Abstract
Recent studies have failed to demonstrate a causal cardioprotective effect of HDL cholesterol levels, shifting focus to the functional aspects of HDL. Phospholipid transfer protein (PLTP) is an HDL-associated protein involved in reverse cholesterol transport. This study sought to determine the genetic and nongenetic predictors of plasma PLTP activity (PLTPa), and separately, to determine whether PLTPa predicted carotid artery disease (CAAD). PLTPa was measured in 1,115 European ancestry participants from a case-control study of CAAD. A multivariate logistic regression model was used to elucidate the relationship between PLTPa and CAAD. Separately, a stepwise linear regression determined the nongenetic clinical and laboratory characteristics that best predicted PLTPa. A final stepwise regression considering both nongenetic and genetic variables identified the combination of covariates that explained maximal PLTPa variance. PLTPa was significantly associated with CAAD (7.90 × 10−9), with a 9% decrease in odds of CAAD per 1 unit increase in PLTPa (odds ratio = 0.91). Triglyceride levels (P = 0.0042), diabetes (P = 7.28 × 10−5), paraoxonase 1 (PON1) activity (P = 0.019), statin use (P = 0.026), PLTP SNP rs4810479 (P = 6.38 × 10−7), and PCIF1 SNP rs181914932 (P = 0.041) were all significantly associated with PLTPa. PLTPa is significantly inversely correlated with CAAD. Furthermore, we report a novel association between PLTPa and PON1 activity, a known predictor of CAAD.
Keywords: phospholipid transfer protein, coronary artery disease, paraoxonase 1
Cholesterol carried on HDL (HDL-C) has long been believed to be cardioprotective, based on consistent epidemiologic findings of an inverse relationship between incident CVD and HDL-C levels in subjects healthy at baseline (1, 2). Estimates from the Framingham Heart Study found that risk of myocardial infarction (MI) increased by 25% for each 5 mg/dl decrease in HDL-C below median values of HDL-C for both healthy men and women (2). Similar findings of the cardioprotective effects of HDL-C have also been reported in subjects with CVD at baseline (3).
In conflict with these epidemiologic findings, recent attempts to establish a causal relationship between HDL-C and CVD have failed. First, in the AIM-HIGH randomized clinical trial, pharmacologic use of niacin resulted in significant increases in HDL-C levels. However, this niacin-induced increase in HDL-C was not found to affect CVD mortality or hospitalization at 2 years of follow-up (4). Second, in a Mendelian randomization study of approximately 20,000 MI cases and 100,000 controls, a genetic variant (LIPGAsn396Ser) that significantly increased HDL-C did not associate with differential MI risk (5). Together, the failure of these two studies to establish a causal link between HDL-C and CVD outcomes has raised new doubts regarding the cardioprotective effects of HDL.
Recent evidence from prospective cohort studies may explain the failure of the aforementioned studies (4, 5) to establish a causal relationship between HDL-C and CVD. In the first study from the European Prospective Investigation into Cancer and Nutrition (EPIC), decreases in incident CVD were significantly associated with increased HDL particle (HDL-P) concentration. Moreover, this protective correlation between HDL-P and incident CVD was independent of apoB and triglyceride levels (6). In a second study from the Multi-Ethnic Study of Atherosclerosis (MESA), HDL-P concentration was significantly associated with decreased carotid intima media thickening. Moreover, although HDL-P and HDL-C were highly correlated with one another (Spearman’s correlation coefficient = 0.69), the association of HDL-P with decreased carotid intima media thickness (cIMT) remained significant even after inclusion of HDL-C levels in the multivariate regression model, suggesting that HDL-P had cardioprotective effects independent of HDL-C levels (7).
The findings in the EPIC (6) and MESA (7) studies suggest that although HDL-C captures a large portion of HDL-P variation, HDL-C measurements do not reflect the functional elements of HDL that are primarily responsible for cardioprotection. To help identify the specific components of HDL responsible for cardioprotection, we have found that the HDL3 subfraction is the superior predictor of carotid artery disease (CAAD) when compared with HDL2, HDL-C, or apoA1 (8), and that this association is independent of the HDL3-associated (9) and cardioprotective (10–13) paraoxonase 1 (PON1) enzyme activity (8). Other work has determined that concentrations of the medium-sized (but not small or large) HDL-Ps differ significantly between CAAD cases and controls, and that medium HDL-P concentration is highly correlated with HDL3 (14, 15).
Phospholipid transfer protein (PLTP) is involved in the transport of cholesterol from the periphery to the liver [reverse cholesterol transport (RCT)], which represents another potentially cardioprotective aspect of HDL that is not reflected by HDL-C. PLTP is ubiquitously expressed (16, 17) and physically associates with apoA1 (18), which facilitates cholesterol and phospholipid transfer from peripheral cells to HDL. Two forms of functionally defined PLTP are found in the plasma (19): an “active” form that can transfer triglyceride-rich phospholipids from VLDLs and chylomicrons to HDL (20, 21), and an “inactive” form that lacks this capability. Between 50 and 90% of total plasma PLTP mass is reported to be in the “inactive” form (19, 22), which is distinguished from active plasma PLTP mass by mean molecular mass (160–210 kDa for active PLTP vs. 340 to >600 kDa for inactive PLTP) and Stokes diameter (7.6–12.0 nM vs. 12 to >17 nM for active and inactive PLTP, respectively). For the remainder of this paper, “PLTPa” will refer to the active form of PLTP in the plasma.
PLTP facilitates the efflux of cholesterol from peripheral cells to HDL for RCT through various mechanisms: first, PLTP is expressed in macrophages, is upregulated in cholesterol-laden macrophage foam cells, and is present in atherosclerotic plaque (16, 17). On peripheral cells or in atherosclerotic plaque, PLTP promotes the binding of HDL to cholesterol-laden macrophages and fibroblasts and assists in HDL remodeling, which improves cholesterol and phospholipid removal (23). Recent evidence demonstrated that in vitro RNA interference silencing of PLTP decreased apoA1- and HDL3-mediated cholesterol efflux by 67 and 30%, respectively, highlighting the potential role of PLTP in RCT (24).
In contrast to these findings, mouse and other animal models have generally supported a proatherogenic role of PLTP [see review in (25)]. Mice deficient in PLTP have decreased atherosclerosis when combined with apoE-deficient or apoB-transgenic backgrounds (26). Increased PLTP expression in mice has been positively associated with atherosclerosis (27, 28) and decreased HDL, LDL, and VLDL levels (27). Moreover, increased PLTP gene expression has been reported to decrease RCT in mice through measurement of the amount of intraperitoneally injected radioactive-labeled cholesterol that reaches the feces (29). In humans, genetic variants in PLTP are associated with decreased HDL-C and increased triglyceride levels (30). Separately, a gene score based on two PLTP SNPs, rs6065904 and rs378114, predicted lower PLTP activity (PLTPa), decreased hepatic PLTP expression, increased HDL3 concentration, and decreased CVD risk in humans (31).
In contrast to these findings that suggest PLTP is atherogenic, it is notable that macrophage PLTP gene expression is cardioprotective in mouse models. First, in transgenic PLTP−/− and APOE−/− mice receiving a bone marrow transplant resulting in APOE expression in macrophages only, the lack of PLTP gene expression resulted in decreased APOE secretion from macrophages, increased cholesterol buildup, and accelerated atherosclerosis development (32). Similarly, in transgenic PLTP−/− and LDLR−/− mice receiving a bone marrow transplant resulting in PLTP gene expression in macrophages only, the mice expressing PLTP had significantly lower LDL cholesterol levels, higher HDL-C levels, and decreased size of atherosclerotic plaque (33). Finally, even in transgenic LDLR−/− mice with normal systemic PLTP and APOAI gene expression, it was found that PLTP-mediated macrophage atheroprotection could still be derived following bone marrow transplant (34). Together, these findings suggest that the physiologic context of PLTP expression is an important factor in determining its effects on atherosclerosis, with atherosclerotic lesion-based PLTPa likely being atheroprotective and systemic PLTPa possibly being atherogenic (35), though conflicting results have also been reported with macrophage PLTPa not being associated with an increase in RCT (29).
Further investigation on the effects of PLTPa and vascular disease in humans is necessary. Several studies have associated PLTPa with increased risk of coronary heart disease (CHD) (31, 36–38). However, cardioprotective effects of PLTPa have also been reported for peripheral artery disease (39). Given this finding in combination with the report that PLTP expression in macrophages is associated with decreased atherosclerosis (33), we hypothesized that PLTPa could potentially be protective against cerebrovascular disease.
We have previously reported the association of PLTPa with PLTP locus SNPs and nongenetic covariates, including HbA1c in a male non-CAAD subset of this present study (n = 87) and a 210 person replication dataset (40). Due to the relatively low throughput of our standardized PLTPa assay, similar data have not been reported in a larger human dataset, nor has the association of PLTPa with other plasma lipid traits been evaluated in a large cohort. Thus, the goal of this larger study was threefold: first, to determine the correlation between fluorescence-based PLTPa assay measures and our standardized PLTPa assay; second, to examine the PLTPa associations with plasma lipid traits, regional SNPs, and other clinical or demographic covariates; and third, to determine the relationship between PLTPa and CAAD, adjusting for potential confounders.
METHODS
Ethics statement
Institutional review boards at the University of Washington, Virginia Mason Medical Center, and Veterans Affairs Puget Sound Health Care approved this study. Written informed consent was obtained from all participants.
