Abstract
All cranial placode progenitors arise from a common precursor field anterior to the neural plate, the pre-placodal region (PPR). We showed that transcription factor Zic1, expressed at the anterior neural plate, is necessary and sufficient to promote placode fate. Here we reveal a non-cell autonomous activity of Zic1 and implicate retinoic acid (RA) signaling as a key player in cranial placode progenitor specification. In a screen for genes activated by Zic1 we identify several factors involved in RA metabolism and function. Among them we show that retinaldehyde dehydrogenase 2 (RALDH2) and lipocalin-type prostaglandin D2 synthase (LPGDS), which respectively regulate the synthesis and transport of RA, directly participate in the establishment of the PPR. We propose that RALDH2 and LPGDS induction by Zic1 at the anterior neural plate allows for the localized production and transport of RA, which in turn activates a cranial placode developmental program in neighboring cells.
Introduction
Cranial sensory placodes are thickenings of the embryonic head ectoderm that give rise to the specialized paired sense organs and sensory cranial ganglia. While they produce very diverse cell types such as sensory neurons, lens fibers and hormone secreting cells 1–3, all placode progenitors arise from a common precursor field that borders the anterior neural plate known as the pre-placodal region (PPR). Subsequently, in response to inductive interactions with surrounding tissues the PPR divides into territories with distinct identities to generate the adenohypophyseal, olfactory, lens, trigeminal, otic, and epibranchial placodes.
Placode progenitors are induced by a combination of inductive signals primarily mediated by FGFs and attenuation of BMP and Wnt signals 4–6. The zinc-finger transcription factor Zic1, is one of the earliest genes activated in response to these signaling events, and in Xenopus Zic1 is both necessary and sufficient to promote placodal fate by regulating the expression of the PPR-specific genes, Six1 and Eya1 7. Interestingly, Zic1 is expressed at the anterior neural plate but does not overlap with the prospective PPR 8,9, suggesting that Zic1 regulates placode formation in a non-cell autonomous manner. To gain insights into the mechanisms by which Zic1 regulates PPR formation, we performed a microarray analysis to identify genes activated by Zic1 in a Xenopus animal explant assay. Among the targets regulated by Zic1 we found a number of genes involved in the synthesis and metabolism of retinoic acid (RA) including lipocalin-type prostaglandin D2 synthase (LPGDS), retinaldehyde dehydrogenase 2 (RALDH2), two members of the cytochrome P450 enzyme family (Cyp26a and Cyp26c) and a cellular retinoic acid binding protein 2 (Crabp2) signifying the importance of this signaling pathway in placode formation. Here we present evidence that Zic1 regulates placode progenitor formation non-cell autonomously by controlling RA production and transport at the anterior neural plate.
Results
Identification of downstream targets of Zic1
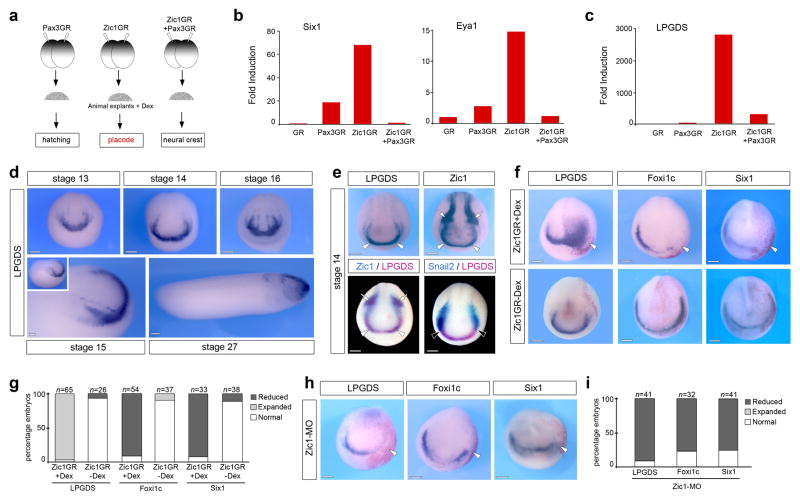
The transcription factor Zic1 is expressed at the anterior neural plate 7–10 and is required for the formation of sensory placode progenitors 7. To gain insights into the mechanisms by which Zic1 regulates sensory placode formation we performed a microarray screen to identify targets of Zic1 11. The screen was based on the observation that while expression of Zic1 favored placode fate in Xenopus animal cap explants, simultaneous expression of Pax3 repressed placode-specific genes to promote neural crest fate 7,11 (Fig. 1a). Among the genes that were both strongly upregulated by Zic1, as compared to Pax3 alone, and repressed by Pax3 co-injection, we found several well-characterized early placode-specific genes, including Six1, Eya1, Xanf1, Ebf2 and Sox11 (Fig 1b; Supplementary Table 1). The recovery of these genes was an important validation of our experimental design. We also found several novel potential regulators of placode formation (Supplementary Table 1). These genes were initially screened by whole-mount in situ hybridization to select factors expressed at the anterior neural plate, in a pattern similar to Zic1. One candidate that fulfilled this criterion was the lipocalin-type prostaglandin D2 synthase (LPGDS; Fig. 1c), also known as Cpl1 12–15. By in situ hybridization LPGDS is first expressed at stage 13 in the anterior region of the neural plate (Fig. 1d). This expression pattern is maintained throughout neurulation and then appears confined to the head region in tailbud stage embryos (Fig. 1d). Double in situ hybridization demonstrates that LPGDS completely overlaps with the anterior expression domain of Zic1 (Fig. 1e), but is excluded from the lateral expression domain of Zic1, which corresponds to the prospective neural crest region. In situ hybridization for the neural crest-specific gene Snail2 16 confirmed that there is no overlap between LPGDS expression domain and neural crest progenitors, LPGDS abuts the anterior expression domain of Snail2 (Fig 1e).
