Abstract
Norovirus protease cleaves the virus-encoded polyprotein into six mature nonstructural proteins, presenting itself as an essential enzyme for the viral replication as well as an attractive target for the antiviral drug development. A deeper understanding of the structural mechanism of the protease-substrates/inhibitors interactions by means of solution NMR methods would facilitate a rational design of the virus protease inhibitor. We here report the backbone and side-chain resonance assignment of the protease from Norwalk virus, which is the prototype strain of norovirus. The assignment data has been deposited in the BMRB database under the accession number 17523.
Keywords: Norovirus, Norwalk virus, Viral protease, NMR, Resonance assignments
Biological context
Noroviruses are now recognized as the major cause of acute nonbacterial gastroenteritis outbreaks in all age groups worldwide. It is estimated that there are 23 million annual infection cases in the US alone. Despite the serious impact to pubic health and highly contagious nature, no vaccine and antiviral targeting norovirus infection are available to date. Norovirus shows high diversity with five genogroups (GI–GV) with Norwalk virus (NV) being the prototype of GI (GI.1). Norovirus genome is a 7.5–7.7 kb, positive-sense, single stranded RNA containing three open reading frames (ORF1–3) (Lambden et al. 1993). ORF-2 and ORF-3 encode major and minor capsid proteins, VP1 and VP2, respectively. ORF1 encodes a ~200 kDa polyprotein that is co- and post-translationally cleaved by a viral protease to produce at least six nonstructural proteins essential for viral infectious cycle (Sosnovtsev et al. 2006), indicating that the norovirus protease can be a potential target for development of antivirals such as substrate peptide-mimetic drugs.
Norovirus protease, which is alternatively called 3C-like protease to indicate the similarity with the picornavirus 3C protease, consists of 181 amino acids. Norovirus protease belongs to the viral cysteine protease family that adopts a serine-protease (chymotrypsin-like) fold hosting the proteolytic machinery with a cysteine residue acting as a nucleophile instead of serine residue (Someya et al. 2002; Zeitler et al. 2006; Nakamura et al. 2005). Overall structure is comprised of two β barrel-like domains separated by a cleft that harbors the active-site catalytic residues, His30, Glu54, and Cys139, although there exists controversy regarding a role of Glu54 (Zeitler et al. 2006; Nakamura et al. 2005). To date, extensive analysis has been carried out in terms of the substrate specificity of norovirus proteases by using mainly in vitro translational assay; the specificity depends on the primary sequence of the cleavage site, where Gln/Gly (P1/P1′), Glu/Ala and Glu/Gly dipeptides of a polyprotein are cleaved (Blackeney et al. 2003; Hardy et al. 2002; Sosnovtsev et al. 2006). Substrate P4 residue adjoining the cleavage site has also been shown to be critical for the substrate recognition by the protease (Hardy et al. 2002). Crystal structures of Norwalk virus protease (NVpro) and Chiba virus protease (CVpro) and the crystal structure-based substrate docking models have suggested several key interactions for the substrate recognition including S1 and S2 binding pockets that accommodate P1 and P2 residues (Zeitler et al. 2006; Nakamura et al. 2005). However, further studies of the structural basis for the substrate specificity is necessary to understand the protease-substrate interactions and to design substrate-based peptidomimetic inhibitors. We here report the backbone and side-chain 1H, 15N and 13C, resonance assignments of NVpro. The assignments will be valuable information for a detailed structural characterization of the enzyme-substrate interactions and facilitate NMR-based structure activity-relationship studies for the antiviral development.