Sample
All data for this study came from the Carotid Lesion Epidemiology and Risk (CLEAR) study (8, 10–12, 40–47). The CLEAR study is a Seattle-based prevalent CAAD case-control study, comprised mostly of veterans, with controls distribution matched by sex and age at diagnosis of CAAD cases. All participants underwent ultrasound assessment of both carotid arteries to quantify atherosclerotic plaque, except for a small number of CAAD cases that had prior carotid endarterectomy for symptomatic obstruction. CAAD was defined as >50% stenosis in either internal carotid artery as determined by ultrasound. Controls had <15% stenosis in both internal carotid arteries and absence of lower extremity peripheral artery disease or of known CHD. Exclusion criteria for the CLEAR study included familial hypercholesterolemia, total fasting cholesterol >400 mg/dl, hypocoagulable state or use of hypocoagulant medication, post-organ transplant, or inability to consent. For the purposes of our study, diabetes status was defined as hemoglobin A1c ≥6.5%, use of oral hypoglycemic therapy or insulin. Medication use was determined via self-report and then matched to pharmacy records.
The study population is a subset (n = 1,115) of the previously described CLEAR study (8, 10–12, 40–47) composed of 493 CAAD cases and 622 controls with data on PLTPa, plasma lipid measures, baseline demographic/clinical characteristics, and genetic variants in the PLTP region. Of the studied 1,115 participants, 462 (41%) were on statin pharmacotherapy; this rate was 72% in CAAD cases and 18% in controls (see supplementary Table 1). Censored age, for analyses with CAAD as the outcome, is defined as the age at CAAD diagnosis for cases and age at enrollment and blood draw for controls. Participants with milder carotid artery stenosis [between 15 and 49% in one (or both) internal carotid artery] were not phenotyped for PLTPa and were therefore excluded from these analyses.
Genotyping and imputation of SNPs
To maintain genetic homogeneity, only European ancestry subjects, as confirmed by use of Illumina HumanCVD BeadChip SNP data (48) and the program STRUCTURE (49), were included in these analyses. Genotyping of 48,742 SNPs relevant to CVD was performed using the Illumina HumanCVD BeadChip (48). Duplicate genotyping in 34 participants showed 99.7% consistency in genotype calls. Participants were filtered for an individual call rate of <97%. SNPs were filtered for a SNP-specific call rate of <97%.
Imputation of the Illumina HumanCVD BeadChip data [post-quality control (QC) measures outlined above] was performed for all autosomes (chromosomes 1-22) using IMPUTE2 (50) with phased haplotype data from the 1000 Genomes Project (March 2012 release) as a reference panel (51). CLEAR participant genotype data was prephased prior to imputation using SHAPEIT (52). Only imputed genotypes with a probability >0.9 were included. Imputed SNPs with >5% missing genotypes or whose genotype distributions were not consistent with Hardy-Weinberg equilibrium at the P < 10−6 level were filtered out of the genetic dataset. SNPs within 50 kb of the PLTP gene and with a minor allele frequency >1% were considered for analyses, a total of 124 SNPs measured in 1,591 European ancestry participants.
Plasma lipid measurements
All plasma lipid measurements were made on fasting whole plasma. Standard methods were used to determine plasma levels of total cholesterol, triglycerides, VLDL, and HDL using an Abbott Spectrum analyzer. HDL subfraction 2 (HDL2), HDL3 (53), and apoA1 (54) levels were measured using previously described methods. The distributions of triglycerides and VLDL were natural log (ln) transformed to reduce left skewness. All plasma lipid levels had approximate normal distributions or were natural log transformed (triglycerides and VLDL).
PON1 arylesterase activity measurement
PON1 enzyme activity was measured by rate of enzymatic degradation of phenylacetate [arylesterase (AREase)] by a continuous spectrophotometric assay with lithium heparin plasma, as previously described (10–12). PON1 AREase activity had an approximate normal distribution.
PLTPa measurement
Radiometric gold-standard liposome assay.
Plasma PLTPa was measured from whole fasting plasma using the standardized radiometric method as previously reported (20, 55), blinded to all other participant data, at the Northwest Lipid Metabolism and Diabetes Research Laboratories. Each plasma sample was stored at −70°C and only thawed once prior to assay. In brief, the transfer of 14C-phosphatidylcholine to a HDL acceptor was measured in the presence of 1 μl of human plasma. All samples were incubated at 37°C for 15 min; the rate of transfer from cultured liposomes to HDL is linear within this period (40, 56). The analysis was performed in three technical replicates and counted four times from each participant’s sample to reduce measurement variation. All assays were performed using the same liposome acceptor preparation to reduce assay variability. QC values varied on average by 4–8%; assays with QC values exceeding 10% variability were repeated. Assay validation was also established by repeating randomly chosen samples in another separate assay, from which 3–5% intra-assay variability in assay yields for triplicate samples was calculated. Similar to the prior step, samples with greater than 10% variability in the intra-assay validation step were re-assayed. After all samples had been assayed, the values of PLTPa were adjusted for the QC variability to remove inter-assay differences.
Fluorescence-based liposome assay.
PLTPa was measured in a subset of 20 male CLEAR participants without CAAD and not taking statin pharmacotherapy using two methods: 1) the previously described radiometric assay; and 2) using a commercial fluorometric assay from Roar Biomedical, Inc, previously used in numerous studies of PLTPa and CVD (36–38). In brief, 3 μl of sample plasma was incubated with kit-provided donor and acceptor particles resulting in the PLTP-mediated transfer of fluorescent phospholipid. Each assay was performed in triplicate and repeated if inter-assay variation exceeded 10%.
Statistical analysis
Determination of nongenetic and genetic (full model) sources of PLTPa.
Using R (R Project for Statistical Computing) with standard regression packages, stepwise linear regression was used to determine the nongenetic covariates that predicted the highest PLTPa variance (n = 1,056). Nongenetic covariates entering the model were: BMI, diabetes status, statin use, current smoking status, total cholesterol levels, ln(triglycerides), ln(VLDL), apoA1, HDL-C, HDL2, HDL3, and PON1 AREase activity. Model comparison was performed using Akaike’s information criterion (AIC), beginning with a base model considering age and sex.
After identification of the nongenetic covariates and PLTP region SNPs that significantly associated with PLTPa, a final stepwise linear regression model considering both genetic and nongenetic covariates was used to determine the regression model that predicted the maximal amount of PLTPa variance (n = 995). All nongenetic covariates and PLTP region SNPs that improved model prediction via AIC were included in the final stepwise linear regression model, with model comparison beginning with a base model with age and sex. SNP genotypes were coded additively.
PLTP region SNP associations with PLTPa.
SNPs with ≥1% frequency identified in the PLTP region between 50 kb upstream of the first codon or downstream of the last (n = 124 SNPs) were included in our analyses of the genetic predictors of PLTPa. Due to a highly dispersed pattern of linkage disequilibrium (LD) among these SNPs (see supplementary Fig. 1), attempts to reduce the number of tests through use of LD patterns to cluster correlated SNPs into a single test (57) were unproductive. PLINK (58) was used to perform linear regression with the 124 identified PLTP region SNPs (with SNP genotypes coded additively) on the outcome of PLTPa, adjusted for the effects of age, sex, diabetes status, statin use, ln(triglyceride levels), current smoking status, and PON1 AREase activity. Only SNPs significantly individually associated with adjusted-PLTPa in univariate analyses (P ≤ 0.05) were included in the final nongenetic and genetic predictors of PLTPa stepwise regression model (see above).
Association of PLTPa and potential confounders with CAAD status.
Using R with standard regression packages available, we performed multivariate logistic regression to determine whether PLTPa was associated with CAAD case status. In this model, age, sex, and all covariates we previously determined to be predictive of PLTPa variance by decreasing model AIC [ln(triglycerides), diabetes status, PON1 AREase activity, current smoking status, and apoA1; all of which are potential confounders of CAAD status] were included in addition to PLTPa.
As CAAD is treated with statins, statin use was confounded for CAAD status and could not be directly included in the model predicting CAAD. Therefore, in the model predicting CAAD status, we adjusted PLTPa for the effects of statin use by subtracting the covariate-adjusted difference in PLTPa from statin use in controls (−1.02, n = 601) using a previously described method (47, 59, 60).
RESULTS
Demographic, clinical, and lipid variables of the studied subset of the CLEAR study are presented in Table 1. The studied subset of CLEAR (n = 1,115) was comprised of two groups: those who had been previously analyzed by Jarvik et al. (40) (n = 87) and those who were newly phenotyped for PLTPa for this study (n = 1,028). The previous analysis examined only non-CAAD control males who were not on statins, while the newly PLTPa phenotyped subset included females (15%), statin users (41%), and CAAD cases (44%). In addition, the newly PLTPa phenotyped subset tended to be older (69.3 years vs. 65.4 years), more likely to be diabetic (16% vs. 8%), and more likely to smoke (12% vs. 7%). With regard to lipid covariates, the newly PLTPa phenotyped subset had significantly lower total cholesterol (mean 187 mg/dl vs. 197 mg/dl) and PON1 AREase activity (mean 137 IU vs. 151 IU), but significantly higher levels of ln(triglycerides) (mean 4.9 vs. 4.7) and ln(VLDL) (mean 3.3 mg/dl vs. 3.2 mg/dl). HDL-related measures (apoA1, HDL-C, HDL2, and HDL3) did not significantly vary between the two subsets of the current data. Finally, the newly PLTPa phenotyped subset of CLEAR had significantly lower mean PLTPa when compared with the previously analyzed subset (13.4 μM/h vs. 15.3 μM/h). Descriptive statistics of the current subset of CLEAR are also presented stratified by CAAD status in supplementary Table 1.