Figure 1. LPGDS is a downstream target of Zic1.
(a) Experimental design for the selection of Zic1 targets. (b) Fold induction of Six1, Eya1 and (c) LPGDS from the microarray data. (d) In situ hybridization for LPGDS (stages 13, 14 and 16 are frontal views; stage 15 and stage 27 are lateral views, anterior to right, dorsal to top). (e) In situ hybridization for LPGDS and Zic1 in stage matched embryos (upper panels). LPGDS and Zic1 co-localize at the anterior neural plate (arrowheads), while Zic1 is also expressed in neural crest progenitors (arrows). Double in situ hybridization (lower panels) shows overlapping expression of LPGDS and Zic1 at the anterior neural plate (arrowheads; left panel), while LPGDS and Snail2 (right panel) have adjacent but non-overlapping expression domains (black arrowheads). Frontal views. (f) In embryos injected with Zic1GR mRNA and treated with dexamethasone (+Dex), LPGDS is dramatically expanded (arrowhead), while Foxi1c and Six1 expression at the PPR are reduced (arrowheads). The same injection in the absence of dexamethasone (-Dex) had no effect on the expression of these genes Frontal views, the injected side is indicated by the lineage tracer (Red-Gal). (g) Quantification of the Zic1GR injection results. Three independent experiments were performed. The number of embryos analyzed (n) is indicated on the top of each bar. (h) Zic1 knockdown (Zic1-MO injection) reduces LPGDS, Foxi1c and Six1 expression. (i) Quantification of the Zic1-MO injection results. Three independent experiments were performed. The number of embryos analyzed (n) is indicated on the top of each bar. Scale bars, 200 μm.
To further establish that LPGDS is a true target of Zic1 we analyzed LPGDS expression pattern in embryos injected with Zic1GR mRNA (a hormone-inducible version of Zic1 fused to human glucocorticoid receptor ligand-binding domain), or a morpholino antisense oligonucleotide that blocks Zic1 function (Zic1-MO) 7,9. In embryos injected with Zic1GR mRNA and treated with dexamethasone, we observed a dramatic upregulation and expansion of the LPGDS expression domain (Fig. 1f, g). The same injection in the absence of dexamethasone had no effect on LPGDS expression (Fig. 1f, g). Conversely, injection of Zic1-MO completely inhibited LPGDS expression on the injected side (Fig. 1h, i). Interestingly, in both situations we observed a reduction of Foxi1c and Six1 expression, two early PPR-specific genes (Fig 1f–i). These observations confirm that LPGDS is a downstream target of Zic1 and indicate that placode formation is sensitive to Zic1 and LPGDS expression levels in the embryos.
LPGDS is required for placode formation
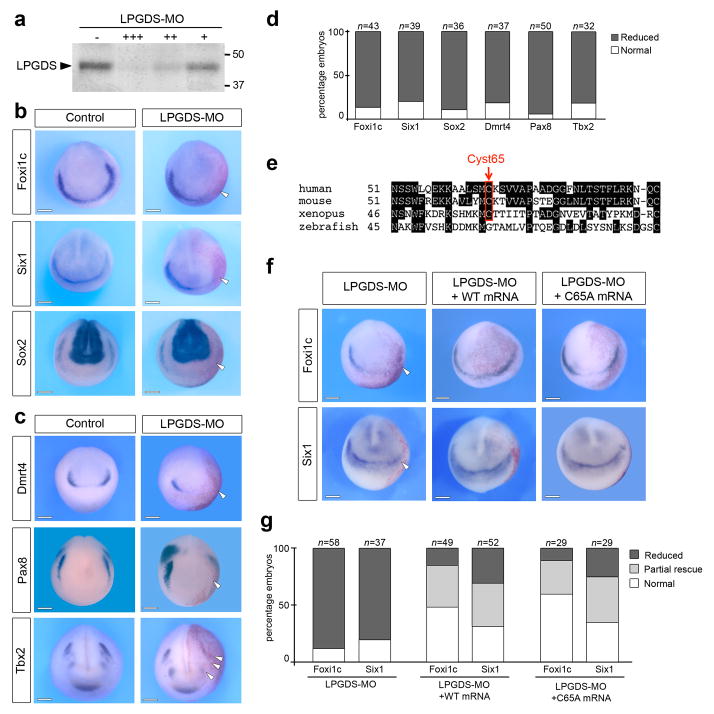
The expression pattern of LPGDS at the anterior neural plate and its regulation by Zic1 suggest a potential role in placode formation. To test this possibility, we used a translation blocking morpholino antisense oligonucleotide (LPGDS-MO) to interfere with LPGDS function. The activity of the morpholino was confirmed in an in vitro transcription/translation assay in which LPGDS-MO blocked LPGDS protein production (Fig 2a; Supplementary Fig. 1). Unilateral injection of LPGDS-MO (40 ng) in the animal pole region of 2-cell stage embryos resulted in a marked decrease expression of Six1 17,18, Foxi1c 18,19 and Sox2 20, three genes broadly expressed at the PPR (Fig 2b, d). We also analyzed the expression of genes restricted to individual placodal domains such as Dmrt4 for the adenohypophyseal and olfactory placodes 21, Pax8 for the otic and lateral line placodes 22 and Tbx2 for otic and trigeminal/profundal placodes 23. For each one of these genes a significant reduction