Materials and experiments
A pET28 (Novagen) plasmid containing NVpro sequence with a stretch of N-terminal six histidine tags was transformed into Escherichia coli BL21(Invitrogen) cells. Uniformly 15N- or 13C/15N-labeled NVpro was expressed in the E. coli cells grown in M9 minimal media supplemented with 1 g/L 15NH4Cl and 2 g/L 13C-d-glucose (Cambridge Isotope Laboratories) for 15N-labeling and 13C-labeling, respectively. Briefly, the cells containing the expression plasmid were grown in a starter culture consisting of 50 mL LB media at 37°C for 6–8 h followed by centrifugation. The cells were resuspended in M9 minimal media, grown to an OD600 value of 1.0, induced with 1.0 mM isopropyl β-d-thiogalactoside, and further grown at 37°C for 5 h. The cells were harvested by centrifugation, and subsequently lysed using sonication. After centrifugation of the cell lysate, NVpro was purified using a Ni–NTA affinity column (QIAGEN). Size exclusion chromatography on a Superdex 75 prep grade (GE healthcare) was applied as a final purification step. NVpro was purified as a monomer and the final protein yield was 35–40 mg from 1 L of growing culture. All NMR samples contained 0.5–0.9 mM uniformly 15N- or 13C/15N-labeled NVpro, 50 mM sodium phosphate pH 6.5, 100 mM NaCl, 5 mM DTT, 3 mM NaN3 in 90% H2O/10% D2O or 99% D2O. In addition, selective amino acid labeling of Val and Ala residues were conducted by growing cells in M9 media containing 150 mg/L of 15N-Ala or 15N-Val, 200 mg/L of the other unlabeled amino acids, and 2 g/L glucose.
NMR measurements were carried out at 25°C on a Varian VNMR 500 MHz, Bruker Avance 700 and 800 MHz spectrometers equipped with cryogenic triple resonance probes. Backbone resonance assignments were achieved using 2D 1H–15N HSQC and 3D HNCA, HN(CO)CA, HNCACB, CBCA(CO)NH, HNCO, HN(CA)CO, 15N-NOESY-HSQC experiments recorded on 13C/15N-labeled NVpro. Following spectra were collected for side chain