TABLE 1.
Demographic and clinical characteristics of the studied CLEAR subset
| Variable | n | Current Subset (n = 1,028) | Prior Subset (n = 87) | Total (n = 1,115) | P |
| Current age, y | 1,115 | 69.3 ± 8.6 | 65.4 ± 9.4 | 69.0 ± 8.7 | <0.001a |
| Female, % | 1,115 | 166 (16%) | 0 (0%) | 166 (15%) | <0.001b |
| Diabetic, % | 1,115 | 169 (16%) | 7 (8%) | 176 (16%) | 0.03b |
| BMI, kg/m2 | 1,115 | 28.1 ± 4.9 | 28.3 ± 4.4 | 28.1 ± 4.9 | 0.88a |
| Statin use, % | 1,115 | 462 (45%) | 0 (0%) | 462 (41%) | <0.001b |
| Current smoking status, % | 1,115 | 123 (12%) | 6 (7%) | 129 (11%) | <0.001b |
| CAAD case, % | 1,115 | 493 (48%) | 0 (0%) | 493 (44%) | <0.001b |
| Total cholesterol, mg/dl | 1,100 | 187 ± 41 | 198 ± 37 | 187 ± 41 | 0.004a |
| apoA1, mg/dl | 1,110 | 144 ± 28 | 143 ± 27 | 144 ± 28 | 0.81a |
| HDL-C, mg/dl | 1,100 | 50 ± 17 | 51 ± 17 | 50 ± 17 | 0.59a |
| HDL2, mg/dl | 1,098 | 9.6 ± 6.3 | 9.7 ± 6.1 | 9.6 ± 6.3 | 0.37a |
| HDL3, mg/dl | 1,099 | 41 ± 11 | 41 ± 12 | 41 ± 11 | 0.75a |
| ln(Triglycerides) | 1,100 | 4.9 ± 0.55 | 4.7 ± 0.55 | 4.9 ± 0.54 | 0.004a |
| ln(VLDL) | 1,100 | 3.3 ± 0.55 | 3.2 ± 0.54 | 3.3 ± 0.54 | 0.003a |
| PON1 AREase activity, IU | 1,077 | 137 ± 51 | 151 ± 46 | 139 ± 50 | <0.001a |
| PLTPa, μM/h | 1,115 | 13.4 ± 4.5 | 15.2 ± 2.7 | 13.5 ± 4.4 | <0.001a |
Wilcoxon rank sum test used for P value calculations for differences between subsets.
Pearson chi-square test used for P value calculations for differences between subsets.
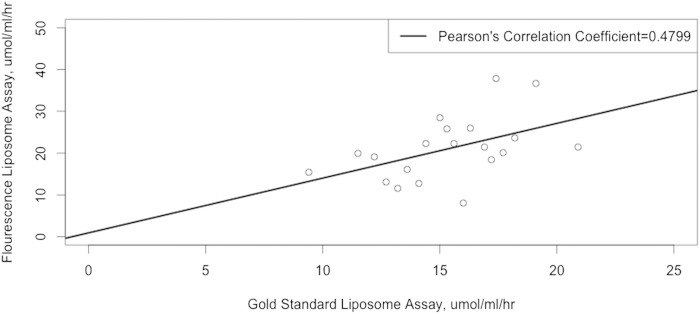
As a separate QC analysis, we performed additional PLTPa assays on 20 male control subjects not on statin pharmacotherapy using the commercial fluorometric PLTPa assay. We found that the Pearson’s pairwise correlation coefficient for this subset of 20 participants was equal to 0.4799 between the gold-standard and low-throughput PLTPa assay (20, 55) used in this study and the commercial fluorescence-based PLTPa assay (see Fig. 1).
Fig. 1.
Correlation between PLTPa measurements from the gold-standard radiometric liposome assay used in the current study and the widely used commercial fluorometric liposome assay. The black line represents the correlation coefficient from a classical linear regression model between the two PLTPa assays.
To determine the predictors of PLTPa in an unbiased method, stepwise linear regression beginning with a base model of age and sex was used in a subset of the cohort (n = 1,056) with full covariate data. Several lipid-related measures and clinical characteristics were found to improve model prediction of PLTPa variance [by decreasing model AIC, see the “Determination of nongenetic and genetic (full model) sources of PLTPa” subsection in the Methods]. Triglyceride levels [β coefficient = 0.88 PLTPa increase per 1 unit increase in ln(triglycerides), P = 0.002] and diabetes status (β coefficient = 1.41 increase in PLTPa if diabetic, P = 0.0003) were positively associated with PLTPa and explained 1.60 and 0.96% of PLTPa variance, respectively. PON1 AREase activity was also associated with an increase in PLTPa (β coefficient = 0.0082 increase in PLTPa per 1 IU increase in PON1 AREase activity, P = 0.01) and explained an additional 0.98% of PLTPa variance. Finally, current smoking status, apoA1, and statin use all met the AIC threshold to remain in the final regression model for PLTPa, but were not significantly associated (P > 0.05) with PLTPa. These results are summarized in Table 2.
TABLE 2.
Predictors of PTLPa from multivariate stepwise linear regression (n = 1,056)
| Variable | Coefficient ± SE | PLTPa (%) | P |
| Intercept | 7.34 ± 2.32 | — | — |
| Current age | −0.014 ± 0.018 | 0.59 | 0.44 |
| Female gender | 0.32 ± 0.44 | 0.00043 | 0.47 |
| ln(Triglycerides) | 0.88 ± 0.28 | 1.60 | 0.0017 |
| Diabetes status | 1.41 ± 0.38 | 0.96 | 0.0003 |
| Statin use | −0.85 ± 0.30 | 0.29 | 0.005 |
| PON1 AREase activity | 0.0082 ± 0.0032 | 0.98 | 0.010 |
| Current smoking status | 0.58 ± 0.44 | 0.19 | 0.19 |
| apoA1 | 0.0090 ± 0.0062 | 0.16 | 0.14 |
The 1,056 participants had complete data on all parameters considered in this stepwise linear regression model. All covariates included in the final model improved model prediction of PLTPa as measured by AIC. Other covariates considered, but not retained in the final model, were: BMI, total cholesterol levels, ln(VLDL), HDL-C, HDL2, and HDL3. Please see the Methods subsection “Determination of nongenetic and genetic (full model) sources of PLTPa” for further information.
Sensitivity analysis evaluating whether the PLTPa relationship with other factors differed for cases versus controls was performed by analyzing each of these groups separately; controls (n = 601, see supplementary Table 2) and CAAD cases (n = 455, see supplementary Table 3). With the exception of statin use, all variables significantly associated with PLTPa in Table 2 had the same direction of effect in both CAAD cases and controls. However, statin use was significantly associated with a decrease in PLTPa of 1.02 μM/h in CAAD controls (P = 0.037, see supplementary Table 2), while it was not associated with a difference in PLTPa in CAAD cases.
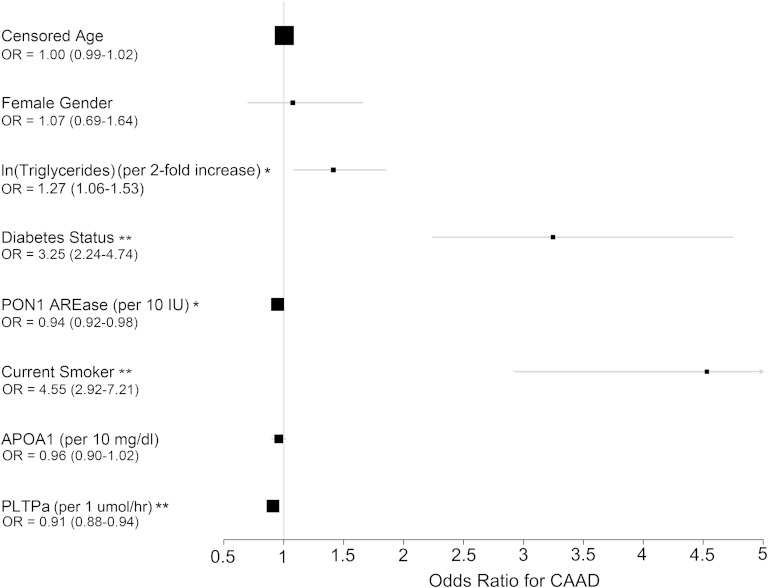
To evaluate the association of PLTPa with CAAD, multivariate logistic regression was used to predict CAAD considering PLTPa, censored age, female gender, ln(triglycerides), diabetes status, PON1 AREase activity, current smoking status, and apoA1 levels (see supplementary Table 4) (n = 1,056 with complete covariate and phenotype information). As discussed in the Methods, PLTPa was pre-adjusted for statin use, due to confounding of statin use and CAAD status. From this analysis, statin-adjusted PLTPa was strongly inversely associated with CAAD status [odds ratio (OR) = 0.91, P = 7.90 × 10−9], with an approximate 9% decrease in odds of CAAD per 1 μM/h increase in statin-adjusted PLTPa. Notably, of the predictors of PLTPa that were also included in the regression model, ln(triglycerides) (OR = 1.27 per 2-fold increase in triglyceride levels, P = 0.0051), diabetes status (OR = 3.25, P = 6.31 × 10−9), PON1 AREase activity (OR = 0.94 per 10 IU increase in activity, P = 0.0011), and current smoking status (OR = 4.55, P = 1.38 × 10−12) were all significantly associated with CAAD status. The sizes of these effects are summarized in Fig. 2.