of expression was observed on the injected side (Fig. 2c) in more than 80% of the embryos (Fig 2d). Injection of a control morpholino (Cont-MO) had no effect on the expression of these genes (Supplementary Fig. 2e). To further establish the specificity of LPGDS knockdown phenotype, we used a second morpholino (LPGDS-MO2) that specifically interfered with LPGDS pre-mRNA splicing by targeting intron1/exon2 junction (Supplementary Fig. 2a), resulting in the production of a transcript of higher size, due to intron retention (Supplementary Fig. 2b). The phenotype of LPGDS-MO2-injected embryos was indistinguishable from the phenotype generated by injection of the translation blocking morpholino, with inhibition of Foxi1c, Six1 and Dmrt4, though at a lower frequency (Supplementary Fig. 2c–d). These results indicate that LPGDS is critically required for the establishment of the PPR and for sensory placode formation.
Figure 2. LPGDS is required for placode development.
(a) Increasing amounts of LPGDS-MO 10 ng (+), 100 ng (++), and 1000 ng (+++) blocks translation directed by LPGDS mRNA in an in vitro coupled transcription/translation reaction. The position of markers of known molecular weight (kDa) is indicated. (b) In situ hybridization for pan-placodal and (c) placode specific genes in control and LPGDS-MO injected embryos (frontal views, dorsal to top). Arrowheads indicate reduced expression on the injected side. (d) Quantification of the results. Four independent experiments were performed. The number of embryos analyzed (n) is indicated on the top of each bar. (e) Amino acid sequences alignment showing the conserved cysteine residue (Cyst65), center of LPGDS enzymatic activity. (f) Foxi1c and Six1 expression domains are rescued in LPGDS-MO-injected embryos by co-injection of either WT or C65A mouse LPGDS mRNA. Frontal views, dorsal to top; injected side is indicated by the lineage tracer (Red-Gal). (g) Quantification of the rescue experiment. Three independent experiments were performed. The number of embryos analyzed (n) is indicated on the top of each bar. LPGDS-MO vs. LPGDS-MO+WT or LPGDS-MO+C65A mRNA injected embryos (p<0.001, Fisher exact test.); LPGDS-MO+WT mRNA vs. LPGDS-MO+C65A mRNA injected embryos show no significant differences. Scale bars, 200 μm.
LPGDS functions independently of its enzymatic activity
LPGDS has a dual function, as an enzyme involved in the biosynthesis of prostaglandin D2 (PGD2) from its precursor, and as a lipophilic ligand-binding protein when secreted 24. The LPGDS molecule contains three cysteine residues, and site-directed mutagenesis has established that the cysteine residue at position 65 (Cyst65) is essential for the enzymatic activity of LPGDS 25, a cysteine highly conserved from human to frogs (Fig 2e). To determine whether LPGDS regulates placode formation through PGD2 signaling, we performed a rescue experiment using mRNA encoding wild type (WT) mouse LPGDS or a mutated version in which Cyst65 has been replaced by an alanine (C65A), thereby completely abolishing the enzymatic activity of LPGDS 25. Embryos at the 2-cell stage were sequentially injected with LPGDS-MO and either with WT or C65A mouse LPGDS mRNA. Both mRNA were equally efficient at rescuing Foxi1c and Six1 expression in morphant embryos (Fig. 2f–g). Complementary to this rescue assay, we found that well-characterized pharmacological agents that specifically block PGD2 signaling pathway did not affect Foxi1c expression at the PPR (Supplementary Fig. 3). Altogether, these results demonstrate that LPGDS regulates placode formation through a mechanism that does not involve its enzymatic activity and the production of PGD2.
