadecadienoic acid produced in the reaction was determined by GC/MS and MNR analyses. Mass spectra of pyrrolidide derivatives of the isolated FA methyl ester identified the isolated FA as 9,13-octadecadienoic acid (Fig. 9B). 1H-NMR and DQF-COSY also suggested one partial structure as -CH2-CH = CH-CH2-CH2-CH = CH-CH2-. The coupling constants were J = 10.8 Hz and J = 12.5 Hz, respectively, indicating that one double bond was in the cis configuration and the other was in the trans configuration. Based on the results of these spectral analyses, one of the dehydrated products was identified as cis-9,trans-13-octadecadienoic acid.
Fig. 9.
Analysis of the dehydration reaction catalyzed by FA-HY1. A: GC chromatogram of the methylated FAs produced by FA-HY1 from 13-hydroxy-cis-9-octadecanoic acid. Dehydration reactions were carried out in 50 mM KPB (pH 6.0) containing 10 mM purified 13-hydroxy-cis-9-octadecenoic acid, 0.3 mg/ml BSA, 0.1 mM FAD, and 0.1 mg/ml FA-HY1 at 37°C for 16 h with shaking (130 strokes/min) under anaerobic conditions. B: GC/MS spectrum of the pyrrolidide derivative of cis-9,trans-13-octadecadienoic acid.
DISCUSSION
The ability to hydrate unsaturated FAs is present in wide range of bacteria because unsaturated FAs may be toxic to many bacteria (25–27). The MCRA protein family is highly conserved among different bacterial species, including gram-positive and gram-negative bacteria, and some of them have FA hydratase activity (12, 13, 16–19, 28).
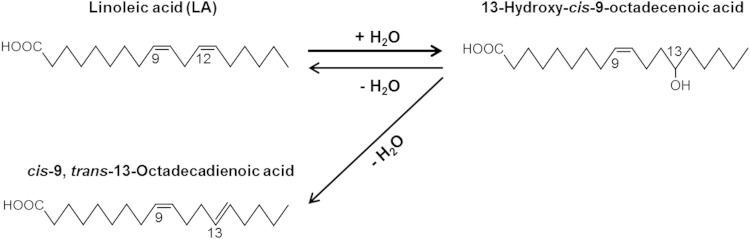
In this study, we revealed that a gut bacterium, L. acidophilus NTV001, has a high ability to produce 13-hydroxy-cis-9-octadecenoic acid from LA. L. acidophilus NTV001 was shown to have two hydratases. One of them, FA-HY1, was responsible for the production of 13-hydroxy-cis-9-octadecenoic acid from LA, and the other, FA-HY2, was responsible for the production of 10-hydroxy-cis-12-octadecenoic acid from LA. FA-HY1 and FA-HY2 belong to the MCRA family of proteins and have FAD binding motifs at their N termini. UV-visible spectra of purified FA-HY1 contain two peaks at ∼350 and 450 nm; FA-HY1 was found to be an FAD-containing enzyme (see Fig. 3). The carbon-chain lengths of the FA substrates of previously reported MCRA protein family hydratases have been limited to C16 and C18. However, FA-HY1 showed broad substrate specificity and could convert not only C16 and C18 FFAs, but also C20 and C22 FFAs into the corresponding hydroxy FAs (see Table 1). To our knowledge, this is the first report of the production of 15-hydroxy-cis-5,cis-11-eicosadienoic acid, 12-hydroxy-cis-14,cis-17-eicosadienoic acid, 15-hydroxy-cis-11,cis-17-eicosadienoic acid, and 14-hydroxy-cis-4,cis-7,cis-10,cis-16,cis-19-docosapentanoic acid from sciadonic acid (cis-5,cis-11,cis-14-eicosatrienoic acid), cis-11,cis-14,cis-17-eicosatrienoic acid, and DHA, respectively, by a bacterial enzyme. The products 13-hydroxy-cis-5,cis-9-octadecadienoic acid, 13-hydroxy-trans-5,cis-9-octadecadienoic acid, 13-hydroxy-cis-9,cis-15-octadecadienoci acid, and 13-hydroxy-cis-6,cis-9,cis-15-octadecatrienoic acid were also newly produced from the C18 FAs pinolenic acid (cis-5,cis-9,cis-12-octadecatrienoic acid), columbinic acid (trans-5,cis-9,cis-12-octadecatrienoic acid), α-linolenic acid (cis-9,cis-12,cis-15-octadecatrienoic acid), and stearidonic acid (cis-6,cis-9,cis-12,cis-15-octadecatetraenoic acid), respectively, by FA-HY1.