Fig. 2.
OR association with CAAD status of PLTPa and predictors from multivariate logistic regression (n = 1,056). *0.05 > P > 0.001. **P < 0.001. See supplementary Table 4 for full regression model coefficients.
To identify the genetic predictors of PLTPa while maximizing statistical power, we used linear regression including PLTPa-associated covariates: age, sex, diabetes status, statin use, ln(triglyceride levels), current smoking status, apoA1 levels, and PON1 AREase activity on 124 SNPs within 50 kb of the PLTP gene on chromosome 20. The analysis included 1,025 CLEAR study participants with genetic and covariate data. Of these 124 PLTP region SNPs, 31 were nominally associated with covariate-adjusted PLTPa in univariate association testing (P ≤ 0.05, supplementary Table 5). An additional 16 of these 31 PLTPa-associated SNPs were also nominally predictive of CAAD status in univariate analyses after adjustment for censored age, sex, diabetes status, ln(triglyceride) levels, current smoking status, apoA1 levels, and PON1 AREase activity (P ≤ 0.05, supplementary Table 5). We then analyzed a subset of nine SNPs previously reported by Jarvik et al. (40) to be significantly associated with PLTPa in the subsample of 87 participants (see Table 3); five of the nine SNPs replicated (significantly associated with PLTPa and same direction of effect) in this larger dataset. The association of these nine SNPs with CAAD was previously reported (40) and is summarized in Table 3. Notably, rs6065904 remains predictive of PLTPa in the larger sample and is also predictive of CAAD status.
TABLE 3.
Comparison of PLTP SNP association with covariate-adjusted PLTPa and in previously reported versus current data
| SNP | Function | MAFa | 2010 Number | 2010 Betab | 2010 b | 2010 CAADc | 2015 Numberd | 2015 Betae | 2015 e | Replicatef |
| rs1736493 | Intron 4 | 0.067 | 87 | 0.09 | 0.011 | + | 1,025 | 0.26 | 0.53 | − |
| rs553359 | Intron 4 | 0.39 | 87 | −0.05 | 0.0083 | − | 1,025 | −0.18 | 0.36 | − |
| rs11086985 | Intron 6 | 0.37 | 87 | 0.07 | 0.0011 | − | 1,025 | 0.23 | 0.27 | − |
| rs6065904 | Intron 8 | 0.22 | 87 | −0.08 | 2.55 × 10−5 | + | 1,025 | −1.05 | 1.35 × 10−5 | + |
| rs6073952 | Intron 11 | 0.19 | 87 | −0.10 | 3.39 × 10−6 | − | 1,025 | −0.89 | 0.00033 | + |
| rs378114 | Intron 12 | 0.27 | 87 | 0.10 | 5.29 × 10−5 | − | 1,025 | 0.51 | 0.024 | + |
| rs11569668 | 5′ of PLTP | 0.026 | 87 | 0.10 | 0.016 | + | 1,025 | 0.51 | 0.39 | − |
| rs4810479 | 5′ of PLTP | 0.25 | 87 | −0.07 | 4.93 × 10−4 | − | 1,025 | −1.18 | 2.63 × 10−7 | + |
| rs7679 | 5′ of PLTP | 0.18 | 87 | −0.11 | 5.39 × 10−7 | − | 987 | −1.10 | 2.84 ×10−5 | + |
MAF, minor allele frequency.
Minor allele frequency within the entire CLEAR cohort.
Analyses adjusted by age, BMI, and HBA1c levels (only men analyzed).
Column indicates whether previously reported PLTP SNP associations with covariate-adjusted CAAD (n = 939) by Jarvik et al (40) are replicated in the current slightly larger dataset (n = 1,118). A (+) symbol indicates that the tested SNP has both P < 0.05 and consistent direction of effect for the ORs in both datasets. We replicate previously reported associations for rs1736493, rs6065904, rs11569668, and CAAD. A (−) symbol indicates that either the tested PLTP SNP was not significantly associated or the direction of effect on the outcome of CAAD was inconsistent between the two datasets.
Number available for linear regression analysis of PLTP region SNP effects on PLTPa dependent on the number of CLEAR participants with complete parameter data (PLTPa, covariates, and genotype data).
Analyses adjusted by age, sex, diabetes status, statin use, ln(triglyceride levels), current smoking status, and PON1 AREase activity.
Replication refers to the PLTP region SNP association with covariate-adjusted PLTPa. To have positive replication (+) a SNP must both be significantly associated with PLTPa (P < 0.05) and have a consistent direction of effect (increase or decrease PLTPa) in both datasets. Negative replication (−) indicates that either the tested PLTP region SNP was not significantly associated with PLTPa in the larger dataset or that the direction of effect on the outcome of PLTPa was not consistent in the two datasets.
To determine the optimal combination of nongenetic factors (from Table 2) and genetic variants (from supplementary Table 5) that best predict PLTPa variance in our data, we used a final stepwise linear regression model in 995 participants with complete genetic and covariate data. In this model, the 31 PLTP region SNPs from supplementary Table 5 were added to the model and compared using AIC to a base model containing all previously identified predictors of PLTPa (from Table 2), such that only the SNPs that further improved model prediction of PLTPa variance were retained. From this process, two SNPs that further improved PLTPa were identified from the list of 31 SNPs, rs4810479 and rs181914932 (see Table 4). PLTP SNP rs4810479 was associated with a decrease in PLTPa (β coefficient = −1.08, P = 6.38 × 10−7) per each minor allele, C, and explained an additional 2.4% PLTPa variance. PCIF1 SNP rs181914932 was positively associated with PLTPa (β coefficient = 1.18, P = 0.041) per each minor allele, C, and explained the 0.31% PLTPa variance.
TABLE 4.
Genetic and nongenetic predictors of PTLP activity from stepwise linear regression (n = 995)
| Variable | Coefficient ± SE | PLTPa (%) | P |
| Intercept | 13.01 ± 1.42 | — | — |
| Current age | −0.018 ± 0.017 | 1.10 | 0.31 |
| Female gender | −0.38 ± 0.40 | 0.0019 | 0.35 |
| Statin use | −0.80 ± 0.33 | 0.29 | 0.026 |
| ln(Triglycerides) | 0.89 ± 0.26 | 1.38 | 0.0042 |
| Diabetes status | 1.55 ± 0.39 | 0.72 | 7.28 × 10−5 |
| PON1 AREase activity | 0.0072 ± 0.0029 | 1.06 | 0.019 |
| Current smoking status | 0.76 ± 0.43 | 0.23 | 0.086 |
| apoA1 | 0.0093 ± 0.0055 | 0.21 | 0.038 |
| rs4810479 | −1.08 ± 0.22 | 2.40 | 6.38 × 10−7 |
| rs181914932 | 1.18 ± 0.66 | 0.31 | 0.041 |
The 995 participants had complete data on all parameters considered in this stepwise linear regression model. All covariates included in the final model improved model prediction of PLTPa as measured by AIC. Other covariates considered, but not retained, in the final model were: BMI, total cholesterol levels, ln(VLDL), HDL-C, HDL2, HDL3, and genotypes (coded additively) of the remaining 29 PLTP region SNPs significantly associated with PLTPa (see supplementary Table 5 for full list). Please see the Methods subsection on “Determination of nongenetic and genetic (full model) sources of PLTPa” for further information.
DISCUSSION
Functional aspects of the HDL-P not captured by HDL-C measures may explain the recent failures of interventions to raise HDL-C to prevent CVD mortality (8, 15). Of the many functional aspects of HDL, one of the most promising is RCT, whereby lipids are removed from peripheral cells and atherosclerotic plaque by HDL-borne proteins, such as PLTP and apoA1, and excreted through the liver (23). The importance of RCT has recently been highlighted in a study that measured cholesterol efflux capacity, the ability of HDL to accept cholesterol from macrophages, in a prospective cohort of 2,924 adults initially free from CVD. After a median of 9.4 years of follow-up, there was a 67% reduction in risk of CVD in the highest quartile of HDL-C efflux capacity compared with the lowest quartile (61), providing some of the first prospective evidence that higher cholesterol efflux capacity prevents CVD.
Within this context, we have provided further evidence that RCT is cardioprotective through our finding that plasma PLTPa is associated with decreased risk of CAAD (OR = 0.91 per 1 μM/h increase in PLTPa). In addition, we have performed the largest to-date analysis of the nongenetic and genetic predictors of plasma PLTPa. For the nongenetic predictors, we confirmed known associations of triglycerides, diabetes, smoking status, apoA1, and statin use, while also discovering the novel and strong association of PON1 AREase activity with PLTPa. With regard to the genetic predictors of PLTPa, we have identified numerous associated PLTP region SNPs, confirmed prior associations reported by Jarvik et al. (40), and also identified PLTP SNP rs4810479 and PCIF1 SNP rs181914932 as the best genetic predictors of PLTPa in these data.
