Abstract
miR-155 is involved in non-coding microRNAs found in humans, mice and chickens of which the sequence is conserved. Historically, miR-155 was identified as a B-cell integration cluster (bic), which induces B-cell leucosis in chickens, by its activation through viral promoter insertion. Subsequent studies have shown that transgenic mice expressing miR-155 in B cells generated lymphoma, showing that miR-155 is oncogenic. Biochemical investigation identifies many substrates of miR-155, and one of them in B cells and macrophages is the SH2-domain containing inositol-5′-phosphatase 1. A deficiency of miR-155 in the immune system causes attenuated immune functions. Clinically, several types of malignancy including diffuse large B-cell lymphoma have high miR-155 expression levels.
Keywords: inflammation, microRNA, SH2-domain containing inositol-5′-phosphatase 1, signal transduction, tyrosine kinase
Introduction
MicroRNAs (miRNAs) are single-strand RNAs that are 18–25 nucleotides in length. Genes of miRNAs found in the genome suppress newly synthesized proteins by either causing mRNA degradation or inhibiting mRNA translation into proteins. This miRNA-mediated modification of post-transcriptionally controlled gene expression was first described in nematodes and was then found in other animals and plants.1
The initial transcripts of miRNA (pri-mRNAs) are several thousand bases in size and are generated in the nucleus by RNA polymerase II (Fig.1). Normally, one newly generated pri-miRNA contains a couple of hairpin-structured RNAs that are subsequently cleaved into each hairpin RNA by an RNase III enzyme Drosha, leading to the precursor miRNAs (pre-miRNAs). They are exported into a cytosol compartment through a protein exportin-5 across the nuclear membrane. The hairpin portions of pre-miRNAs are excised by another RNase III enzyme Dicer, producing duplexed miRNAs, which subsequently bind to an RNA-induced silencing complex (RISC) with Argonaute protein to produce a complex with miRNAs (miRISCs). In most cases, one strand of miRNA is stable and binds to target mRNA in miRISC, followed by protein synthesis.
Figure 1.
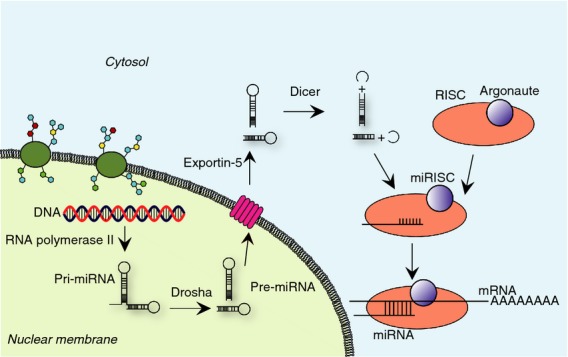
Biotransformation of microRNAs (miRNAs). First, precursor miRNA is transcribed by RNA polymerase II from DNA in the nucleus, followed by hydrolysis by an RNase III enzyme Drosha to form multiple hairpin-shaped pre-miRNAs. The generated pre-miRNA is transported into the cytosol by a nuclear membrane protein exportin-5. Subsequently, the loop structure in the hairpin RNA is cleaved by another RNase III enzyme Dicer. This reaction results in a duplexed RNA that is incorporated into an RNA-induced silencing complex (RISC) constituted with Argonaute protein to form an miRISC. Finally, mRNA to be processed is incorporated in this protein–RNA machinery.
The miRNAs can act in two ways. First, they degrade mRNA of targets followed by attenuating the accumulation of newly synthesized proteins. In this case, miRNAs bind to the 3′-untranslated region of target mRNA, leading to either mRNA degradation or protein synthesis suppression. A specific heptameric nucleotide sequence between nucleotides 2 and 8 of miRNAs determines target specificity; so this is occasionally called a ‘seed sequence’ (Fig.2a). Second, in rare cases, miRNA promotes the translation of target genes. The best characterized example includes human miR-122 that binds to the 5′-untranslated region of hepatitis C virus, leading to disease initiation. Clinically, anti-human miR-122 agents to prevent this viral activation are under development for therapeutic use with favourable results.2
Figure 2.
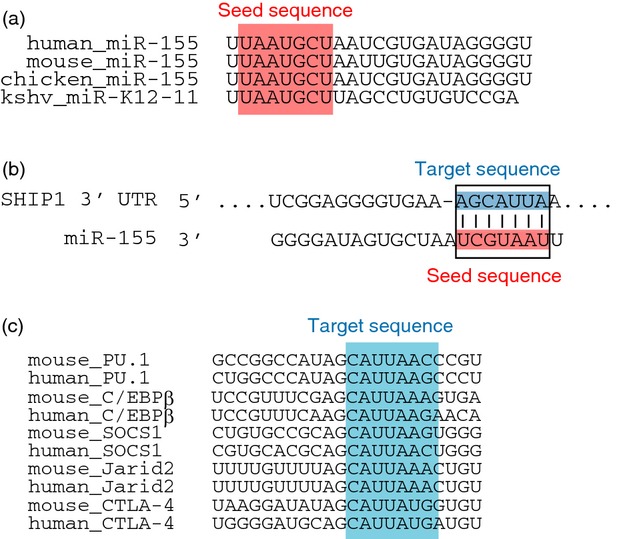
Comparison of target sequences in mammalian genes and the seed sequence of miR-155. (a) Human, mouse and chicken miR-155 seed sequences. The Kaposi sarcoma-associated herpesvirus (KSHV) -derived miR-K12-11 seed sequence is also shown. (b) Complementarity of the target sequences of human SHIP1 3′-UTR and the seed sequence of human miR-155. (c) Target sequences of PU.1, C/EBPβ, SOCS1, and Jarid2 in mice and humans. C/EBPβ, CCAAT/enhancer-binding protein beta; Jarid2, jumonji, AT rich interactive domain 2; PU.1, transcription factor that binds to the PU-box, a purine-rich DNA sequence; SOCS1, suppressor of cytokine signalling 1; UTR, untranslated region.
It is important to note that miRNA has a wide range of targets. Alternatively, the 3′-untranslated region of mRNA often, but not always, contains multiple miRNA binding sites. Normally, miRNAs induce the degradation of mRNA or the inhibition of protein synthesis, so that miRNAs usually attenuate, rather than diminish, new protein accumulation. For this reason, some researchers hypothesize that miRNAs finely tune the regulation of protein expression. Essentially, miRNAs are meant to target endogenous mRNAs to regulate their translation properly, whereas small interfering RNAs (siRNAs) normally target exogenous RNAs such as viral RNAs to protect against infection. In any case, RISCs commonly provide a platform for the reaction of both miRNAs and siRNAs.
miR-155
MicroRNA-155 (miR-155) was originally identified as a gene B-cell integration cluster (bic), which induces leucosis by its activation in the chicken genome through viral promoter insertion.3 Subsequent investigation identified a homologous gene to bic in humans and mice.4,5 The expression profile of miR-155 revealed that it is mainly expressed in the thymus and spleen; little expression can be detected in other tissues. Hence, there are many studies describing the direct correlation between the expression of miR-155 and tumour occurrence.6–8 In particular, accumulating clinical evidence has shown that poor prognosis is correlated with high miR-155 expression. To understand the underlying molecular mechanisms, many studies have investigated the target proteins of miR-155 to understand the mechanism of how miR-155 acts (Table1). In the initial stage of research, typically, a database search is performed according to the target mRNA sequence to find the seed sequence of specific miRNA. The effect of the miRNA-mediated suppression of target protein expression is experimentally examined either by the transfection of chemically synthesized miRNA into cells or by the expression of miRNA under the control of an appropriate expression promoter. This method has been applied to the investigation of targets of miR-155, and identified SH2-domain containing inositol-5′-phosphatase 1 (SHIP1) as the target in macrophages in response to lipopolysaccharide (LPS) (Fig.2b).9 Interestingly, the retrovirally induced expression of miR-155 in mice through bone marrow cell transfer revealed myeloproliferative disorder and splenomegaly, consistent with the phenotype of SHIP1-deficient mice.10 As the expression of SHIP1 in both B cells and myeloid cells is high, its physiological role through miR-155-mediated regulation in these cells has been characterized accordingly.