assignments: HBHA(CO)NH, H(C)(CO)NH, (H)C(CO)NH, HCCH-TOCSY, (H)CCH-TOCSY. Validation of the assignments has also been conducted with 13C-NOESY-HSQC spectrum. All NMR spectra were processed using NMRPipe (Delaglio et al. 1995), and analyzed with Sparky (Goddard and Kneller 2006) and CARA (http://www.nmr.ch) (Keller 2004).
Extent of assignments and data deposition
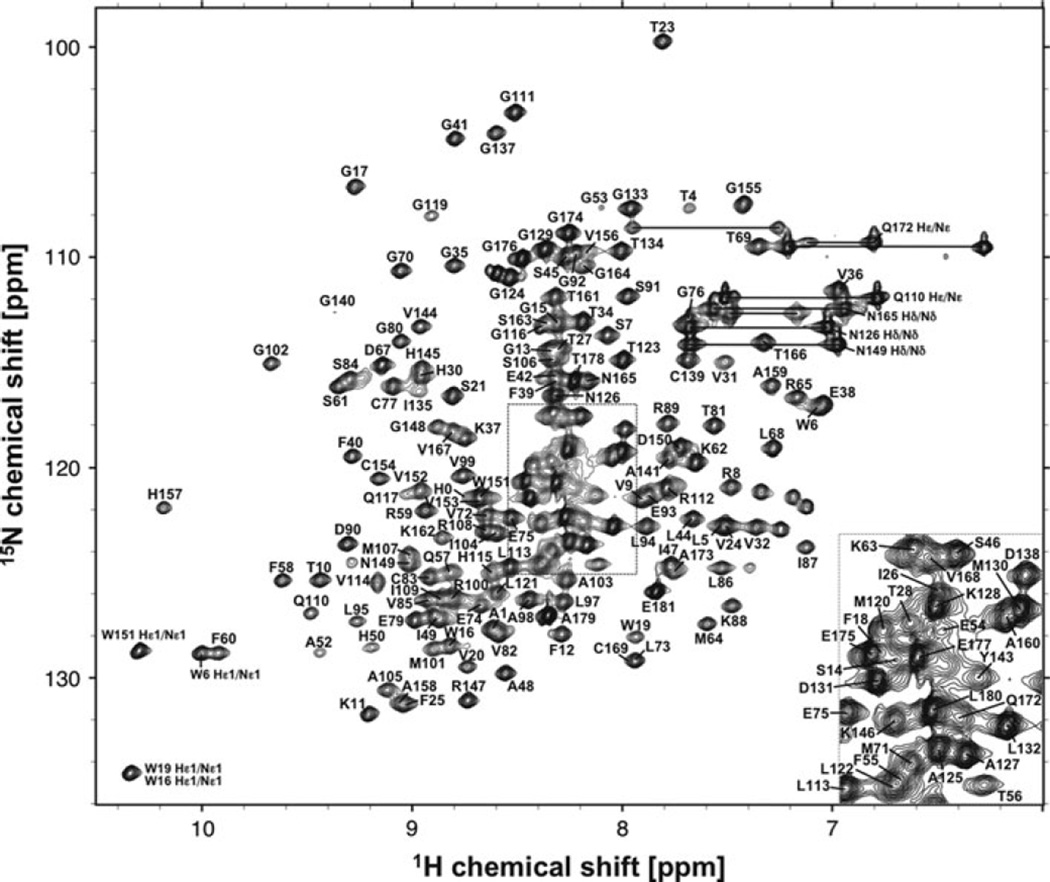
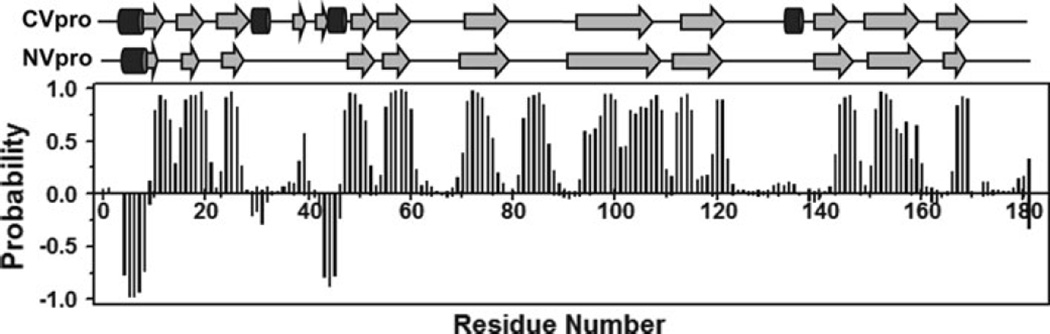
2D 1H–15N HSQC measurement of NVpro resulted in a well-dispersed spectrum (Fig. 1). Amino acid numbering is made on the authentic NVpro sequence, which excludes N-terminal His tag residues. A total of 94% of backbone 1HN and 15N resonances of 177 non-proline residues, 95% of all 13Cα resonances, all 13Cβ resonances, and 13C′ resonances have been unambiguously assigned based on a standard set of triple resonance spectra described above. These assignments were also aided by selective amino acid labeling of Val and Ala residues. The backbone amide resonances that could not be assigned include five N-terminal His tags, T29, Q51, S118, and A170–V171. Based on crystal structure of NVpro, most of the unassigned residues are located on loops connecting β-strands and are due to severe overlap, broadening and missing 13Cα and 13Cβ resonances. It should also be noted that resonance assignments were achieved for a region of L122-G133 for which no electron density was observed in NVpro crystal structure (Zeitler et al. 2006). Side-chain 1H and 13C resonance assignments were ~90% complete with the exceptions of aromatic rings. The secondary structures of NVpro were predicted by TALOS+ program (Shen et al. 2009) using the resonance assignment of 13Cα, 13Cβ, and 13C′ resonances (Fig. 2), which were in good agreement with that of the crystal structure of NVpro (Zeitler et al. 2006). The assignment has been deposited in BioMagResBank (http://www.bmrb.wisc.edu) under the accession number 17523.
Fig. 1.