The 13-hydroxy-cis-9-octadecenoic acid produced by FA-HY1 from LA was of the (R)-configuration. It was distinct from that of the 13-hydroxy-cis-9-octadecenoic acid produced by L. acidophilus IFO 13951, which had the (S)-configuration (14). The 10-hydroxy-cis-12-octadecenoic acid produced by FA-HY2 has the (S)-configuration (data not shown), which is the same as that of the 10-hydroxy-cis-12-octadecenoic acid produced by CLA-HY (12).
FA-HY1 catalyzed a dehydration reaction that produced LA and cis-9,trans-13-octadecadienoic acid from 13-hydroxy-cis-9-octadecenoic acid (see Fig. 9 and Fig. 10). The same reaction was observed when 13-hydroxy-cis-6,cis-9-octadecadienoic acid was used as a substrate (data not shown). The dehydration products from 13-hydroxy-cis-6,cis-9-octadecadienoic acid were γ-linolenic acid and cis-6,cis-9, trans-13-octadecatrienoic acid.
Fig. 10.
Hydration/dehydration reaction catalyzed by FA-HY1. FA-HY1 hydrates LA at its cis-12 double bond, yielding 13-hydroxy-cis-9-octadecenoic acid. FA-HY1 dehydrates 13-hydroxy-cis-9-octadecenoic acid, yielding LA and cis-9,trans-13-octadecadienoic acid.
Hydroxy FAs are widely used as starting materials in the chemical, medical, and cosmetic industries because of their increased reactivity and viscosity compared with nonhydroxy unsaturated FAs (29). The bioactivities of hydroxy FAs have recently attracted considerable attention and have been intensively studied (1–4). In our previous study, 13-hydroxy-cis-9-octadecenoic acid was detected at higher levels in SPF mice than in germ-free mice (6). Further investigation of the bioactivities of 13-hydroxy-cis-9-octadecenoic acid is needed. Various 13-hydroxy FAs produced by FA-HY1 will be very useful to elucidate the physiological functions of endogenous 13-hydroxy-cis-9-octadecenoic acid.
10-Hydroxy-cis-9-octadecenoic acid, a derivative of LA, suppresses intestinal inflammation via GPR40. In contrast, 10-hydroxyoctadecanoic acid, a derivative of oleic acid that lacks a carbon-carbon double bond, did not exert a colitis-suppressive activity (3). These results suggest that the presence of carbon-carbon double bonds in hydroxy FAs is related to their physiological functions. In this study, we succeeded in production of various hydroxy FAs by using FA-HY1, and these are varying in the position of the hydroxyl group and the number of carbon-carbon double bonds. These products will contribute to advancing studies on bioactivities of hydroxy FAs. Evaluation of the biological properties of 13-hydroxy FAs and 15-hydroxy FAs generated by FA-HY1 will open new industrial and medicinal applications.
Footnotes
Abbreviations:
- CLA-HY
- linoleic acid hydratase
- DQF-COSY
- 1H-1H double-quantum-filtered chemical-shift correlation spectroscopy
- FAD
- flavin adenine dinucleotide
- FA-HY1
- fatty acid hydratase 1
- FMN
- flavin mononucleotide
- 1H-NMR
- proton NMR
- KPB
- potassium phosphate buffer
- LA
- linoleic acid
- MCRA
- myosin-cross-reactive antigen
This work was supported by the Bio-Oriented Technology Research Advancement Institution of Japan (J.O.); the Advanced Low Carbon Technology Research and Development Program of Japan (S.K.); the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry from the Ministry of Agriculture, Forestry and Fisheries of Japan (J.O.); and the NEDO Innovation Commercialization Venture Support Project (collaboration of NITTO PHARMA and J.O.).