Table 1.
Target proteins of miR-155
| Target proteins | References |
|---|---|
| Cytosolic proteins | |
| FADD | 24 |
| IKKε | 24 |
| MyD88 | 58 |
| Ripk | 24 |
| SHIP1 | 9,18,59 |
| SOCS1 | 38,60 |
| TAB 2 | 29 |
| Nuclear proteins | |
| AID | 61,62 |
| Bach1 | 54 |
| Bcl-6 | 63 |
| C/EBPβ | 18,64 |
| Ets1 | 31 |
| Fos | 54 |
| HDAC4 | 19 |
| Jarid2 | 36 |
| PU.1 | 62,65 |
| Smad2 | 66 |
| Smad5 | 67 |
| κB-Ras1 | 68 |
| Membrane proteins | |
| CTLA-4 | 44 |
| IL-13 receptor α1 | 69 |
| Sphingosine-1-phosphate receptor 1 | 35 |
AID, activation-induced cytidine deaminase; CTLA-4, cytotoxic T-lymphocyte-associated protein; C/EBPβ, CCAAT/enhancer-binding protein β; FADD, Fas-Associated protein with Death Domain; HDAC4, Histone Deacetylase 4; IKKε, IκB kinase ε; IL, interleukin; MyD88, Myeloid differentiation primary response gene 88; SOCS1, suppressor of cytokine signaling protein 1; SHIP1, SH2 domain-containing inositol 5′-phosphatase 1; TAB 2, TAK1-binding protein 2.
Role of miR-155 in immune cells
Among the many miRNAs, current article primarily focuses on the roles of miR-155 in immune cells in terms of its physiological functions and potential signalling mechanisms (Tables1 and 2). A large number of subsequent studies has revealed how miR-155 regulates specific signalling pathways in these cells.
Table 2.
Signalling pathways involving miR-155
| Cells | Receptors or ligands | Action | References |
|---|---|---|---|
| B | BCR/anti-IgM-F(ab')2 | ERK activation→ | 14 |
| Cell proliferation→ | 14 | ||
| BCR+FcγRIIB/anti-IgM | ERK activation↑ | 14 | |
| Cell proliferation↑ | 14 | ||
| Ca2+↑ | 7 | ||
| Macrophages | LPS | TNF-α↑, G-CSF↑, SHIP1↑ | 9,23,64 |
| DCs | LPS | TNF-α↑, IL-6↑, IL-12↑, IL-23↑ | 25–27 |
| LPS | IL-1β↑, caspase 1↑ | 29 | |
| R837 | IFN-α/β↑, TNF-α↑ | 28 | |
| Th1 | TCR | IFN-γ→ | 25 |
| Th2 | TCR | IL-4/5/13↑ | 33,35 |
| Th17 | TCR | IL-17↑ | 31 |
| Tregs | IL-2 | STAT5 activation↑ | 38 |
| CD4 T | TGF-β+IL-6 | Th17↑ | 26,36 |
| TGF-β+IL-6 + IL-1β | Th17→ | 36 | |
| CD8 T | TCR | IFN-γ↑ | 32 |
| AKT activation↑ | 47 | ||
| Cell proliferation↑ | 46 |
BCR, B-cell receptor; DC, dendritic cell; G-CSF, granulocyte cell-stimulating factor; IFN-γ, interferon-γ; IL-6, interleukin-6; LPS, lipopolysaccharide; SHIP1, SH2-domain containing inositol-5′-phosphatase 1; STAT, signal transducer and activator of transcription; TCR, T-cell receptor; TGF, transforming growth factor; Th, T helper; TNF, tumour necrosis factor; Treg, regulatory T cell.
B cells
In B cells, miR-155 positively regulates antibody-mediated signalling. For example, an initial study reported that both the number of germinal centres and antibody production have been greatly reduced in miR-155-deficient mice.11 Similarly, in a collagen-induced arthritis model, the production of specific antibodies was severely impaired.12,13 The defective effect of miR-155 on antibody production in B cells seems to be potent, as even in the genetic background of Faslpr mice, which readily causes autoimmune diseases due to impaired apoptotic activity, the accumulation of autoantibody against endogenous DNA is low.14 In Faslpr mice, miR-155 expression was easily detected in B cells, suggesting that these cells have been strongly activated by endogenously generated antibody-mediated stimulation at the basal level. Other biologically relevant markers in Faslpr mice essentially correlated with antibody production. For example, tumour necrosis factor-α (TNF-α) production in response to anti-IgM antibody was decreased in miR-155-deficient mice.15 Similarly, the expression of miR-155 caused enhanced Ca2+ accumulation in response to anti-IgM antibody, whereas treatment with a miR-155-specific inhibitor effectively attenuated this in human B cells.7 Although the basal expression of miR-155 is normally very low, the activation of B cells induced markedly high expression in response not only to anti-IgM antibody, but also to LPS, CpG and anti-CD40.15
From a mechanistic point of view, autoimmune-prone Faslpr mice have high miR-155 expression with concomitantly impaired SHIP1 expression at the basal level. The stimulation of these B cells induced maximal activation of ERK (extracellular signal-regulated kinase) in response to B-cell receptor (BCR) because the F(ab')2 of antibody selectively activates the BCR-mediated positive, but not Fcγ receptor IIB (FcγRIIB) -mediated inhibitory, signalling pathway (Fig.3). Alternatively, the stimulation of both BCR and FcγRIIB with intact anti-IgM antibody suppressed ERK activation in miR-155-deficient B cells due to their co-ligation. Under these conditions, B cells from miR-155-deficient mice on a Faslpr genetic background exhibited impaired ERK phosphorylation in response to anti-IgM antibody, but not to F(ab')2, compared with those from control Faslpr mice. Importantly, in B cells from SHIP1-deficient mice, there is clear enhancement of ERK activation in response to anti-IgM antibody resulting from simultaneous stimulation with both BCR and FcγRIIB.16 This is due to the immunoreceptor tyrosine-based inhibition motif within the cytoplasmic region of FcγRIIB, where SHIP1 as well as other negative regulatory molecules are to be recruited upon tyrosine phosphorylation. Therefore, SHIP1 has been regarded as a negative regulator of the tyrosine kinase-mediated signalling pathway. In sum, the above results collectively support the idea that SHIP1 acts as a functional target of miR-155 in B cells.
Figure 3.