LPGDS functions as a carrier for retinoic acid
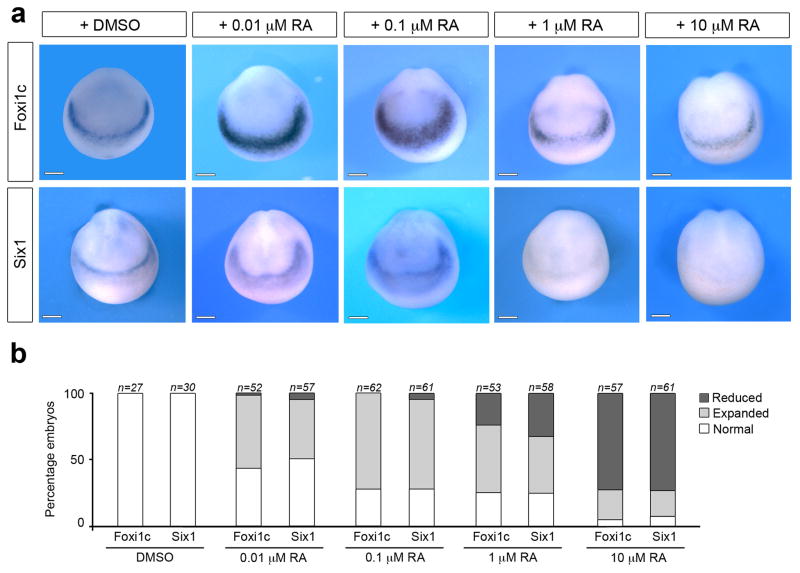
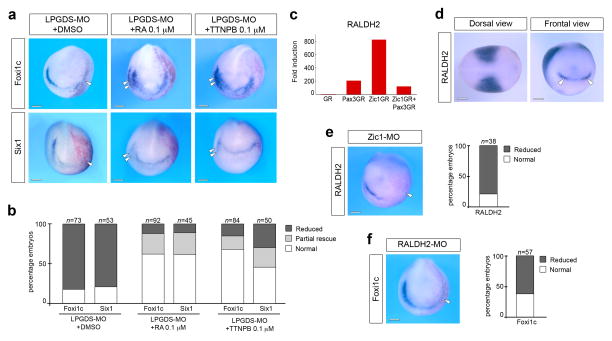
Since LPGDS binds retinoids with high affinity 14,26 we wished to determine whether LPGDS regulated placode formation through this mechanism. As a preliminary evaluation we treated intact embryos at the gastrula stage (stage 11) with increasing doses of retinoic acid (RA; 0.01 μM to 10 μM) and analyze the consequence on PPR formation. We found that the lower doses of RA (0.01 μM and 0.1 μM) resulted in an expansion of Foxi1c and Six1 expression domains, while the higher doses of RA (1 μM and 10 μM) inhibited the expression of both genes (Fig. 3), indicating that PPR formation is very sensitive to RA levels in the embryo. Based on these observations, we performed a rescue experiment in which LPGDS-MO-injected embryos were exposed at stage 11 to 0.1 μM of RA (the dose of RA that promotes strong expansion of PPR genes; Fig. 3) or a RA receptor agonist, TTNPB 27. Both treatments efficiently rescued Foxi1c and Six1 expression in morphant embryos and occasionally expanded Foxi1c and Six1 expression on the control side (Fig. 4a–b). LPGDS can also bind retinaldehyde (retinal), the RA precursor, with high affinity 14,26, however this compound was unable to rescue Foxi1c expression domain in LPGDS-depleted embryos (Supplementary Fig. 4), indicating that in the context of PPR formation LPGDS is functioning primarily as a RA carrier, and that RA is the active compound promoting PPR formation.
Figure 3. Retinoic acid affects PPR formation in a dose dependent manner.
(a) Treatment of intact embryos at stage 11 with increasing doses of RA (0.01 μM, 0.1 μM, 1 μM and 10 μM) disrupts Fox1ic and Six1 expression at the PPR. DMSO was used as a control. Frontal views, dorsal to top. (b) Quantification of the results. Four independent experiments were performed. The number of embryos analyzed for each condition (n) is indicated on the top of each bar. Scale bars, 200 μm.
Figure 4. Retinoic acid signaling regulates placode formation.
(a) Treatment at stage 11 of LPGDS-depleted embryos (LPGDS-MO) with 0.1 μM RA or with the RA receptor agonist, TTNPB, restored completely or partially Foxi1c and Six1 expression on the injected side. Frontal views, dorsal to top. Injected side is indicated by the lineage tracer (Red-Gal). Double arrowheads indicate Foxi1c or Six1 expansion on the control side. (b) Quantification of the rescue experiment. Four independent experiments were performed. The number of embryos analyzed for each condition (n) is indicated on the top of each bar. DMSO vs. RA or TTNPB treated embryos (p< 0.001, Fisher exact test) RA vs. TTNPB treated embryos show no significant differences. (c) Fold induction of RALDH2 from the microarray data. (d) By in situ hybridization RALDH2 is detected in the trunk mesoderm (dorsal view, anterior to right) and at the anterior neural plate (frontal view; arrowheads). (e) RALDH2 expression is lost in Zic1-depleted embryos. The graph is a quantification of the results. Three independent experiments were performed. The number of embryos analyzed (n) is indicated on the top of each bar. (f) Foxi1c expression is reduced in RALDH2-MO injected embryos. The graph is a quantification of the results. Three independent experiments were performed. The number of embryos analyzed (n) is indicated on the top of each bar. (e–f) Arrowheads indicate reduced expression on the injected side. Scale bars, 200 μm.