In this work we also report on the relatively poor concordance (Pearson’s pairwise correlation coefficient (r) of 0.4799) between the gold-standard radiometric PLTPa assay used in the current study and the commercially available fluorometric PLTPa assay previously used for numerous studies of PLTPa and CVD. This finding of poor correlation in male controls not on statin pharmacotherapy is in contrast with prior reports of high concordance (r = 0.90) between the two methods (36–38). Similar to the high correlation between HDL-C and HDL-P in MESA (r = 0.69) but the lack of HDL-C to predict cIMT in a multivariate model including HDL-P (7), the relative lack of correlation observed between the two PLTPa assays (see Fig. 1) in the current work may underlie the differences in conclusions drawn from our study and others in the published literature. To begin, in contrast to our findings of an inverse association of PLTPa and CAAD, cross-sectional human epidemiologic CVD studies that have reported increased PLTPa associated with CHD (37) and cIMT (62). Additionally, one prospective study of 1,085 participants with angiographically documented CHD at baseline has reported a positive association between PLTPa and the composite outcome of incident MI and death (38). A separate prospective study of 2,679 participants without known CVD at baseline from the Framingham Heart Study found that PLTP higher than the median was a risk factor for incident CVD in males only (36).
However, the cross-sectional study of CHD (37) and the prospective studies of CVD outcomes (36, 38) both used fluorescence-based assay kits to measure PLTPa rather than the radiometric-based standardized direct transfer method of assessing PLTPa. As previously discussed, we have found very poor correlation between the standardized method of PLTPa assessment and the commercially available fluorescence PLTPa assay (see Fig. 1). Thus, assay differences and the unexplained variation not captured by the fluorescence-based PLTPa measures (r = 0.4799) may explain inconsistency in the direction of the PLTPa-CVD association. Additionally, as PLTPa is upregulated in atherosclerosis (16, 17), it is possible that reports of positive association between PLTPa and acute CVD (37, 62) reflect sampling of subjects already with significant atherosclerotic disease, and therefore, higher PLTPa, which may be a consequence of the underlying pathophysiological processes or a compensatory change. In addition, it may be that the cardioprotective effects of PLTPa have increased importance in primary prevention (i.e., before development of sizable atherosclerotic plaque) due to its key role in RCT. Notably, prospective work by Robins et al. (36) did find that higher levels of PLTPa were associated with incident CVD; however, the measures of PLTPa were from the fluorometric-based assay and did not specifically assess cerebrovascular disease, instead using the composite outcome (CHD, hemorrhagic stroke, or ischemic stroke) of all incident CVD. As a result, we believe that further prospective studies beginning with disease-free participants and measuring PLTPa with the radiometric-based standardized direct transfer method are needed to firmly establish whether PLTPa is causally protective against cerebrovascular disease (including CAAD) in particular.
In this work, we present what is, to the best of our knowledge, the first report of the association of PON1 AREase activity with plasma PLTPa. In our data, PON1 AREase activity and PLTPa were positively associated with each other and were each inversely correlated with increased CAAD risk. In contrast to the inconsistent direction of the PLTPa-CVD associations, PON1 activity has been consistently reported to be cardioprotective [(10, 11), as reviewed in (63)], including in a well-described and large prospective cohort for incident heart disease (64).
PON1 is an antioxidant glycoprotein enzyme physically associated with the HDL-P (65) and is enriched in the smaller HDL3 subfraction (9). PON1 has previously been reported to prevent both HDL (66) and LDL oxidation (67), leading to theories that LDL-oxidized phospholipids are one of the numerous targets of PON1 enzymatic action (68). Use of transgenic mice demonstrated a key role for PON1 in atherosclerosis, as mice lacking PON1 had accelerated atherosclerosis when fed a high-fat diet (69), a process that was further exacerbated in double knockout PON1/APOE mice (70). More recent evidence has shown that rare coding variation in PON1 is associated with ischemic stroke in a sample of approximately 5,000 human participants (13). With regard to PLTP, it is known that PON1 protein is one of 24 different proteins that associate with PLTP in plasma (71). It is not known whether PON1 directly interacts with PLTP in this plasma macromolecular complex, but its association with this complex may relate to its known interaction with apoA1 and clusterin, two major components of PLTP-containing plasma complexes (56). It is notable that PLTP interacts with both HDL2 and HDL3 and is critically involved in the maturation processes of the HDL-P (23). Furthermore, PLTP has been shown to reduce secretion of the pro-inflammatory markers and mediators in human primary macrophages and differentiated THP1 cells in vitro by modulation of the signal transduction pathways (72), and PLTPa was inversely correlated with the extent of tissue damage in a model of chronic inflammation in humans (73). Given the enrichment of acute phase reactants in PLTP complexes (71), further molecular work could elucidate a unique role for PON1 and PLTP relationship in inflammation.
Triglyceride and apoA1 levels were both positively associated with plasma PLTPa in our cohort. For triglyceride levels, this finding validates numerous prior reports (56, 74) that have previously proposed that this is due to the enhanced ability of triglyceride-rich HDL to accept additional phospholipids via the actions of PLTP (75). PLTP physically associates with apoA1 in large protein complexes (71); moreover, apoA1 stabilizes recombinant PLTP lipid transfer activity (71). It should be noted that both PLTP and apoA1 are able to elicit ABCA1-dependent lipid efflux for RCT, but with different lipoprotein efflux acceptors; PLTP-mediated ABCA1 lipid efflux requires the presence of mature HDL-Ps, while apoA1 uses lipid-poor HDL-Ps (23, 76). Moreover, it is notable that in this process, PLTP binds to ABCA1 and can competitively inhibit apoA1 binding, indicating a shared protein-interaction region (76). Finally PLTP is ubiquitously expressed (16, 17), while apoA1 is largely restricted to the liver and intestines (77), thereby highlighting the importance of plasma PLTPa in RCT involving macrophage foam cells at peripheral sites with atherosclerotic plaque (78).
In this work, we report that 31 total regional SNPs significantly associated with PLTPa when evaluated separately; moreover, five of nine previously reported SNPs (40) replicated in this dataset. Of the 31 significantly associated SNPs, two were significantly and additively predictive of plasma PLTPa, when considering other covariates, indicating that there were likely two distinct regions independently associated with PLTPa. The first of these SNPs is a 5′ PLTP SNP, rs4810479. Our report that rs4810479 is negatively associated with plasma PLTPa replicates prior findings in a smaller subset of the current data (40). Moreover, rs4810479 has previously been associated with decreased mean HDL-P size (79), increased small HDL-P concentration (80), and decreased large LDL cholesterol concentrations (80). As rs4810479 is located 5′ of PLTP and is not protein coding, it likely represents a regulatory region for the PLTP gene; it should be noted that it is not in strong LD (r2 < 0.8) with rs7679, a PLTP SNP previously reported to affect PLTP expression (81). However, in our data, rs4810479 was the PLTP region SNP most strongly associated with plasma PLTPa, and once it was in the final regression model, no other PLTP SNP (including rs7679) explained additional PLTPa variance. The second unique genetic signal associating with PLTPa was PCIF1 5′ SNP rs181914932, which was positively associated with PLTPa. We were unable to ascribe this association to LD with a PLTP SNP. Comparatively little is known about PCIF1 or rs181914932. PCIF1 encodes a WW-domain interacting protein that has been shown to affect RNA polymerase II activity (82) and MODY4 expression (83), a gene that has been implicated in diabetes pathogenesis. Further work is needed to elaborate the role that PCIF1 plays in PLTPa.
We also report in this work that 16 of the 31 SNPs significantly associated with PLTPa are also predictive of CAAD status (see supplementary Table 5), replicating three SNPs from prior report (40) and addressing Mendelian randomization evidence (5) of the potential causal role of PLTPa in atherogenesis. We note that of the two SNPs that were retained in the final model predicting PLTPa (see Table 4), PLTP SNP rs4810479 was only marginally predictive of CAAD (P = 0.097), while PCIF1 SNP rs181914932 was not (P = 0.62). Moreover, it is notable that those SNPs that significantly predict both decreased PLTPa and CAAD in our univariate data are associated with decreased CAAD risk. For example, rs6065904 is associated with a decrease in PLTPa of 1.054 μM/h per minor allele (P = 1.35 × 10−5) and is also associated with decreased CAAD odds (OR = 0.74, 0.59–0.94, P = 0.012). Such findings are not consistent with PLTPa being cardioprotective and suggest a complex relationship between genotype, PLTPa, and atherosclerotic end-organ damage. As a potential explanation, it is possible that the PLTP SNPs associated with CAAD in this work are more strongly correlated with increased atherogenic PLTP gene expression that is hepatic or systemic (27, 28), resulting in the finding that PLTP SNPs are predictive of CAAD. In contrast, our gold-standard assay measure of PLTPa may better reflect peripheral tissue and atherosclerotic plaque PLTPa, thereby confirming our finding that PLTPa measured by the radiometric assay used in this study is atheroprotective (33, 34). As a whole, such a complex relationship with high correlation between systemic and peripheral PLTP gene expression that is differentially reflected in our PLTPa measurements could result in conflicting findings, which are overall reflective of the immense complexity of PLTP and its role in atherosclerosis. Further work on the tissue-specific gene expression changes in PLTP with varying genotype are needed, both in peripheral tissue and atherosclerotic lesions where PLTP expression is expected to be protective against atherosclerosis, in addition to measurements of systemic and hepatic elevations of PLTP expression that are likely proatherogenic, to firmly elucidate the role of PLTPa and PLTP genetic variants on risk of CAAD.