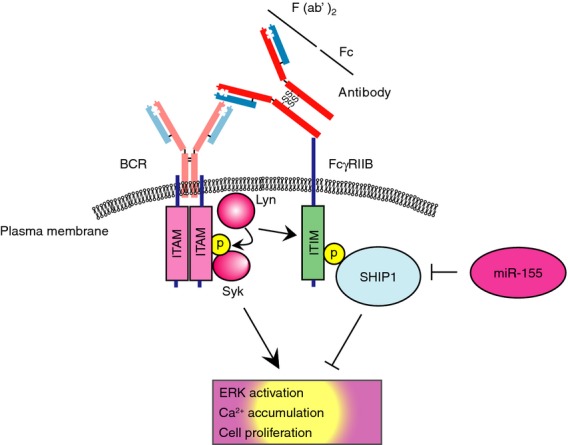
Activation of a B-cell and its regulation by miR-155. When B-cell receptor is activated by the F(ab')2 of antibody in general, a cytoplasmic protein tyrosine kinase Lyn activates a downstream tyrosine kinase Syk, followed by the initiation of the ITAM-mediated signalling pathway. When B cells are activated by the whole molecule of antibody, but not by F(ab')2, the activated Lyn phosphorylates Syk as well as ITIM in the cytoplasmic region of FcγRIIB. Tyrosine-phosphorylated ITIM then recruits SHIP1 as well as other negative regulators, leading to cell inactivation. miR-155 impairs SHIP1 expression, leading to enhanced B-cell activation in response to the whole molecule of antibody. Note that an intact antibody in general can be cleaved by pepsin protease into an F(ab')2 fragment and an Fc fragment. ITAM, immunoreceptor tyrosine-based activation motif; ITIM, immunoreceptor tyrosine-based inhibitory motif.
In contrast to miR-155-deficient mice, a transgenic mouse model expressing miR-155 under the control of Eμ enhancer and VH promoter (Eμ-miR-155) has been investigated.17 At 3–4 weeks after birth, dysregulated B-cell development caused the marked accumulation of preB cells in these mice. When these animals were housed beyond this time-point, they developed acute lymphoma due to the abnormal proliferation of B cells. The targets of miR-155 in Eμ-miR-155 mice were first identified as SHIP1 and CCAAT-enhance binding protein β (C/EBPβ) in lymphoma using a biochemical method (Fig.2b,c).18 Subsequent research further demonstrated several proteins as targets including HDAC4 in naive B cells through the use of gene expression cluster analysis.19 The latter study also showed that signalling pathways involving BCR, Toll-like receptor (TLR), ERK, c-Jun N-terminal kinase, as well as interferon regulatory factor, those directly or indirectly regulated by miR-155, were similarly impaired, whereas the downstream targets of Bcl6 were activated in these mice.19
Ectopic miR-155 expression in haematopoietic progenitors in human CD34+ cord blood cells revealed an enhanced B-cell proliferation in humanized interleukin-2 (IL-2) receptor γ-deficient NOD/scid mice.20 This careful examination revealed that the mild regulation of B-cell proliferation was observed in this animal model, showing the critical role of miR-155 on B-cell development. Moreover, this study demonstrated that malignant B-cell proliferation requires aberrant B-cell activation in vivo.
Macrophages
The expression of miR-155 in macrophages increases in response to LPS, TNF-α and interferon-β (IFN-β).21 As anticipated in B cells, accumulating evidence has shown that SHIP1 is a prominent target of miR-155 in macrophages.9 For example, the expression of miR-155 and SHIP1 was inversely correlated in human CD14+ monocytes and CD68+ macrophages in patients with rheumatoid arthritis.12 Similarly, the forced expression of miR-155 repressed SHIP1 expression in peripheral blood-derived CD14+ cells. In terms of the molecular basis of miR-155 expression, biochemical studies showed that this expression is transcriptionally regulated by AP-1 through c-Jun N-terminal kinase activation.21,22
Macrophages play an important role in the production of inflammatory cytokines under infectious settings. Essentially, miR-155 acts as a positive regulator of cytokine production in macrophages. A study has shown that miR-155 expression spontaneously induced the enhanced production of TNF-α and IL-6 in peripheral blood-derived macrophages in humans.12 Similarly, forced miR-155 expression in human monocyte-like THP1 cells produced more TNF-α and IL-1β in response to sodium monourate.23 An enhanced inflammatory response to macrophages in the absence of miR-155 was also observed in vivo, as Eμ-miR-155 mice produced more TNF-α in the serum soon after LPS/d-galactosamine administration.24 Hence, miR-155 positively regulates the nuclear factor-κB-mediated signalling pathway in macrophages as in B cells, raising the possibility that a previously unappreciated negative regulatory mechanism might be controlled by miR-155.
Dendritic cells
In dendritic cells, miR-155 has also been implicated as a positive regulator of inflammatory cytokine production similar to macrophages. Essentially, miR-155 is induced in the TLR-mediated signalling pathway. For example, in miR-155-deficient mice, bone marrow-derived dendritic cells produced a smaller amount of inflammatory cytokines, including IL-12 and IL-6, in response to LPS, which is indicative of miR-155 induction.25–27 The TLR7 agonist R837 induces less IFN-α/β and TNF-α production in miR-155-transfected human plasmacytoid dendritic cells.28 Similar to inflammatory cytokines, sterile inflammation is also positively regulated by miR-155, as the expression of IL-1β and caspase 1 decreased when miR-155 expression was attenuated by biologically stable miR-155-specific oligonucleotides in human monocyte-derived dendritic cells.29 Evidence suggests that miR-155 regulates the function of dendritic cells in animal models. One example includes a Helicobacter pylori infection model, where miR-155-deficient mice showed impaired T helper type 1 (Th1) differentiation and defective bacterial eradication, consistent with attenuated IFN-γ production from T cells stimulated by dendritic cells with LPS.27
T cells
T cells play a major role in immune homeostasis, and their biological behaviours are both positively and negatively controlled by miR-155 (Table3). T-cell-intrinsic properties regulated by miR-155 are summarized below, and the outcomes of in vivo results are to be interpreted as an integration of functionally regulated T cells and antigen-presenting cells.
Table 3.
Phenotypes of T-cell-mediated experimental animal models in miR-155-deficient mice
| Model | Effector cells | Phenotype | Cytokines | References |
|---|---|---|---|---|
| CD4-dependent | ||||
| Tumour transplantation | Th1 | Enhanced tumour growth | IFN-γ↓ | 32 |
| Salmonella typhimurium | Th1 | Reduced optimal response | IFN-γ↓ | 11 |
| Delayed-type hypersensitivity | Th1, Th17 | Less severe | IL-17↓, IFN-γ↓, IL-6↓ | 26 |
| Helicobacter pyroli | Th1, Th17 | Bacteria not eliminated | IFN-γ↓, IL-17↓ | 27 |
| Asthma/HDM | Th2 | Less severe | IL-13↓ | 35 |
| Asthma/OVA | Th2 | Less severe | IL-4/5/13↓ | 33 |
| Heligmosomoides polygyrus | Th2 | Helminth not eliminated | IL-13↓ | 35 |
| Collagen-induced arthritis | Th17, Th1 | Less severe | IL-17↓, IFN-γ↓ | 12,13 |
| EAE | Th17, Th1 | Less severe | IL-17↓, IFN-γ↓ | 25,26 |
| Toxoplasma gondii | Th17, Th1 | Reduced optimal response | IL-17↓ | 36 |
| CD8-dependent | ||||
| Influenza | CD8 | Virus not eliminated | IFN-γ↓ | 46 |
| Listeria monocytogenes | CD8 | Reduced optimal response | IFN-γ↓ | 46,47 |
| Lymphocytic choriomeningitis | CD8 | Reduced optimal response | IFN-γ↓ | 47 |
| Murid herpesvirus 68 | CD8 | Virus not eliminated | IFN-γ↓ | 45 |
| Tumour transplantation | CD8 | Enhanced tumour growth | IFN-γ↓ | 32 |
EAE, experimental autoimmune encephalomyelitis; HDM, house dust mite; IFN-γ, interferon-γ; IL-6, interleukin-6; OVA, ovalbumin; Th, T helper.