Zic1 regulates RALDH2 expression
Our results suggest that LPGDS activity at the anterior neural plate is linked to RA activity. Consistent with this observation we found several components of RA metabolism and function strongly upregulated by Zic1 in the microarray screen (Supplementary Table 1). These include retinaldehyde dehydrogenase 2 (RALDH2) the enzyme responsible for the synthesis of RA from its precursor retinal, two RA-degrading enzymes (Cyp26a and Cyp26c) and a cellular retinoic acid binding protein 2 (Crabp2). In the microarray samples, RALDH2 induction levels followed the same pattern as LPDGS (Fig 4c). RALDH2 is expressed in the trunk mesoderm, as well as in a discrete U-shaped ectodermal domain around the anterior neural plate, similar to LPGDS (Fig 4d) 28. Zic1 knockdown reduced RALDH2 expression in the embryo (Fig 4e), confirming that RALDH2 is functioning downstream of Zic1. Moreover, in the absence of RALDH2 function Foxi1c was severely reduced at the PPR (Fig 4f), further demonstrating the link between Zic1, RALDH2 activity and placode formation.
Zic1 induces placode fate non-cell autonomously
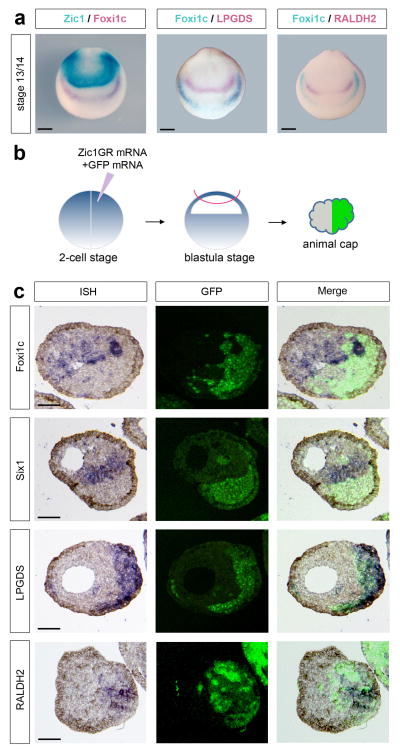
Our data indicate that Zic1 may control RA signaling at the anterior neural plate through the activation of RALDH2 to produce RA and LPGDS to transport RA extracellularly. Zic1, RALDH2 and LPGDS are all confined to the anterior neural plate, at a distance from the prospective PPR (Foxi1c-expressing cells; Fig 5a) suggesting that Zic1 regulates placode formation non-cell autonomously. To test this possibility, embryos were injected in one blastomere at the 2-cell stage with Zic1GR mRNA and mRNA encoding the lineage tracer GFP, animal explants isolated at the blastula stage, treated with dexamethasone and cultured for 8 hours (Fig 5b). Injection of GR mRNA was used as a negative control (Supplementary Fig. 5). Explants were then processed for in situ hybridization on sections. In these explants, Six1 and Foxi1c expression was exclusively confined to GFP-negative cells, derived from the uninjected blastomere (Fig 5c; upper panels). By contrast, LPGDS and RALDH2 expression was always closely associated with Zic1GR/GFP-positive cells (Fig 5c; lower panels). Altogether, these results demonstrate that Zic1 is inducing placode fate by a non-cell autonomous mechanism involving activation of RA signaling.
Figure 5. Zic1 regulates placode formation non-cell autonomously.
(a) Double in situ hybridization for Zic1/Foxi1c, Foxi1c/LPGDS and Foxi1c/RALDH2 showing that Zic1, LPGDS and RALDH2 are expressed at a distance from the PPR (Foxi1c-expressing cells). Frontal views, dorsal to top. Scale bars, 200 μm. (b) Animal explants dissected from embryos injected in one blastomere at the 2-cell stage with Zic1GR and GFP mRNAs were cultured for 8 hours in dexamethasone and analyzed by In situ hybridization. (c) In situ hybridization (ISH) for Foxi1c, Six1, LPGDS and RALDH2 on sections of animal explants derived from embryos injected with Zic1GR and GFP mRNA in one blastomere at the 2-cell stage (left panels). In each case the Zic1GR-expressing cells (GFP-positive) are shown (middle panels). Merge of fluorescence and in situ hybridization images (right panels). Three independent experiments were performed with similar results for each probe as pictured (Foxi1c, n=14; Six1, n=13; LPGDS, n=13 and RALDH2, n=11). Scale bars, 100 μm.
Zic1-activated RA signaling uses non-canonical RA receptors
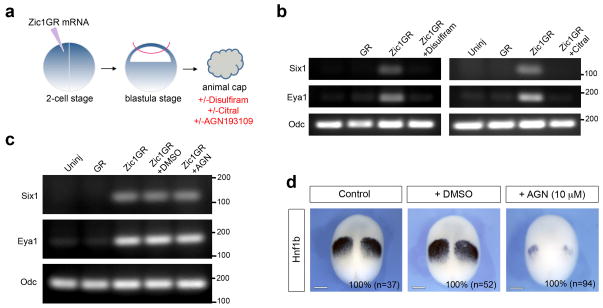
To further establish that the placode-inducing activity of Zic1 depends on active RA signaling, we analyzed by RT-PCR the expression Six1 and Eya1 in Zic1GR-injected animal explants treated with 100 μM of Disulfiram or Citral (Fig 6a), two general inhibitors of alcohol and aldehyde dehydrogenases 29,30, the group of enzymes that sequentially catalyze the oxidation of retinol and retinaldehyde to produce RA. In these animal explants treated with the inhibitors, the induction of Six1 and Eya1 by Zic1GR was significantly reduced (Fig. 6b; Supplementary Fig. 6a, b) consistent with the view that placode induction by Zic1 depends on RA production.