Several limitations of our study should be considered. First, this subset of the CLEAR study was composed entirely of European ancestry subjects, limiting inferences from our data to participants of other races. Second, due to the cross-sectional nature of this study, no inferences can be made on causality, although the association of PLTP SNPs with CAAD suggests a causal role. Future follow-up with prospective design, PLTP genotype data, and baseline plasma PLTPa measured using the gold-standard assay is required to better understand the etiological role that PLTPa has in CVD. Third, we only examined SNPs in the PLTP region. Other genetic predictors of PLTPa likely exist. Finally, we could not directly adjust for the effects of statins in our analysis of CAAD, as statin use is confounded with CAAD status. Instead, we adjusted the levels of plasma PLTPa by the mean change in controls on statins (−1.02), a method previously used to adjust for the effects of statins on lipid levels (47, 59), leading to a more conservative estimate of the effect of PLTPa on CAAD status.
In summary, we have performed the largest known study of the genetic and nongenetic factors predicting plasma PLTPa. In our data, we confirm the associations of triglycerides, diabetes, smoking status, apoA1, statin use, and numerous PLTP region SNPs on PLTPa. We also report two novel associations: first, the positive association of PON1 AREase activity with PLTPa and second, the association of PCIF1 SNPs. Due to the emerging focus on the process of RCT (61), an increased focus on PLTPa, which we have shown to be cardioprotective in this data, is warranted.
Supplementary Material
Acknowledgments
The authors would like to thank all CLEAR participants.
Footnotes
Abbreviations:
- AIC
- Akaike’s information criterion
- AREase
- arylesterase
- CAAD
- carotid artery disease
- CHD
- coronary heart disease
- cIMT
- carotid intima media thickness
- CLEAR
- Carotid Lesion Epidemiology and Risk
- EPIC
- European Prospective Investigation into Cancer and Nutrition
- HDL-C
- HDL cholesterol
- HDL-P
- HDL particle
- LD
- linkage disequilibrium
- MESA
- Multi-Ethnic Study of Atherosclerosis
- MI
- myocardial infarction
- OR
- odds ratio
- PLTP
- phospholipid transfer protein
- PLTPa
- phospholipid transfer protein activity (the active form of phospholipid transfer protein in the plasma)
- PON1
- paraoxonase 1
- QC
- quality control
- RCT
- reverse cholesterol transport
This work was funded in part by National Institutes of Health Grant RO1 HL67406 and a State of Washington Life Sciences Discovery Award (265508) to the Northwest Institute of Genetic Medicine. D.S.K. was supported in part by the Benjamin and Margaret Hall Endowed Fellowship in Genome Sciences, a Markey Foundation award, and National Institutes of Health Grant 1F31MH101905-01. The authors declare no conflicts of interest.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one figure and five tables.
REFERENCES
- 1.Rosenson R. S. 2005. Low HDL-C: a secondary target of dyslipidemia therapy. Am. J. Med. 118: 1067–1077. [DOI] [PubMed] [Google Scholar]
- 2.Castelli W. P. 1983. Cardiovascular disease and multifactorial risk: challenge of the 1980s. Am. Heart J. 106: 1191–1200. [DOI] [PubMed] [Google Scholar]
- 3.Acharjee S., Boden W. E., Hartigan P. M., Teo K. K., Maron D. J., Sedlis S. P., Kostuk W., Spertus J. A., Dada M., Chaitman B. R., et al. 2013. Low levels of high-density lipoprotein cholesterol and increased risk of cardiovascular events in stable ischemic heart disease patients: a post-hoc analysis from the COURAGE trial (Clinical Outcomes Utilizing Revascularization and Aggressive Drug Evaluation). J. Am. Coll. Cardiol. 62: 1826–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boden W. E., Probstfield J. L., Anderson T., Chaitman B. R., Desvignes-Nickens P., Koprowicz K., McBride R., Teo K., Weintraub W.; AIM-HIGH Investigators. 2011. Niacin in patients with low HDL cholesterol levels receiving intensive statin therapy. N. Engl. J. Med. 365: 2255–2267. [DOI] [PubMed] [Google Scholar]
- 5.Voight B. F., Peloso G. M., Orho-Melander M., Frikke-Schmidt R., Barbalic M., Jensen M. K., Hindy G., Hólm H., Ding E. L., Johnson T., et al. 2012. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet. 380: 572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El Harchaoui K., Arsenault B. J., Franssen R., Després J-P., Hovingh G. K., Stroes E. S. G., Otvos J. D., Wareham N. J., Kastelein J. J. P., Khaw K-T., et al. 2009. High-density lipoprotein particle size and concentration and coronary risk. Ann. Intern. Med. 150: 84–93. [DOI] [PubMed] [Google Scholar]
- 7.Mackey R. H., Greenland P., Goff D. C., Lloyd-Jones D., Sibley C. T., Mora S. 2012. High-density lipoprotein cholesterol and particle concentrations, carotid atherosclerosis, and coronary events: MESA (multi-ethnic study of atherosclerosis). J. Am. Coll. Cardiol. 60: 508–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim D. S., Burt A. A., Rosenthal E. A., Ranchalis J. E., Eintracht J. F., Hatsukami T. S., Furlong C. E., Marcovina S., Albers J. J., Jarvik G. P. 2014. HDL-3 is a superior predictor of carotid artery disease in a case-control cohort of 1725 participants. J. Am. Heart Assoc. 3: e000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kontush A., Chantepie S., Chapman M. J. 2003. Small, dense HDL particles exert potent protection of atherogenic LDL against oxidative stress. Arterioscler. Thromb. Vasc. Biol. 23: 1881–1888. [DOI] [PubMed] [Google Scholar]
- 10.Jarvik G. P., Rozek L. S., Brophy V. H., Hatsukami T. S., Richter R. J., Schellenberg G. D., Furlong C. E. 2000. Paraoxonase (PON1) phenotype is a better predictor of vascular disease than is PON1192 or PON155 genotype. Arterioscler. Thromb. Vasc. Biol. 20: 2441–2447. [DOI] [PubMed] [Google Scholar]
- 11.Jarvik G. P., Hatsukami T. S., Carlson C., Richter R. J., Jampsa R., Brophy V. H., Margolin S., Rieder M., Nickerson D., Schellenberg G. D. 2003. Paraoxonase activity, but not haplotype utilizing the linkage disequilibrium structure, predicts vascular disease. Arterioscler. Thromb. Vasc. Biol. 23: 1465–1471. [DOI] [PubMed] [Google Scholar]
- 12.Kim D. S., Burt A. A., Ranchalis J. E., Richter R. J., Marshall J. K., Eintracht J. F., Rosenthal E. A., Furlong C. E., Jarvik G. P. 2012. Additional common polymorphisms in the PON gene cluster predict PON1 activity but not vascular disease. J. Lipids. 2012: 476316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim D. S., Crosslin D. R., Auer P. L., Suzuki S. M., Marsillach J., Burt A. A., Gordon A. S., Meschia J. F., Nalls M. A., Worrall B. B., et al. ; on behalf of the NHLBI Exome Sequencing Project. 2014. Rare coding variation in paraoxonase-1 is associated with ischemic stroke in the NHLBI Exome Sequencing Project. J. Lipid Res. 55: 1173–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutchins P. M., Ronsein G. E., Monette J. S., Pamir N., Wimberger J., He Y., Anantharamaiah G. M., Kim D. S., Ranchalis J. E., Jarvik G. P., et al. 2014. Quantification of HDL particle concentration by calibrated ion mobility analysis. Clin. Chem. 60: 1393–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim D. S., Hutchins P. M., Jarvik G. P. 2014. The HDL particle: frontiers for new discovery in cardioprotection. Clin. Lab. Int. 38: 16–19. [Google Scholar]
- 16.O’Brien K. D., Vuletic S., McDonald T. O., Wolfbauer G., Lewis K., Tu A-Y., Marcovina S., Wight T. N., Chait A., Albers J. J. 2003. Cell-associated and extracellular phospholipid transfer protein in human coronary atherosclerosis. Circulation. 108: 270–274. [DOI] [PubMed] [Google Scholar]
- 17.Desrumaux C. M., Mak P. A., Boisvert W. A., Masson D., Stupack D., Jauhiainen M., Ehnholm C., Curtiss L. K. 2003. Phospholipid transfer protein is present in human atherosclerotic lesions and is expressed by macrophages and foam cells. J. Lipid Res. 44: 1453–1461. [DOI] [PubMed] [Google Scholar]
- 18.Cheung M. C., Albers J. J. 2006. Active plasma phospholipid transfer protein is associated with apoA-I-but not apoE-containing lipoproteins. J. Lipid Res. 47: 1315–1321. [DOI] [PubMed] [Google Scholar]
- 19.Oka T., Kujiraoka T., Ito M., Egashira T., Takahashi S., Nanjee M. N., Miller N. E., Metso J., Olkkonen V. M., Ehnholm C., et al. 2000. Distribution of phospholipid transfer protein in human plasma: presence of two forms of phospholipid transfer protein, one catalytically active and the other inactive. J. Lipid Res. 41: 1651–1657. [PubMed] [Google Scholar]
- 20.Tall A. R., Krumholz S., Olivecrona T., Deckelbaum R. J. 1985. Plasma phospholipid transfer protein enhances transfer and exchange of phospholipids between very low density lipoproteins and high density lipoproteins during lipolysis. J. Lipid Res. 26: 842–851. [PubMed] [Google Scholar]