REFERENCES
- 1.Kim K. R., Oh D. K. 2013. Production of hydroxy fatty acids by microbial fatty acid-hydroxylation enzymes. Biotechnol. Adv. 31: 1473–1485. [DOI] [PubMed] [Google Scholar]
- 2.Bergamo P., Luongo D., Miyamoto J., Cocca E., Kishino S., Ogawa J., Tanabe S., Rossi M. 2014. Immunomodulatory activity of a gut microbial metabolite of dietary linoleic acid, 10-hydroxy-cis-12-octadecenoic acid, associated with improved antioxidant/detoxifying defences. J. Funct. Foods. 11: 192–202. [Google Scholar]
- 3.Miyamoto J., Mizukure T., Park S. B., Kishino S., Kimura I., Hirano K., Bergamo P., Rossi M., Suzuki T., Arita M., et al. 2015. A gut microbial metabolite of linoleic acid, 10-hydroxy-cis-12-octadecenoic acid, ameliorates intestinal epithelial barrier impairment partially via GPR40-MEK-ERK pathway. J. Biol. Chem. 290: 2902–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yore M. M., Syed I., Moraes-Vieira P. M., Zhang T., Herman M. A., Homan E. A., Patel R. T., Lee J., Chen S., Peroni O. D., et al. 2014. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell. 159: 318–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goto T., Kim Y. I., Furuzono T., Takahashi N., Yamakuni K., Yang H. E., Li Y., Ohue R., Nomura W., Sugawara T., et al. 2015. 10-oxo-12(Z)-octadecenoic acid, a linoleic acid metabolite produced by gut lactic acid bacteria, potently activates PPARγ and stimulates adipogenesis. Biochem. Biophys. Res. Commun. 459: 597–603. [DOI] [PubMed] [Google Scholar]
- 6.Kishino S., Takeuchi M., Park S. B., Hirata A., Kitamura N., Kunisawa J., Kiyono H., Iwamoto R., Isobe Y., Arita M., et al. 2013. Polyunsaturated fatty acid saturation by gut lactic acid bacteria affecting host lipid composition. Proc. Natl. Acad. Sci. USA. 110: 17808–17813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogawa J., Matsumura K., Kishino S., Omura Y., Shimizu S. 2001. Conjugated linoleic acid accumulation via 10-hydroxy-12-octadecaenoic acid during microaerobic transformation of linoleic acid by Lactobacillus acidophilus. Appl. Environ. Microbiol. 67: 1246–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamada Y., Uemura H., Nakaya H., Sakata K., Takatori T., Nagao M., Iwase H., Iwadate K. 1996. Production of hydroxy fatty acid (10-hydroxy-12(Z)-octadecenoic acid) by Lactobacillus plantarum from linoleic acid and its cardiac effects to guinea pig papillary muscles. Biochem. Biophys. Res. Commun. 226: 391–395. [DOI] [PubMed] [Google Scholar]
- 9.Kishino S., Ogawa J., Omura Y., Matsumura K., Shimizu S. 2002. Conjugated linoleic acid production from linoleic acid by lactic acid bacteria. J. Am. Oil Chem. Soc. 79: 159–163. [Google Scholar]
- 10.Hudson J. A., Morvan B., Joblin K. N. 1998. Hydration of linoleic acid by bacteria isolated from ruminants. FEMS Microbiol. Lett. 169: 277–282. [DOI] [PubMed] [Google Scholar]
- 11.Yu I. S., Yeom S. J., Kim H. J., Lee J. K., Kim Y. H., Oh D. K. 2008. Substrate specificity of Stenotrophomonas nitritireducens in the hydroxylation of unsaturated fatty acid. Appl. Microbiol. Biotechnol. 78: 157–163. [DOI] [PubMed] [Google Scholar]
- 12.Takeuchi M., Kishino S., Hirata A., Park S. B., Kitamura N., Ogawa J. 2015. Characterization of the linoleic acid Δ9 hydratase catalyzing the first step of polyunsaturated fatty acid saturation metabolism in Lactobacillus plantarum AKU 1009a. J. Biosci. Bioeng. 119: 636–641. [DOI] [PubMed] [Google Scholar]