Figure 6. Zic1 regulates placode fate independently of canonical RA receptors.
(a) Animal explants dissected from embryos injected in one blastomere at the 2-cell stage with Zic1GR mRNA and cultured for 8 hours in dexamethasone, with or without the pharmacological inhibitors Disulfiram (100 μM), Citral (100 μM) or AGN193109 (10 μM). (b) RT-PCR analysis of Six1 and Eya1 expression in animal explants expressing Zic1GR treated with Disulfiram or Citral. Odc (Ornithine decarboxylase) is shown as a loading control. Controls are uninjected (Uninj) and GR mRNA injected (GR) animal explants. Similar results were obtained in four independent experiments for each inhibitor. The position of markers of known size is indicated (bp). (c) RT-PCR analysis of Six1 and Eya1 expression in Zic1GR injected animal explants treated with the pan-RAR antagonist, AGN193109. Controls are uninjected (Uninj), GR mRNA injected (GR) and Zic1GR mRNA injected treated with DMSO (+DMSO) animal explants. Similar results were obtained in six independent experiments. The position of markers of known size is indicated (bp). (d) AGN193109 treatment blocks Hnf1b expression in the posterior hindbrain (100% of the embryos; n=94), while in DMSO-treated embryos Hnf1b expression is similar to that of control embryos (100% of the embryos; n=52 and n=37, respectively). Three independent experiments were performed. Stage 13 embryos, dorsal views, anterior to top. Scale bars, 200 μm.
To determine whether the placode-inducing activity of Zic1 is mediated through canonical RA receptors (RARs), Zic1GR-injected animal explants were treated with a well-characterized pan-RAR antagonist, AGN193109 31,32 (Fig. 6a). Surprisingly, we found that AGN193109 was unable to block the expression of Six1 and Eya1 (Fig 6c; Supplementary Fig. 6c), while the same concentration of antagonist (10 μM) was extremely efficient at repressing Hnf1b expression in the whole embryo (Fig 6d), a gene directly regulated by RAR signaling 33. These results suggest that Zic1-activated RA signaling mediates its activity through a mechanism that does not involve canonical RARs, and may signal through other nuclear receptors that also bind RA, such as peroxisome proliferator-activated receptor β (PPARβ) 34, and members of the RAR-related orphan receptors (RORs) 35,36.
Discussion
Our findings uncover a non-cell autonomous activity of Zic1 in the control of placode fate, and implicate RA signaling as a major player in cranial placode progenitor formation. We have identified several genes activated by Zic1 that are involved in RA metabolism and function. Among them we demonstrate that RALDH2 and LPGDS, respectively, are responsible for the synthesis and the transport of RA, and directly participate in the establishment of the PPR. We propose that the activation of RALDH2 and LPGDS at the anterior neural plate by Zic1 allows for the localized production and transport of RA, which in turn activates a placode developmental program in neighboring cells (Fig 7).
Figure 7. Model for the regulation of PPR formation by Zic1 and RA signaling.
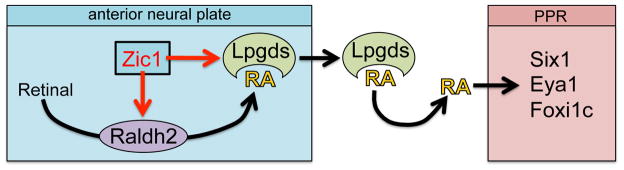
Zic1 controls RA signaling at the anterior neural plate through the activation of RALDH2 to produce RA, and LPGDS to transport RA extracellularly. As a consequence RA induces the expression of Six1, Eya1 and Foxi1c in neighboring cells (PPR).
Xenopus LPGDS, also known as cpl1 (choroid plexus lipocalin 1), was first isolated in a screen for genes activated at gastrulation and specifically expressed in the nervous system 15, and has been primarily used as a forebrain marker 12,13,37. Its specific role at the anterior neural plate has not been studied. Cpl1/LPGDS was also isolated in another screen designed to identify direct targets of Zic1 38, however further studies are needed to confirm that Zic1 directly activates LPGDS expression. LPGDS is a bifunctional protein that acts as a prostaglandin D2-producing enzyme and a lipophilic ligand-binding protein 39. More specifically LPGDS can bind retinaldehyde and RA with high affinities 13,14,26. In a rescue assay, using WT and mutant versions of mouse LPGDS we completely ruled out the involvement of its enzymatic activity, and demonstrated that LPGDS functions during placode formation through a mechanism implicating RA signaling. By exposing the entire embryo to RA, the expression of PPR genes was efficiently rescued in LPGDS-depleted embryos (Fig 4a, b). In these experiments, the exogenous source of RA bypasses the requirement for an LPGDS-mediated transport of RA to the PPR. We posit that LPGDS is essential to transport RA outside the producing cells. LPGDS knockdown in the embryo leads to a loss of genes expressed at the PPR, as well as genes that are more specific for individual cranial placodes, suggesting a broad requirement for LPGDS and RA signaling in placode progenitor formation.