- 21.Bailey D., Ruel I., Hafiane A., Cochrane H., Iatan I., Jauhiainen M., Ehnholm C., Krimbou L., Genest J. 2010. Analysis of lipid transfer activity between model nascent HDL particles and plasma lipoproteins: implications for current concepts of nascent HDL maturation and genesis. J. Lipid Res. 51: 785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheung M. C., Wolfbauer G., Albers J. J. 2011. Different phospholipid transfer protein complexes contribute to the variation in plasma PLTP specific activity. Biochim. Biophys. Acta. 1811: 343–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolfbauer G., Albers J. J., Oram J. F. 1999. Phospholipid transfer protein enhances removal of cellular cholesterol and phospholipids by high-density lipoprotein apolipoproteins. Biochim. Biophys. Acta. 1439: 65–76. [DOI] [PubMed] [Google Scholar]
- 24.Chirackal Manavalan A. P., Kober A., Metso J., Lang I., Becker T., Hasslitzer K., Zandl M., Fanaee-Danesh E., Pippal J. B., Sachdev V., et al. 2014. Phospholipid transfer protein is expressed in cerebrovascular endothelial cells and involved in high density lipoprotein biogenesis and remodeling at the blood-brain barrier. J. Biol. Chem. 289: 4683–4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albers J. J., Vuletic S., Cheung M. C. 2012. Role of plasma phospholipid transfer protein in lipid and lipoprotein metabolism. Biochim. Biophys. Acta 1821: 345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang X. C., Qin S., Qiao C., Kawano K., Lin M., Skold A., Xiao X., Tall A. R. 2001. Apolipoprotein B secretion and atherosclerosis are decreased in mice with phospholipid-transfer protein deficiency. Nat. Med. 7: 847–852. [DOI] [PubMed] [Google Scholar]
- 27.Lie J., de Crom R., van Gent T., van Haperen R., Scheek L., Sadeghi-Niaraki F., van Tol A. 2004. Elevation of plasma phospholipid transfer protein increases the risk of atherosclerosis despite lower apolipoprotein B-containing lipoproteins. J. Lipid Res. 45: 805–811. [DOI] [PubMed] [Google Scholar]
- 28.Yang X. P., Yan D., Qiao C., Liu R. J., Chen J-G., Li J., Schneider M., Lagrost L., Xiao X., Jiang X-C. 2003. Increased atherosclerotic lesions in apoE mice with plasma phospholipid transfer protein overexpression. Arterioscler. Thromb. Vasc. Biol. 23: 1601–1607. [DOI] [PubMed] [Google Scholar]
- 29.Samyn H., Moerland M., van Gent T., van Haperen R., Grosveld F., van Tol A., de Crom R. 2009. Elevation of systemic PLTP, but not macrophage-PLTP, impairs macrophage reverse cholesterol transport in transgenic mice. Atherosclerosis. 204: 429–434. [DOI] [PubMed] [Google Scholar]
- 30.Teslovich T. M., Musunuru K., Smith A. V., Edmondson A. C., Stylianou I. M., Koseki M., Pirruccello J. P., Ripatti S., Chasman D. I., Willer C. J., et al. 2010. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 466: 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vergeer M., Boekholdt S. M., Sandhu M. S., Ricketts S. L., Wareham N. J., Brown M. J., de Faire U., Leander K., Gigante B., Kavousi M., et al. 2010. Genetic variation at the phospholipid transfer protein locus affects its activity and high-density lipoprotein size and is a novel marker of cardiovascular disease Susceptibility. Circulation. 122: 470–477. [DOI] [PubMed] [Google Scholar]
- 32.Liu R., Hojjati M. R., Devlin C. M., Hansen I. H., Jiang X. C. 2007. Macrophage phospholipid transfer protein deficiency and ApoE secretion: impact on mouse plasma cholesterol levels and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 27: 190–196. [DOI] [PubMed] [Google Scholar]
- 33.Valenta D. T., Bulgrien J. J., Bonnet D. J., Curtiss L. K. 2008. Macrophage PLTP is atheroprotective in LDLr-deficient mice with systemic PLTP deficiency. J. Lipid Res. 49: 24–32. [DOI] [PubMed] [Google Scholar]
- 34.Valenta D. T., Ogier N., Bradshaw G., Black A. S., Bonnet D. J., Lagrost L., Curtiss L. K., Desrumaux C. M. 2006. Atheroprotective potential of macrophage-derived phospholipid transfer protein in low-density lipoprotein receptor-deficient mice is overcome by apolipoprotein AI overexpression. Arterioscler. Thromb. Vasc. Biol. 26: 1572–1578. [DOI] [PubMed] [Google Scholar]
- 35.Curtiss L. K. 2006. What is so special about apolipoprotein AI in reverse cholesterol transport? Arterioscler. Thromb. Vasc. Biol. 26: 12–19. [DOI] [PubMed] [Google Scholar]
- 36.Robins S. J., Lyass A., Brocia R. W., Massaro J. M., Vasan R. S. 2013. Plasma lipid transfer proteins and cardiovascular disease. The Framingham Heart Study. Atherosclerosis. 228: 230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlitt A., Bickel C., Thumma P., Blankenberg S., Rupprecht H. J., Meyer J., Jiang X-C. 2003. High plasma phospholipid transfer protein levels as a risk factor for coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 23: 1857–1862. [DOI] [PubMed] [Google Scholar]
- 38.Schlitt A., Blankenberg S., Bickel C., Lackner K. J., Heine G. H., Buerke M., Werdan K., Maegdefessel L., Raaz U., Rupprecht H. J., et al. 2009. PLTP activity is a risk factor for subsequent cardiovascular events in CAD patients under statin therapy: the AtheroGene Study. J. Lipid Res. 50: 723–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schgoer W., Mueller T., Jauhiainen M., Wehinger A., Gander R., Tancevski I., Salzmann K., Eller P., Ritsch A., Haltmayer M., et al. 2008. Low phospholipid transfer protein (PLTP) is a risk factor for peripheral atherosclerosis. Atherosclerosis. 196: 219–226. [DOI] [PubMed] [Google Scholar]
- 40.Jarvik G. P., Rajagopalan R., Rosenthal E. A., Wolfbauer G., McKinstry L., Vaze A., Brunzell J., Motulsky A. G., Nickerson D. A., Heagerty P. J., et al. 2010. Genetic and nongenetic sources of variation in phospholipid transfer protein activity. J. Lipid Res. 51: 983–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jarvik G. P., Tsai N. T., McKinstry L. A., Wani R., Brophy V. H., Richter R. J., Schellenberg G. D., Heagerty P. J., Hatsukami T. S., Furlong C. E. 2002. Vitamin C and E intake is associated with increased paraoxonase activity. Arterioscler. Thromb. Vasc. Biol. 22: 1329–1333. [DOI] [PubMed] [Google Scholar]
- 42.Jarvik G. P., Jampsa R., Richter R. J., Carlson C. S., Rieder M. J., Nickerson D. A., Furlong C. E. 2003. Novel paraoxonase (PON1) nonsense and missense mutations predicted by functional genomic assay of PON1 status. Pharmacogenetics. 13: 291–295. [DOI] [PubMed] [Google Scholar]
- 43.Kim D. S., Burt A. A., Ranchalis J. E., Richter R. J., Marshall J. K., Nakayama K. S., Jarvik E. R., Eintracht J. F., Rosenthal E. A., Furlong C. E., et al. 2012. Dietary cholesterol increases paraoxonase 1 enzyme activity. J. Lipid Res. 53: 2450–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim D. S., Maden S. K., Burt A. A., Ranchalis J. E., Furlong C. E., Jarvik G. P. 2013. Dietary fatty acid intake is associated with paraoxonase 1 activity in a cohort-based analysis of 1,548 subjects. Lipids Health Dis. 12: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim D. S., Burt A. A., Ranchalis J. E., Jarvik L. E., Eintracht J. F., Furlong C. E., Jarvik G. P. 2014. Effects of dietary components on high-density lipoprotein measures in a cohort of 1,566 participants. Nutr. Metab. (Lond). 11: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim D. S., Burt A. A., Crosslin D. R., Robertson P. D., Ranchalis J. E., Boyko E. J., Nickerson D. A., Furlong C. E., Jarvik G. P. 2013. Novel common and rare genetic determinants of paraoxonase activity: FTO, SERPINA12, and ITGAL. J. Lipid Res. 54: 552–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim D. S., Burt A. A., Ranchalis J. E., Jarvik E. R., Rosenthal E. A., Hatsukami T. S., Furlong C. E., Jarvik G. P. 2013. Novel gene-by-environment interactions: APOB and NPC1L1 variants affect the relationship between dietary and total plasma cholesterol. J. Lipid Res. 54: 1512–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gunderson K. L., Steemers F. J., Lee G., Mendoza L. G., Chee M. S. 2005. A genome-wide scalable SNP genotyping assay using microarray technology. Nat. Genet. 37: 549–554. [DOI] [PubMed] [Google Scholar]
- 49.Pritchard J. K., Stephens M., Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics. 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Howie B. N., Donnelly P., Marchini J. 2009. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5: e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abecasis G. R., Auton A., Brooks L. D., DePristo M. A., Durbin R. M., Handsaker R. E., Kang H. M., Marth G. T., McVean G. A.; 1000 Genomes Project Consortium. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature. 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Delaneau O., Zagury J-F., Marchini J. 2013. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods. 10: 5–6. [DOI] [PubMed] [Google Scholar]