Th1
Historically, effector CD4 T cells have been classified into Th1 and Th2 cells, and their differentiation mechanisms and immunological properties have been studied in detail. Th1 cells are IFN-γ-producing helper CD4 T cells expressing T-bet as a master transcription factor. Most bacterial infection causes IFN-γ production, so the Th1-mediated process contributes to disease progression.30 A milestone study of miR-155 in a bacterial infection model was performed in Salmonella infection.11 This pathogenesis is initially caused by the accumulation of bacteria in the reticulo-endothelial system in the host, which relates to bacterial virulence. During the bacterial eradication process, innate immune cells play a major role in an earlier phase whereas Th1 cells contribute in the later phase, respectively. Deficiency of miR-155 in this infection model showed an impaired survival rate beyond 2 weeks after infection whereas there was no difference in the survival of acute phase induced by a higher dose of bacteria, suggesting that miR-155 plays an important role in the host defence mechanism in Th1 cells in the late phase. Consistent with animal studies, IFN-γ production from miR-155-deficient splenocytes was impaired in immunized mice. Hence, these results indicate that miR-155 is required for bacterial eradication in the Salmonella infection model. In an H. pylori infection model where Th1 cells play a key role in disease progression, bacterial eradication was impaired in miR-155-deficient mice.27 A more detailed examination demonstrated that IFN-γ expression was severely impaired at the late stage of disease in these infected mice. Adoptively transferred T cells isolated from infected miR-155-deficient mice also failed to eliminate infected bacteria properly, and so T-cell-intrinsic IFN-γ production controlled by miR-155 seems to be important. Furthermore, depressed Th1 differentiation in this model might be associated with the impaired secretion of inflammatory cytokines from dendritic cells in response to LPS. In an experimental autoimmune encephalomyelitis (EAE) model, both Th1 and Th17 were found to be important for disease progression. The absence of miR-155 in this model showed a less severe disease phenotype, demonstrating that this miRNA positively regulates disease development.25,26,31 Consistently, CD4 T cells isolated from immunized animals produced less IFN-γ in miR-155-deficient mice. As anticipated, the reason was attributed to the secretion of a smaller amount of IL-12, a potent Th1-skewing cytokine, in miR-155-deficient dendritic cells.25
Another Th1 example described in detail is a tumour transplantation model. In miR-155-deficient mice, transplanted tumour cells originating from both lymphoma and melanoma were not effectively eliminated.32 In this case, the generation of IFN-γ from CD4 T cells in the spleen was significantly impaired. Hence, these results suggest that miR-155 plays a critical role in anti-tumour immunity.
Th2 cells
Histological observation demonstrated that the lungs of miR-155-deficient mice exhibit a thickening of the airway epithelial layer and an accumulation of collagen, both indicative of lung airway remodelling, which is commonly observed in patients with asthma.11 This pulmonary phenotype is similarly observed in IL-13 and IL-4 transgenic mice, suggesting a strong link between Th2 skewing and miR-155. This notion was supported by the enhanced secretion of IL-4 and IL-5, but not IFN-γ and IL-2, from Th2 cells in vitro. On the other hand, in an antigen-sensitized asthma model, miR-155-deficient mice, similar to SHIP1-deficient mice, show milder disease symptoms.33,34 Eosinophilia, a hallmark of asthma, was markedly attenuated because of repressive IL-13 accumulation in bronchoalveolar lavage fluid in miR-155-deficient mice. Further research revealed that miR-155-deficiency in this asthma model led to the impaired accumulation of Th2 cells, whereas Th1 cells remained unchanged. The underlying molecular mechanism of this observation was explained by the accumulation of PU.1, a transcription factor that negatively regulates Th2 differentiation, in airway lymph node cells. This indicates that miR-155 might directly regulate the expression of PU.1 to repress Th2 (Fig.2c). Separately, a recent study also showed the suppression of Th2-dependent asthma in the absence of miR-155 using bone marrow chimeric mice. This study further identified the sphingosine 1-phosphate receptor 1 as a novel target of miR-155 in Th2 models, and its efficacy was studied in an antigen-sensitized asthma model.35
Th17
Th17 cells are newly appreciated inflammatory cells in which a transcription factor Retinoic acid-related orphan receptor ct (Rorγt) plays an essential role in their differentiation. The expression of miR-155 in Th17 is highest among Th1, Th2, Th17 and regulatory T cells. In Th17 cells, transcription factors signal transducer and activator of transcription 3 (STAT3), interferon regulatory factor 4, basic leucine zipper transcription factor, and Rorγt are upstream regulators for miR-155.36 It is well known that Th17-mediated diseases involve EAE and collagen-induced arthritis. The initiation of EAE is induced by inflammation and Th17 differentiation followed by the development of the disease with IL-23 receptor expression on Th17 cells for its maintenance. In EAE models, miR-155 deficiency is associated with milder disease symptoms.25,26,31 Essentially, the production of IL-17 and IFN-γ from T cells is impaired in miR-155-deficient mice. Such an attenuated disease progression includes the defective production of IL-6 and IL-12 from dendritic cells, recessive Th17 skewing, and attenuated IL-23 receptor expression. Hence, it is conceivable that one would expect miR-155 to have some type of therapeutic role in Th17-mediated inflammatory disorders. In fact, the administration of anti-miR-155 oligonucleotide synthesized by a recently developed locked nucleotide acid showed promising results in vivo.25
The production of IL-17 was also impaired in miR-155-deficient mice in another Th17 model. One example includes a delayed hypersensitivity model in which the immunization of antigen into the footpad in mice caused IL-17 production. In this model, reduced IL-17 expression in miR-155-deficient mice demonstrated that miR-155 positively regulates IL-17 production.26 Although potential targets of miR-155 in Th17 have been explored for some time, a recent study has shown that a transcription factor Ets1 is directly regulated by miR-155 and is considered a key target in Th17.31
Th17 cells specifically induce il17a, il17f, il1r1 and il22 as signature cytokine genes. Previous studies have suggested that not all of these Th17-type genes are regulated equally. For example, il22 has been known to be differently regulated compared with il17a. However, exactly how the il22 gene is regulated in Th17 has remained elusive. A recent paper has shown an augmented association of another miR-155 target Jarid2, a DNA-binding protein that recruits an epigenetic modulator Polycomb repressive complex 2 (PRC2), to chromatin in miR-155-deficient cells (Figs2c and 4).36 In this case, the absence of miR-155 consistently showed an increased accumulation of H3K27, Jarid2 and Suz12, a core component of PRC2, at distal, proximal and promoter regions of the il22 locus. This recruitment of PRC2 to chromatin increases H3K27 methylation followed by impaired il22 expression. Similar to il22, the expression of il10, il9 and Atf3 were regulated by this mechanism in Th17 cells.