A recent study has proposed that signaling through RA receptors (RARα2), which are expressed in the caudal half of the embryo 40, was critical to set up the posterior boundary of the PPR 41. Our experiments using the pan-RAR antagonist (AGN193109) suggest that the placode-inducing activity of Zic1 may use an RAR-independent pathway, pointing to the intriguing possibility that another set of receptors may mediate this activity. A similar activity of RA independent of canonical RARs has been described in the context of the developing mouse neural retina 42. Several classes of nuclear receptors have been reported to bind RA, including peroxisome proliferator-activated receptor β (PPARβ) 34 and members of the RAR-related orphan receptors (RORs) 35,36, and are therefore potential candidate to mediate the placode-inducing activity of Zic1. Future work will test this hypothesis.
During embryonic development, RA signaling is essential for the regionalization of the embryo along the anteroposterior axis. Studies in several organisms have shown that RA is an important posteriorizing signal in all three germ layers, acting in concert with molecules of the Wnt and FGF families 43–45. However, RA signaling may have a more direct role in PPR specification, independent of its posteriorizing activity, in a similar manner as the posteriorizing activity of FGF and canonical Wnt can be uncoupled from their role in neural crest induction 10,46. Consistent with this possibility, RALDH2 has two major expression domains in the early embryo, the paraxial and lateral plate mesoderm in the trunk region, as well as a discrete U-shaped ectodermal domain around the anterior neural plate 28 (Fig 4d). While the former has been linked to the posteriorizing activity of RA, the later represents an independent source of RA, and we propose that this anterior domain is essential to pattern the cranial region of the embryo. In a recent study using novel transgenic RA sensor lines, RA activity has been visualized at the very anterior region of the zebrafish embryo and at the polster, and a candidate source of this activity is RALDH3 47. In the mouse, RALDH2 is responsible for RA signaling in the early embryonic head, and knockout embryos die around E9.5–10.5 with severe forebrain and craniofacial defects 48. In birds, RALDH3 is expressed anteriorly at early stages 49, and vitamin A-deficient quail embryos have defective otic vesicles and lack the pituitary gland, two derivatives of cranial placodes 50,51.
Zic1 gain-of-function resulted in a dramatic upregulation of LPGDS in the embryo, which was surprisingly associated with a loss of early placode markers (Six1 and Foxi1c; Fig. 1f). However, in light of our results linking Zic1 to RA signaling, this result is not completely unexpected and indicates that PPR formation is very sensitive to RA levels. As in other RA-regulated developmental processes, excess or insufficient RA signaling often result in similar outcomes 45. Excess RA can shut down its own production through the downregulation of RALDH2 52. Indeed, in embryos treated with RA, RALDH2 is preferentially lost anteriorly, leading to severe head truncation 28. Furthermore, exposure of intact embryos to varying doses of RA affects PPR genes expression in a concentration dependent manner (Fig 3). Altogether these observations support the view that RA levels at the anterior neural plate are critical for placode formation.
The non-cell autonomous activity of Zic1 in the control of placode fate was especially evident in the animal explant system (Fig 5). Interestingly, we never observed the activation of Six1 or Foxi1c in Zic1GR-expressing cells, rather the expression Six1 and Foxi1c was always confined to adjacent cells, derived from the uninjected blastomere. A possible explanation is related to the activity of Cyp26c and Cyp26a, the RA metabolizing enzymes, which were strongly upregulated by Zic1GR in the microarray. Typically, RA is released by RA-producing cells and can enter adjacent cells where it faces two scenarios: (i) in cells expressing Cyp26, RA is degraded preventing RA signaling; and (ii) in cells that do not express Cyp26, RA enters the nucleus and initiates the transcription of target genes. It is possible that Cyp26 expression renders Zic1-expressing cells refractory to RA signaling, thereby allowing for the unidirectional propagation of the RA signal, and the establishment of sharp boundaries of gene expression. Consistent with this interpretation, in Xenopus Cyp26c is expressed in the neural plate but is excluded from the most anterior region of the embryo 53.
While the placode inducing activity of Zic1 requires active RA signaling, RA is likely to act in concert with other signals to impart placode fate. Work in zebrafish suggests that Zic1 is functioning not only upstream of RA, but also by regulating Nodal and Hedgehog signaling to control midline tissue development in the head 54,55. These factors are therefore good candidates to synergize with RA to promote cranial placode fate.
Methods
Plasmids, constructs and oligonucleotides
Xenopus laevis Pax3GR and Zic1GR, the hormone-inducible versions of Pax3 and Zic1, fused to the human glucocorticoid receptor (GR) ligand-binding domain 56, wild type (WT) and mutated (C65A) mouse LPGDS 7,25 were subcloned into pCS2+ expression plasmid. Pax3GR, Zic1GR, GR, WT and C65A mouse LPGDS, green fluorescent protein (GFP), and β-galactosidase mRNAs were synthesized in vitro using the Message Machine Kit (Ambion, Austin, TX). Zic1 (Zic1-MO) 9, RALDH2 (RALDH2-MO) 29 and LPGDS (LPGDS-MO: TCAGAGCAAGCAAAAT CCTCATCAT; LPGDS-MO2: AGACCTAGAGGCAGAGAGAGGAATT) morpholino antisense oligonucleotides were purchased from GeneTools (Philomath, OR). The specificity of the LPGDS-MO was tested in an in vitro transcription/translation coupled rabbit reticulocyte lysate assay (Promega, Madison, WI). LPGDS-MO was designed to interfere with both LPGDS-a and LPGDS-b function (accession numbers: NM_001092825 and NM_001088044), while LPGDS-MO2 interfered only with LPGDS-a. A standard morpholino (Cont-MO; CCTCTTACCTCAGTTACAATTTATA) was used as control.