2D 1H-15N HSQC spectrum of 0.8 mM 13C/15N-labeled NVpro recorded at 298 K on a Bruker 800 MHz spectrometer equipped with a cryogenic triple resonance probe. Sequence specific assignments are indicated
Fig. 2.
Secondary structure prediction for NVpro based on TALOS+ program with obtained chemical shift values. β-strand probabilities are given by positive values and those of α-helix are by negative for clarity. Shown in top of the chart are the secondary structure topology obtained from the crystal structure of NVpro and CVpro (PDB ID: 2FYQ, 1WQS) with α-helix shown as cylinder and β-sheet as arrow, respectively. The predicted secondary structure corresponds well with that of the crystal structures
Acknowledgements
We would like to thank Drs Thallapuranam Krishnaswamy S. Kumar, Haribabu Arthanari, and Gianluigi Veglia for their supports and discussion with NMR experiments. This work was supported by NIH grants 1S10RR025441, RR17686, RR17708 and U01AI081891, Johnson Center for Basic Cancer Research, and Targeted Excellence Programs of Kansas State University.
Contributor Information
Daisuke Takahashi, Department of Biochemistry, Kansas State University, 141 Chalmers Hall, Manhattan, KS 66506, USA.
Yunjeong Kim, Department of Diagnostic Medicine and Pathobiology, College of Veterinary Medicine, Kansas State University, 1800 Denison Avenue, Manhattan, KS 66506, USA.
Kyeong-Ok Chang, Department of Diagnostic Medicine and Pathobiology, College of Veterinary Medicine, Kansas State University, 1800 Denison Avenue, Manhattan, KS 66506, USA.
Asokan Anbanandam, Structural Biology Center, The University of Kansas, Lawrence, KS 66047, USA.
Om Prakash, Email: omp@ksu.edu, Department of Biochemistry, Kansas State University, 141 Chalmers Hall, Manhattan, KS 66506, USA.
References
- Blackeney SJ, Cahill A, Reilly PA. Processing of Norwalk virus nonstructural proteins by a 3C-like cysteine proteinase. Virology. 2003;308:216–224. doi: 10.1016/s0042-6822(03)00004-7. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG. SPARKY 3.113. University of California, San Francisco: 2006. [Google Scholar]
- Hardy ME, Crone TJ, Brower JE, Ettayebi K. Substrate specificity of the Norwalk virus 3C-like proteinase. Virus Res. 2002;89:29–39. doi: 10.1016/s0168-1702(02)00114-4. [DOI] [PubMed] [Google Scholar]
- Keller R. The computer aided resonance assignment tutorial. Verlag Goldau, Switzerland: 2004. [Google Scholar]
- Lambden PR, Caul EO, Ashley CR, Clarke IN. Sequence and genome organization of a human small round-structured (Norwalk-like) virus. Science. 1993;259:516–519. doi: 10.1126/science.8380940. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Someya Y, Kumasaka T, Ueno G, Yamamoto M, Sato T, Takeda N, Miyamura T, Tanaka N. A norovirus protease structure provides insights into active and substrate binding site integrity. J Virol. 2005;79:13685–13693. doi: 10.1128/JVI.79.21.13685-13693.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS +: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Someya Y, Takeda N, Miyamura T. Identification of active-site amino acid residues in the Chiba virus protease. J Virol. 2002;76:5949–5958. doi: 10.1128/JVI.76.12.5949-5958.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovtsev SV, Belliot G, Chang KO, Prikhodko VG, Thackray LB, Wobus CE, Karst SM, Virgin HW, Green KY. Cleavage map and proteolytic processing of the murine norovirus nonstructural polyprotein in infected cells. J Virol. 2006;80:7816–7831. doi: 10.1128/JVI.00532-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitler CE, Estes MK, Prasad BVV. X-ray crystallographic structure of the Norwalk virus protease at 1.5-Å resolution. J Virol. 2006;80:5050–5058. doi: 10.1128/JVI.80.10.5050-5058.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]