Figure 4.
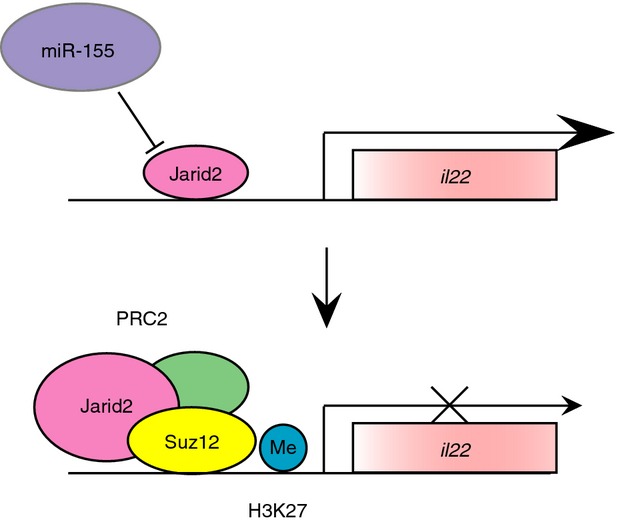
Mode of action of Jarid2 on il22 expression and its regulation by miR-155. Under basal conditions, the transcription of il22 occurs normally in response to physiological stimuli (upper). In the absence of miR-155, when Jarid2 expression increases, the recruitment of PRC2 complex including Suz12 occurs. The formed PRC2 induces H3K27 methylation in the il22 locus, leading to its inactivation (lower).
Regulatory T cells
The major function of regulatory T (Treg) cells is to suppress the growth of CD4 T cells in the presence of antigen-presenting cells. One study showed that the suppression activity of Treg cells remained unchanged in miR-155-deficient mice.37 In contrast, accumulating evidence has suggested that the number of Treg cells in the spleen and thymus is reduced in miR-155-deficient mice, and a bone marrow cell transfer experiment confirmed the results.37,38 There are several potential reasons for this. First, the decreased Treg cell number in vivo could be linked to impaired IL-6 production from dendritic cells under inflammatory conditions, as this cytokine is essential for Treg cell differentiation. Second, both apoptosis and proliferation are major factors determining the number of cells. Experimental evidence has supported the suppressed Treg cell proliferation in miR-155-deficient mice, as STAT5 phosphorylation in response to IL-2 was impaired in Treg cells isolated from them.39 In this context, an inhibitor of cytokine signalling – suppressor of cytokine signalling 1 (SOCS-1) protein – has been suggested as attenuating STAT5 phosphorylation. A database search identified that the 3′-untranslated region of SOCS-1 mRNA contains a target sequence of miR-155, leading to augmented SOCS-1 expression with enhanced Treg cell proliferation in miR-155-deficient mice (Fig.2c). In contrast, susceptibility to apoptosis in response to T-cell receptor was similar in these animals compared with wild-type controls, demonstrating clearly that there is no defect in Treg cell survival.38
Atopic dermatitis, known as a frequently observed skin disease in childhood, is caused by disorder of various signalling pathways involving Treg cells.40 The best characterized part of its aetiology is its inappropriate control of Treg cells via cytotoxic T-lymphocyte antigen 4 (CTLA-4) (Table1). This hypothesis is supported by evidence that CTLA-4 expression in T cells of patients with atopic dermatitis was elevated.41,42 Consistently, CTLA-4 polymorphism is highly detected in infants with atopic dermatitis.43 A recent study revealed that miR-155 expression was augmented in patients with atopic dermatitis.44 This study reported that CTLA-4 mRNA has a target sequence against an miR-155 seed sequence, therefore CTLA-4 expression can be modulated by miR-155 (Fig.2). These authors also showed that forced expression of pre-miR-155 in CD4 T cells reduced intracellular CTLA-4 protein expression. Hence, at least in some way, miR-155 is involved in atopic dermatitis through CTLA-4 expression in humans.
CD8 T cells
In CD8 T cells, miR-155 expression increases in response to T-cell receptor and is maintained with IL-2 and IL-15.45 Upon viral infection, miR-155 expression is elevated in effector and effector memory CD8 T-cell compartments. Animal studies have shown that the antiviral response in CD8 T cells decreases and virus eradication does not occur properly in miR-155-deficient mice.45–47 In these experiments, the number of CD8 T cells in miR-155-deficient mice was lower than in wild-type mice, suggesting that either cell proliferation or cell death may have been abnormally regulated. In Listeria monocytogenes infection, one study has shown that the phosphorylation of Akt in response to T-cell receptor in CD8 T cells in miR-155-deficient mice was impaired, whereas the number of cell divisions was unaltered, demonstrating that defective survival, but not proliferation, was attributed to the impaired viral response.47 In influenza virus infection, another study showed that STAT1-mediated signalling plays an important role in defective CD8 T-cell accumulation, whereas the contributions of IL-2 and IL-15 could be minor in these mice.46 To support this putative molecular mechanism, complementation of a dominant-negative form of either STAT1 or interferon regulatory factor 7 in miR-155-deficient CD8 T cells partially restored the number of them in reconstituted mice, confirming the hypothesis that the IFN–STAT1 signalling pathway is regulated by miR-155 in influenza virus infection. In a murid herpesvirus 68 infection model, a viral titre was enhanced by reduced optimal response due to an impaired effector response with a defective virus-specific CD8 T-cell number in miR-155-deficient mice.45 Further examination revealed that these virus-specific CD8 T cells have preferably differentiated into memory cells with enhanced Bcl-2 and IL-2 expression. In this model, there was no apparent alteration in the secondary response in these animals, indicating that miR-155 plays a critical role in their primary response in this infection model.
CD8 T cells play a key role in tumour eradication. Transplanted myeloma-derived B16-F10 cells grow faster in miR-155-deficient mice.32 In this mouse model, the population of IFN-γ-producing CD8 T cells and the expression of IFN-β itself were markedly decreased, showing the relevance of miR-155 to anti-tumour immunity.
Interaction between miR-155 and other miRNA in immune cells
The question of how miR-155 and other miRNAs are involved in immune cells is an important one. First, a transcriptome-wide study reported that the deficiency of miR-155 in CD4 T cells has little effect on the expression of other miRNAs, as determined by the RNA-sequence analysis of the immunoprecipitates of anti-Argonaute antibody.48 Inversely, miR-155 has a complimentary miRNA (miR-155-3p, previously known as miR-155*) that is generated from the opposite strand from hairpin-structured pre-miRNA by Dicer enzyme (Fig.1). One study has shown that miR-155-3p targets IL-1 receptor-associated kinase M (IRAK M) in dendritic cells,28 and so this complementary miRNA might modulate the stability of miR-155. In most cases, however, it is conceivable that such a complimentary strand is less stable; so, it is widely accepted that its biological role might be limited. Third, it is possible that other miRNAs could compete with miR-155, leading to interference with the mRNA degradation caused by it. In the precursor of human miR-155, which is 65 nucleotides in length, a database search identified that a family of miR-548s has similar nucleotide sequence to miR-155. However, they have more than four mismatched bases in matched 15 bases (see Supporting information, Fig. S1). One experimental result shows that a mutation at one nucleotide in the sequence of miRNA has almost no effect on the suppression of mRNA expression, whereas four nucleotides in the sequence cause insufficient suppression.48 Hence, these miRNAs are unlikely to be involved in the miR-155-dependent mRNA regulation mechanism.