- 13.Volkov A., Liavonchanka A., Kamneva O., Fiedler T., Goebel C., Kreikemeyer B., Feussner I. 2010. Myosin cross-reactive antigen of Streptococcus pyogenes M49 encodes a fatty acid double bond hydratase that plays a role in oleic acid detoxification and bacterial virulence. J. Biol. Chem. 285: 10353–10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kishimoto N., Yamamoto I., Toraishi K., Yoshioka S., Saito K., Masuda H., Fujita T. 2003. Two distinct pathways for the formation of hydroxy FA from linoleic acid by lactic acid bacteria. Lipids. 38: 1269–1274. [DOI] [PubMed] [Google Scholar]
- 15.Takeuchi M., Kishino S., Tanabe K., Hirata A., Park S-B., Shimizu S., Ogawa J. 2013. Hydroxy fatty acid production by Pediococcus sp. Eur. J. Lipid Sci. Technol. 115: 386–393. [Google Scholar]
- 16.O’Connell K. J., Motherway M. O., Hennessey A. A., Brodhun F., Ross R. P., Feussner I., Stanton C., Fitzgerald G. F., van Sinderen D. 2013. Identification and characterization of an oleate hydratase-encoding gene from Bifidobacterium breve. Bioengineered. 4: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Volkov A., Khoshnevis S., Neumann P., Herrfurth C., Wohlwend D., Ficner R., Feussner I. 2013. Crystal structure analysis of a fatty acid double-bond hydratase from Lactobacillus acidophilus. Acta Crystallogr. D Biol. Crystallogr. 69: 648–657. [DOI] [PubMed] [Google Scholar]
- 18.Bevers L. E., Pinkse M. W., Verhaert P. D., Hagen W. R. 2009. Oleate hydratase catalyzes the hydration of a nonactivated carbon-carbon bond. J. Bacteriol. 191: 5010–5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joo Y. C., Jeong K. W., Yeom S. J., Kim Y. S., Kim Y., Oh D. K. 2012. Biochemical characterization and FAD-binding analysis of oleate hydratase from Macrococcus caseolyticus. Biochimie. 94: 907–915. [DOI] [PubMed] [Google Scholar]
- 20.Okuda T., Ando A., Negoro H., Muratsubaki T., Kikukawa H., Sakamoto T., Sakuradani E., Shimizu S., Ogawa J. J. Biosci. Bioeng. In press. [DOI] [PubMed] [Google Scholar]
- 21.Kishino S., Ogawa J., Ando A., Yokozeki K., Shimizu S. 2010. Microbial production of conjugated gamma-linolenic acid from gamma-linolenic acid by Lactobacillus plantarum AKU 1009a. J. Appl. Microbiol. 108: 2012–2018. [DOI] [PubMed] [Google Scholar]
- 22.Ohtani I., Kusumi T., Kashman Y., Kakisawa H. 1991. High-field FT NMR application of Mosher’s method. The absolute configurations of marine terpenoids. J. Am. Chem. Soc. 113: 4092–4096. [Google Scholar]
- 23.Bowden K., Heilbron I. M., Jones E. R. H., Weedon B. C. L. 1946. 13. Researches on acetylenic compounds. Part I. The preparation of acetylenic ketones by oxidation of acetylenic carbinols and glycols. J. Chem. Soc. 1946: 39–45. [Google Scholar]
- 24.Dalla V., Catteau J. P., Pale P. 1999. Mechanistic rationale for the NaBH4 reduction of α-keto esters. Tetrahedron Lett. 40: 5193–5196. [Google Scholar]
- 25.Greenway D. L., Dyke K. G. 1979. Mechanism of the inhibitory action of linoleic acid on the growth of Staphylococcus aureus. J. Gen. Microbiol. 115: 233–245. [DOI] [PubMed] [Google Scholar]
- 26.Raychowdhury M. K., Goswami R., Chakrabarti P. 1985. Effect of unsaturated fatty acids in growth inhibition of some penicillin-resistant and sensitive bacteria. J. Appl. Bacteriol. 59: 183–188. [DOI] [PubMed] [Google Scholar]
- 27.Zheng C. J., Yoo J. S., Lee T. G., Cho H. Y., Kim Y. H., Kim W. G. 2005. Fatty acid synthesis is a target for antibacterial activity of unsaturated fatty acids. FEBS Lett. 579: 5157–5162. [DOI] [PubMed] [Google Scholar]
- 28.Yang B., Chen H., Song Y., Chen Y. Q., Zhang H., Chen W. 2013. Myosin-cross-reactive antigens from four different lactic acid bacteria are fatty acid hydratases. Biotechnol. Lett. 35: 75–81. [DOI] [PubMed] [Google Scholar]
- 29.Metzger J. O., Bornscheuer U. 2006. Lipids as renewable resources: current state of chemical and biotechnological conversion and diversification. Appl. Microbiol. Biotechnol. 71: 13–22. [DOI] [PubMed] [Google Scholar]