- 53.Bachorik P. S., Albers J. J. 1986. Precipitation methods for quantification of lipoproteins. Methods Enzymol. 129: 78–100. [DOI] [PubMed] [Google Scholar]
- 54.Marcovina S. M., Albers J. J., Henderson L. O., Hannon W. H. 1993. International Federation of Clinical Chemistry standardization project for measurements of apolipoproteins A-I and B. III. Comparability of apolipoprotein A-I values by use of international reference material. Clin. Chem. 39: 773–781. [PubMed] [Google Scholar]
- 55.Cheung M. C., Brown B. G., Marino Larsen E. K., Frutkin A. D., O’Brien K. D., Albers J. J. 2006. Phospholipid transfer protein activity is associated with inflammatory markers in patients with cardiovascular disease. Biochim. Biophys. Acta. 1762: 131–137. [DOI] [PubMed] [Google Scholar]
- 56.Cheung M. C., Wolfbauer G., Albers J. J. 1996. Plasma phospholipid mass transfer rate: relationship to plasma phospholipid and cholesteryl ester transfer activities and lipid parameters. Biochim. Biophys. Acta. 1303: 103–110. [DOI] [PubMed] [Google Scholar]
- 57.Carlson C. S., Eberle M. A., Rieder M. J., Yi Q., Kruglyak L., Nickerson D. A. 2004. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet. 74: 106–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A. R., Bender D., Maller J., Sklar P., de Bakker P. I. W., Daly M. J., et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ronald J., Rajagopalan R., Ranchalis J. E., Marshall J. K., Hatsukami T. S., Heagerty P. J., Jarvik G. P. 2009. Analysis of recently identified dyslipidemia alleles reveals two loci that contribute to risk for carotid artery disease. Lipids Health Dis. 8: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mihaylova B., Emberson J., Blackwell L., Keech A., Simes J., Barnes E. H., Voysey M., Gray A., Collins R., Baigent C.; Cholesterol Treatment Trialists’ (CTT) Collaborators. 2012. The effects of lowering LDL cholesterol with statin therapy in people at low risk of vascular disease: meta-analysis of individual data from 27 randomised trials. Lancet. 380: 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rohatgi A., Khera A., Berry J. D., Givens E. G., Ayers C. R., Wedin K. E., Neeland I. J., Yuhanna I. S., Rader D. R., de Lemos J. A., et al. 2014. HDL cholesterol efflux capacity and incident cardiovascular events. N. Engl. J. Med. 371: 2383–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Vries R., Dallinga-Thie G. M., Smit A. J., Wolffenbuttel B. H. R., van Tol A., Dullaart R. P. F. 2006. Elevated plasma phospholipid transfer protein activity is a determinant of carotid intima-media thickness in type 2 diabetes mellitus. Diabetologia. 49: 398–404. [DOI] [PubMed] [Google Scholar]
- 63.Kim D. S., Marsillach J., Furlong C. E., Jarvik G. P. 2013. Pharmacogenetics of paraoxonase activity: elucidating the role of high-density lipoprotein in disease. Pharmacogenomics. 14: 1495–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bhattacharyya T., Nicholls S. J., Topol E. J., Zhang R., Yang X., Schmitt D., Fu X., Shao M., Brennan D. M., Ellis S. G. 2008. Relationship of paraoxonase 1 (PON1) gene polymorphisms and functional activity with systemic oxidative stress and cardiovascular risk. JAMA. 299: 1265–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vaisar T., Pennathur S., Green P. S., Gharib S. A., Hoofnagle A. N., Cheung M. C., Byun J., Vuletic S., Kassim S., Singh P., et al. 2007. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Invest. 117: 746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aviram M., Rosenblat M., Bisgaier C. L., Newton R. S., Primo-Parmo S. L., La Du B. N. 1998. Paraoxonase inhibits high-density lipoprotein oxidation and preserves its functions. A possible peroxidative role for paraoxonase. J. Clin. Invest. 101: 1581–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mackness M. I., Arrol S., Abbott C., Durrington P. N. 1993. Protection of low-density lipoprotein against oxidative modification by high-density lipoprotein associated paraoxonase. Atherosclerosis. 104: 129–135. [DOI] [PubMed] [Google Scholar]
- 68.Watson A. D., Berliner J. A., Hama S. Y., La Du B. N., Faull K. F., Fogelman A. M., Navab M. 1995. Protective effect of high density lipoprotein associated paraoxonase. Inhibition of the biological activity of minimally oxidized low density lipoprotein. J. Clin. Invest. 96: 2882–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shih D. M., Gu L., Xia Y-R., Navab M., Li W-F., Hama S., Castellani L. W., Furlong C. E., Costa L. G., Fogelman A. M., et al. 1998. Mice lacking serum paraoxonase are susceptible to organophosphate toxicity and atherosclerosis. Nature. 394: 284–287. [DOI] [PubMed] [Google Scholar]
- 70.Shih D. M., Xia Y-R., Wang X-P., Miller E., Castellani L. W., Subbanagounder G., Cheroutre H., Faull K. F., Berliner J. A., Witztum J. L., et al. 2000. Combined serum paraoxonase knockout/apolipoprotein E knockout mice exhibit increased lipoprotein oxidation and atherosclerosis. J. Biol. Chem. 275: 17527–17535. [DOI] [PubMed] [Google Scholar]
- 71.Cheung M. C., Vaisar T., Han X., Heinecke J. W., Albers J. J. 2010. Phospholipid transfer protein in human plasma associates with proteins linked to immunity and inflammation. Biochemistry. 49: 7314–7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vuletic S., Dong W., Wolfbauer G., Tang C., Albers J. J. 2011. PLTP regulates STAT3 and NFκB in differentiated THP1 cells and human monocyte-derived macrophages. Biochim. Biophys. Acta. 1813: 1917–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vuletic S., Taylor B. A., Tofler G. H., Chait A., Marcovina S. M., Schenck K., Albers J. J. 2008. SAA and PLTP activity in plasma of periodontal patients before and after full-mouth tooth extraction. Oral Dis. 14: 514–519. [DOI] [PubMed] [Google Scholar]
- 74.Jänis M. T., Siggins S., Tahvanainen E., Vikstedt R., Silander K., Metso J., Aromaa A., Taskinen M-R., Olkkonen V. M., Jauhiainen M., et al. 2004. Active and low-active forms of serum phospholipid transfer protein in a normal Finnish population sample. J. Lipid Res. 45: 2303–2309. [DOI] [PubMed] [Google Scholar]
- 75.Settasatian N., Duong M., Curtiss L. K., Ehnholm C., Jauhiainen M., Huuskonen J., Rye K. A. 2001. The mechanism of the remodeling of high density lipoproteins by phospholipid transfer protein. J. Biol. Chem. 276: 26898–26905. [DOI] [PubMed] [Google Scholar]
- 76.Oram J. F., Wolfbauer G., Vaughan A. M., Tang C., Albers J. J. 2003. Phospholipid transfer protein interacts with and stabilizes ATP-binding cassette transporter A1 and enhances cholesterol efflux from cells. J. Biol. Chem. 278: 52379–52385. [DOI] [PubMed] [Google Scholar]
- 77.Sliwkowski M. B., Windmueller H. G. 1984. Rat liver and small intestine produce proapolipoprotein A-I which is slowly processed to apolipoprotein A-I in the circulation. J. Biol. Chem. 259: 6459–6465. [PubMed] [Google Scholar]
- 78.Lee-Rueckert M., Vikstedt R., Metso J., Ehnholm C., Kovanen P. T., Jauhiainen M. 2006. Absence of endogenous phospholipid transfer protein impairs ABCA1-dependent efflux of cholesterol from macrophage foam cells. J. Lipid Res. 47: 1725–1732. [DOI] [PubMed] [Google Scholar]
- 79.Kaess B. M., Tomaszewski M., Braund P. S., Stark K., Rafelt S., Fischer M., Hardwick R., Nelson C. P., Debiec R., Huber F., et al. 2011. Large-scale candidate gene analysis of HDL particle features. PLoS ONE. 6: e14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chasman D. I., Paré G., Mora S., Hopewell J. C., Peloso G., Clarke R., Cupples L. A., Hamsten A., Kathiresan S., Mälarstig A., et al. 2009. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genet. 5: e1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kathiresan S., Voight B. F., Purcell S., Musunuru K., Ardissino D., Mannucci P. M., Anand S., Engert J. C., Samani N. J., Schunkert H., et al. 2009. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 41: 334–341. [Erratum. 2009. Nat. Genet. 41: 762.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fan H., Sakuraba K., Komuro A., Kato S., Harada F., Hirose Y. 2003. PCIF1, a novel human WW domain-containing protein, interacts with the phosphorylated RNA polymerase II. Biochem. Biophys. Res. Commun. 301: 378–385. [DOI] [PubMed] [Google Scholar]
- 83.Liu A., Desai B. M., Stoffers D. A. 2004. Identification of PCIF1, a POZ domain protein that inhibits PDX-1 (MODY4) transcriptional activity. Mol. Cell. Biol. 24: 4372–4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.