Epistasis is an occasion in which two genes separately modulate downstream events. Such examples were reported in miR-155 as well. One well-characterized example includes miR-146a, which, together with miR-155, is also regulated by nuclear factor-κB.49 In innate immune cells, miR-155 impairs TLR-mediated signalling molecules including TAB 2 and MyD88 in addition to SHIP1 through AGCAUUAA as a target sequence. Alternatively, miR-146a attenuates TRAF6 and IL-1 receptor-associated kinase 1 (IRAK 1) expression through CAGTTCTC as its target.49,50 Apparently, the targets of these miRNAs might differ, whereas these miRNAs obviously interact on the same innate immune signalling pathway. A similar example can be found in T cells in tumour immunity. In a tumour transplantation model, miR-155 and miR-146a are cooperatively involved.32 In this case, miR-155 promotes and miR-146a suppresses IFN-γ responses in T cells in tumours, and overall, mice deficient in both miRNAs showed a tumour growth phenotype similar to that in miR-155-deficient mice. In this model, at least in part, SHIP1 is involved in the miR-155-mediated process, whereas entire mechanisms beyond this remain uncertain.
Importantly, some viruses have miRNAs with sequences that are almost identical to mammalian miR-155 (Fig.2a). The details of an example can be found in the section on Oncogenic viruses.
miR-155 in tumour cells
Transgenic mice
The first reported Eμ-miR-155 mice become oncogenic at 4–6 months after birth, and their survival rate was lower than that of wild-type mice. This lethality was transplantable because all of the transplanted mice receiving Eμ-miR-155 mouse-derived cells succumb to malignancy. These leukaemic B cells express CD79a and CD20 as surface markers, indicative of pre-B cells as determined by immunohistochemistry. More detailed FACS analysis revealed that these B cells had two populations expressing both B220low IgM− IgD− and IgM+ IgD+. Additional phenotypes in these transgenic mice include T lymphopenia with a normal CD4/CD8 ratio and myeloproliferation. Clonal analysis of BCR revealed that tumorigenic B cells contain mostly a polyclonal population, suggesting that B-cell malignancy in this miR-155 transgenic mouse strain was induced by multiple genetic mutations with enhanced proliferation. In these mice, the expression of SHIP1 and C/EBPβ, both well-described target proteins of miR-155, was markedly impaired. Using these transgenic mice as a source, additional targets of miR-155 have been intensively explored.
More recently, another line of transgenic mice has been established by a different group.51 The transgene was designed as a tetracycline-controlled Cre/loxP-driven knockin model that was homologously recombined into ROSA26 locus. Hence, in these transgenic mice, the administration of doxycycline attenuates miR-155 expression at the site where Cre-recombinase is activated. The expression of miR-155 was apparent in the spleen, thymus, and bone marrow, and the expression level in the spleen was approximately 45-fold compared with wild-type mice. Upon doxycycline administration, such an elevation of miR-155 was suppressed significantly; therefore, the expression of miR-155 in these transgenic mice could be experimentally controlled. Similar to the previous Eμ-miR-155 mouse model, the developed tumour contained pre-B cells, was transplantable, and was regressed by the withdrawal of doxycycline administration. Importantly, this transgenic model generated clonal, rather than polyclonal, tumour cells, which was not reported in the previous Eμ-miR-155 mice, suggesting that this conditionally regulated expression system would be suitable for investigating the initial stage of B-cell lymphoma. From a therapeutic point of view, the same authors reported that the administration of nanoparticles modified with polylysine-conjugated anti-miR-155 peptide nucleic acid effectively inhibited doxycycline-induced tumour formation in mice, showing nicely that miR-155 could be a promising prototype for nucleic acid-based medicine.
Diffuse large B-cell lymphoma
Diffuse large B-cell lymphoma (DLBCL) is a clinically, morphologically and genetically heterogeneous group of malignant proliferations of large lymphoid B cells that accounts for approximately 40% of adult non-Hodgkin lymphoma.52 An earlier study has described the accumulation of miR-155 in DLBCL-derived B-cell lines and in clinical samples.6 DLBCLs are further classified into two groups. One group involves germinal centre B cell-like DLBCL (GC-DLBCL), which usually has a good prognosis. The other group includes the rest of the GC-DLBCLs, which mainly contain activated B-cell-like DLBCL (ABC-DLBCL). The determination of miR-155 in ABC-DLBCL by laser capture dissection identified unusually high miR-155 expression compared with GC-DLBCL, supporting the notion that miR-155 acts in this tumour as an oncogene. In these clinical samples, augmented miR-155 expression was inversely correlated with SHIP1 expression and a patient group of ABC-DLBCL with low SHIP1 expression exhibited a significantly low survival rate, indicating that aberrantly high miR-155 expression is apparently linked to a poor prognosis. Emerging evidence has shown that the expression of miR-155 in some other, but not all, tumour cell types was also high, suggesting that miR-155 could be a promising oncogenic marker. Of note, whether such a high expression might represent the cause or result of oncogenesis remains to be individually characterized.6,52 In cases where miR-155 plays a causative role, anti-miR-155 was used for therapeutic purposes with beneficial results in B-cell lymphoma in mice.53
Oncogenic virus
Kaposi sarcoma-associated herpesvirus (KSHV) is a causative oncogenic virus that is occasionally activated in opportunistic infection or circumstances of immunocompromise. KSHV is lymphotropic and is associated with two B-cell lymphomas: primary effusion lymphoma and multicentric Castleman's disease. This oncogenic virus has 12 miRNAs, one of which, miR-K12-11, has an identical seed sequence to miR-155 (Fig.2).54,55 A study has reported that this miR-155 orthologue miR-K12-11 effectively suppressed its target proteins such as Bach1 and Fos in B cells, raising the possibility that KSHV might modulate B-cell function and ultimately induce B-cell lymphoma.48 In mice, a complementation experiment that expresses miR-K12-11 in miR-155-deficient mice rescued their defective phenotypes at the physiological level, demonstrating that viral miRNA could be functional in the maintenance of immune regulation in a host.56 Transgenic mice expressing a viral latency locus that contains all viral miRNAs including miR-K12-11 induces B-cell lymphoma, suggesting that this viral miRNA can be oncogenic.57
Conclusions
The role of miR-155 includes the proper homeostasis of immune regulation and the suppression of oncogenesis. Among miRNAs found in mice and humans, miR-155 forms a distinct group because its expression is regulated by immunologically relevant stimuli induced by cytokines. miR-155 positively regulates inflammatory cytokines in macrophages and dendritic cells, suggesting that previously unrecognized targets that negatively regulate the nuclear factor-κB-mediated signalling pathway could be potential candidates.
Most autoimmune diseases are implicated in the failure of proper T-cell regulation. Due to the wide range of target proteins, dysregulated miRNA expression can affect multiple signalling pathways, which are not usually linked with each other. Hence, the discovery of such a readily unappreciated interaction between cell signalling pathways might lead to the development of a novel therapeutic agent.
Disclosure
I declare that there are no competing interests in this study.
Supporting Information
Figure S1. Sequence alignment of human miR-155 precursor microRNA and mature forms of miR-155 and related microRNAs.
References
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Janssen HL, Reesink HW, Lawitz EJ, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–94. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- Tam W, Ben-Yehuda D, Hayward WS. bic, a novel gene activated by proviral insertions in avian leukosis virus-induced lymphomas, is likely to function through its noncoding RNA. Mol Cell Biol. 1997;17:1490–502. doi: 10.1128/mcb.17.3.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam W. Identification and characterization of human BIC, a gene on chromosome 21 that encodes a noncoding RNA. Gene. 2001;274:157–67. doi: 10.1016/s0378-1119(01)00612-6. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–9. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:3627–32. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui B, Chen L, Zhang S, et al. MicroRNA-155 influences B-cell receptor signaling and associates with aggressive disease in chronic lymphocytic leukemia. Blood. 2014;124:546–54. doi: 10.1182/blood-2014-03-559690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrajoli A, Shanafelt TD, Ivan C, et al. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood. 2013;122:1891–9. doi: 10.1182/blood-2013-01-478222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci USA. 2009;106:7113–8. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason CD, Damen JE, Rosten P, et al. Targeted disruption of SHIP leads to hemopoietic perturbations, lung pathology, and a shortened life span. Genes Dev. 1998;12:1610–20. doi: 10.1101/gad.12.11.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–11. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurowska-Stolarska M, Alivernini S, Ballantine LE, et al. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc Natl Acad Sci USA. 2011;108:11193–8. doi: 10.1073/pnas.1019536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluml S, Bonelli M, Niederreiter B, et al. Essential role of microRNA-155 in the pathogenesis of autoimmune arthritis in mice. Arthritis Rheum. 2011;63:1281–8. doi: 10.1002/art.30281. [DOI] [PubMed] [Google Scholar]
- Thai TH, Patterson HC, Pham DH, Kis-Toth K, Kaminski DA, Tsokos GC. Deletion of microRNA-155 reduces autoantibody responses and alleviates lupus-like disease in the Faslpr mouse. Proc Natl Acad Sci USA. 2013;110:20194–9. doi: 10.1073/pnas.1317632110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–8. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Liu Q, Oliveira-Dos-Santos AJ, Mariathasan S, et al. The inositol polyphosphate 5-phosphatase ship is a crucial negative regulator of B cell antigen receptor signaling. J Exp Med. 1998;188:1333–42. doi: 10.1084/jem.188.7.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in Eµ-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–9. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costinean S, Sandhu SK, Pedersen IM, et al. Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein beta are targeted by miR-155 in B cells of Eµ-MiR-155 transgenic mice. Blood. 2009;114:1374–82. doi: 10.1182/blood-2009-05-220814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu SK, Volinia S, Costinean S, et al. miR-155 targets histone deacetylase 4 (HDAC4) and impairs transcriptional activity of B-cell lymphoma 6 (BCL6) in the Eµ-miR-155 transgenic mouse model. Proc Natl Acad Sci USA. 2012;109:20047–52. doi: 10.1073/pnas.1213764109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss IW, Nadeau PE, Abbott JR, Yang Y, Mergia A, Renne R. A Kaposi's sarcoma-associated herpesvirus-encoded ortholog of microRNA miR-155 induces human splenic B-cell expansion in NOD/LtSz-scid IL2Rγnull mice. J Virol. 2011;85:9877–86. doi: 10.1128/JVI.05558-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604–9. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer TJ, Fatehchand K, Shah P, et al. MiR-155 induction by microbes/microbial ligands requires NF-κB-dependent de novo protein synthesis. Front Cell Infect Microbiol. 2012;2:73. doi: 10.3389/fcimb.2012.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin HM, Kim TJ, Choi JH, et al. MicroRNA-155 as a proinflammatory regulator via SHIP-1 down-regulation in acute gouty arthritis. Arthritis Res Ther. 2014;16:R88. doi: 10.1186/ar4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tili E, Michaille JJ, Cimino A, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-α stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–9. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- Murugaiyan G, Beynon V, Mittal A, Joller N, Weiner HL. Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. J Immunol. 2011;187:2213–21. doi: 10.4049/jimmunol.1003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Kahn D, Gibson WS, et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–19. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertli M, Engler DB, Kohler E, Koch M, Meyer TF, Muller A. MicroRNA-155 is essential for the T cell-mediated control of Helicobacter pylori infection and for the induction of chronic gastritis and colitis. J Immunol. 2011;187:3578–86. doi: 10.4049/jimmunol.1101772. [DOI] [PubMed] [Google Scholar]
- Zhou H, Huang X, Cui H, Luo X, Tang Y, Chen S, Wu L, Shen N. miR-155 and its star-form partner miR-155* cooperatively regulate type I interferon production by human plasmacytoid dendritic cells. Blood. 2010;116:5885–94. doi: 10.1182/blood-2010-04-280156. [DOI] [PubMed] [Google Scholar]
- Ceppi M, Pereira PM, Dunand-Sauthier I, Barras E, Reith W, Santos MA, Pierre P. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Natl Acad Sci USA. 2009;106:2735–40. doi: 10.1073/pnas.0811073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroeni P. Immunity to systemic Salmonella infections. Curr Mol Med. 2002;2:393–406. doi: 10.2174/1566524023362492. [DOI] [PubMed] [Google Scholar]
- Hu R, Huffaker TB, Kagele DA, Runtsch MC, Bake E, Chaudhuri AA, Round JL, O'Connell RM. MicroRNA-155 confers encephalogenic potential to Th17 cells by promoting effector gene expression. J Immunol. 2013;190:5972–80. doi: 10.4049/jimmunol.1300351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker TB, Hu R, Runtsch MC, et al. Epistasis between microRNAs 155 and 146a during T cell-mediated antitumor immunity. Cell Rep. 2012;2:1697–709. doi: 10.1016/j.celrep.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmhall C, Alawieh S, Lu Y, Sjostrand M, Bossios A, Eldh M, Radinger M. MicroRNA-155 is essential for TH2-mediated allergen-induced eosinophilic inflammation in the lung. J Allergy Clin Immunol. 2014;133:1429–38. doi: 10.1016/j.jaci.2013.11.008. , 38 e1–7. [DOI] [PubMed] [Google Scholar]
- Roongapinun S, Oh SY, Wu F, Panthong A, Zheng T, Zhu Z. Role of SHIP-1 in the adaptive immune responses to aeroallergen in the airway. PLoS ONE. 2010;5:e14174. doi: 10.1371/journal.pone.0014174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoye IS, Czieso S, Ktistaki E, et al. Transcriptomics identified a critical role for Th2 cell-intrinsic miR-155 in mediating allergy and antihelminth immunity. Proc Natl Acad Sci USA. 2014;111:E3081–90. doi: 10.1073/pnas.1406322111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar TM, Kanellopoulou C, Kugler DG, et al. miR-155 activates cytokine gene expression in Th17 cells by regulating the DNA-binding protein Jarid2 to relieve polycomb-mediated repression. Immunity. 2014;40:865–79. doi: 10.1016/j.immuni.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. J Immunol. 2009;182:2578–82. doi: 10.4049/jimmunol.0803162. [DOI] [PubMed] [Google Scholar]
- Lu LF, Thai TH, Calado DP, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R, Ma YL, Liang W, Li HH, Ma ZJ, Yu X, Liao YH. MicroRNA-155 modulates Treg and Th17 cells differentiation and Th17 cell function by targeting SOCS1. PLoS ONE. 2012;7:e46082. doi: 10.1371/journal.pone.0046082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auriemma M, Vianale G, Amerio P, Reale M. Cytokines and T cells in atopic dermatitis. Eur Cytokine Netw. 2013;24:37–44. doi: 10.1684/ecn.2013.0333. [DOI] [PubMed] [Google Scholar]
- Choi SY, Sohn MH, Kwon BC, Kim KE. CTLA-4 expression in T cells of patients with atopic dermatitis. Pediatr Allergy Immunol. 2005;16:422–7. doi: 10.1111/j.1399-3038.2005.00274.x. [DOI] [PubMed] [Google Scholar]
- Garfias Y, Rojas-Ramos E, Jimenez MdelC, Martinez-Cairo S, Chavez R, Gorocica P, Zenteno E, Lascurain R. Comparative analysis of mononuclear cell surface markers in atopic processes – a preliminary study. Immunol Invest. 2003;32:95–104. doi: 10.1081/imm-120019211. [DOI] [PubMed] [Google Scholar]
- Jones G, Wu S, Jang N, Fulcher D, Hogan P, Stewart G. Polymorphisms within the CTLA4 gene are associated with infant atopic dermatitis. Br J Dermatol. 2006;154:467–71. doi: 10.1111/j.1365-2133.2005.07080.x. [DOI] [PubMed] [Google Scholar]
- Sonkoly E, Janson P, Majuri ML, et al. MiR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte-associated antigen 4. J Allergy Clin Immunol. 2010;126:581–9. doi: 10.1016/j.jaci.2010.05.045. e1–20. [DOI] [PubMed] [Google Scholar]
- Tsai CY, Allie SR, Zhang W, Usherwood EJ. MicroRNA miR-155 affects antiviral effector and effector memory CD8 T cell differentiation. J Virol. 2013;87:2348–51. doi: 10.1128/JVI.01742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracias DT, Stelekati E, Hope JL, et al. The microRNA miR-155 controls CD8+ T cell responses by regulating interferon signaling. Nat Immunol. 2013;14:593–602. doi: 10.1038/ni.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind EF, Elford AR, Ohashi PS. Micro-RNA 155 is required for optimal CD8+ T cell responses to acute viral and intracellular bacterial challenges. J Immunol. 2013;190:1210–6. doi: 10.4049/jimmunol.1202700. [DOI] [PubMed] [Google Scholar]
- Loeb GB, Khan AA, Canner D, Hiatt JB, Shendure J, Darnell RB, Leslie CS, Rudensky AY. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol Cell. 2012;48:760–70. doi: 10.1016/j.molcel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LF, Boldin MP, Chaudhry A, et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–29. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babar IA, Cheng CJ, Booth CJ, Liang X, Weidhaas JB, Saltzman WM, Slack FJ. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci USA. 2012;109:E1695–704. doi: 10.1073/pnas.1201516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen IM, Otero D, Kao E, et al. Onco-miR-155 targets SHIP1 to promote TNFα-dependent growth of B cell lymphomas. EMBO Mol Med. 2009;1:288–95. doi: 10.1002/emmm.200900028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Roccaro AM, Rombaoa C, et al. LNA-mediated anti-miR-155 silencing in low-grade B-cell lymphomas. Blood. 2012;120:1678–86. doi: 10.1182/blood-2012-02-410647. [DOI] [PubMed] [Google Scholar]
- Gottwein E, Mukherjee N, Sachse C, et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–9. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, Renne R. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol. 2007;81:12836–45. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin SH, Kim YB, Dittmer DP. Latency locus complements MicroRNA 155 deficiency in vivo. J Virol. 2013;87:11908–11. doi: 10.1128/JVI.01620-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin SH, Dittmer DP. Viral latency locus augments B-cell response in vivo to induce chronic marginal zone enlargement, plasma cell hyperplasia, and lymphoma. Blood. 2013;121:2952–63. doi: 10.1182/blood-2012-03-415620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B, Xiao B, Liu Z, et al. Identification of MyD88 as a novel target of miR-155, involved in negative regulation of Helicobacter pylori-induced inflammation. FEBS Lett. 2010;584:1481–6. doi: 10.1016/j.febslet.2010.02.063. [DOI] [PubMed] [Google Scholar]
- Trotta R, Chen L, Ciarlariello D, et al. miR-155 regulates IFN-γ production in natural killer cells. Blood. 2012;119:3478–85. doi: 10.1182/blood-2011-12-398099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Huang X, Zhang X, et al. miR-221 and miR-155 regulate human dendritic cell development, apoptosis, and IL-12 production through targeting of p27kip1, KPC1, and SOCS-1. Blood. 2011;117:4293–303. doi: 10.1182/blood-2010-12-322503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng G, Hakimpour P, Landgraf P, Rice A, Tuschl T, Casellas R, Papavasiliou FN. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–9. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett Y, McBride KM, Jankovic M, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–8. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazari-Jahantigh M, Wei Y, Noels H, et al. MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in macrophages. J Clin Invest. 2012;122:4190–202. doi: 10.1172/JCI61716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worm J, Stenvang J, Petri A, et al. Silencing of microRNA-155 in mice during acute inflammatory response leads to derepression of C/EBPβ and down-regulation of G-CSF. Nucleic Acids Res. 2009;37:5784–92. doi: 10.1093/nar/gkp577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigorito E, Perks KL, Abreu-Goodger C, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–59. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louafi F, Martinez-Nunez RT, Sanchez-Elsner T. MicroRNA-155 targets SMAD2 and modulates the response of macrophages to transforming growth factor-β. J Biol Chem. 2010;285:41328–36. doi: 10.1074/jbc.M110.146852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai D, Kim SW, McKeller MR, Dahia PL, Aguiar RC. Targeting of SMAD5 links microRNA-155 to the TGF-β pathway and lymphomagenesis. Proc Natl Acad Sci USA. 2010;107:3111–6. doi: 10.1073/pnas.0910667107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang H, Rodriguez S, et al. Notch-dependent repression of miR-155 in the bone marrow niche regulates hematopoiesis in an NF-κB-dependent manner. Cell Stem Cell. 2014;15:51–65. doi: 10.1016/j.stem.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Nunez RT, Louafi F, Sanchez-Elsner T. The interleukin 13 (IL-13) pathway in human macrophages is modulated by microRNA-155 via direct targeting of interleukin 13 receptor α1 (IL13Rα1) J Biol Chem. 2011;286:1786–94. doi: 10.1074/jbc.M110.169367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Sequence alignment of human miR-155 precursor microRNA and mature forms of miR-155 and related microRNAs.