Embryos, injections and explants culture
Xenopus laevis embryos were staged according to Nieuwkoop and Faber 57 and raised in 0.1X NAM (Normal Amphibian Medium) 58. This study was performed in accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The procedures were approved by the Institutional Animal Care and Use Committee of New York University under animal protocol #120311. Embryos were injected in one blastomere at the 2-cell stage and analyzed by in situ hybridization at stage 15. Morpholino antisense oligonucleotides (40–50 ng) were injected together with 0.5 ng of β-galactosidase mRNA as a lineage tracer. Rescue experiments were performed by sequential injection of LPGDS-MO and 0.5 ng of WT or C65A mouse LPGDS mRNA. For all-trans RA and TTNPB (Sigma-Aldrich, St.Louis, MO) treatments control and LPGDS-MO-injected embryos were incubated at stage 11 in the retinoids (0.01 μM, 0.1 μM, 1 μM and 10 μM diluted in 0.1X NAM) and collected at stage 15 for in situ hybridization. Control and LPGDS-MO-injected embryos treated with DMSO were used as control. For animal explant experiments, one blastomere at the 2-cell stage was injected in the animal pole region with GR or Zic1GR mRNA (0.5–1 ng), explants were dissected at the late blastula stage and immediately cultured for several hours in NAM 0.5X plus 10 μM of dexamethasone (Sigma-Aldrich, St.Louis, MO). Coinjection of 1 ng of GFP mRNA was used as a lineage tracer to identify the progeny of the injected blastomere. Dissected animal explants injected with Zic1GR mRNA were treated with 100 μM Disulfiram (tetraethylthiuram disulfide) or Citral (Sigma-Aldrich, St.Louis, MO), two general inhibitors of alcohol and aldehyde dehydrogenases, or with 10 μM of AGN193109 (Santa Cruz Biotechnology, Tallas, TX), an RAR antagonist.
In situ hybridization
Embryos were fixed in MEMFA and prior to in situ hybridization the β-galactosidase activity was revealed using Red-Gal (Research Organics, Cleveland, OH). Whole-mount in situ hybridization 59 was performed in 4 ml glass vials using digoxygenin (DIG)- and fluorescein isothiocyanate (FITC)-labeled antisense RNA probes (Genius Kit; Roche, Indianapolis, IN) synthesized from template cDNA encoding Zic1 20, Foxi1c 18,19, Six1 17, Sox2 20, Dmrt4 21, Pax8 22, Tbx2 23, LPGDS (pSPORT6-LPGDS; Open Biosystems), RALDH2 29 and Snail2 16. Embryos were hybridized overnight at 60°C. RNA probes were detected using an anti-DIG antibody conjugated to alkaline phosphatase (Roche, Indianapolis, IN) at a 1:2000 dilution overnight at 4°C. After several washes, the chromogenic reaction was performed overnight by incubation in BM purple (Roche, Indianoapolis, IN). The reaction was then stopped by fixation in MEMFA and the embryos were bleached in 10% hydrogen peroxide in methanol for 48 h. For double in situ hybridization DIG- and FITC-labeled probes were hybridized simultaneously and sequentially detected using anti-FITC and anti-DIG alkaline phosphatase conjugated antibodies (Roche, Indianapolis, IN; 1:10000 and 1:2000 dilution, respectively). FITC-labeled probe was visualized first using Magenta Phosphate (5-bromo-6chloro-3indoxyl phosphate; Biosynth, Itasca, IL) and after inactivation of the anti-FITC antibody by treatment with Glycine (0.1M, pH 2.2) for 30 min, the color reaction for the DIG-labeled probe was performed using 4-toluidine salt (BCIP; Roche, Indianapolis, IN). The reaction was then stopped by fixation in MEMFA and the embryos were bleached in 10% hydrogen peroxide in PBS for 48 h. For in situ hybridization on sections, animal explants were fixed in 4% paraformaldehyde in phosphate buffer saline (PBS; Gibco, Grand Island, NY) for 1 hour, embedded in Paraplast+ and 12 μm sections collected on a glass slide, hybridized overnight at 60°C in a humidified chamber with Foxi1c, Six1, LPGDS or RALDH2 DIG-labeled probes. Sections were then washed in 2x SSPE (saline sodium phosphate EDTA; Invitrogen, Grand Island, NY). Probes were detected using an anti-DIG antibody conjugated to alkaline phosphatase at a 1:2000 dilution for 2 h at room temperature. After several washes, the chromogenic reaction was performed overnight by incubation in BM purple (Roche, Indianoapolis, IN). The reaction was stopped by fixation in MEMFA. The slides were then dehydrated and mounted in Permount (Fisher Scientific, Pittsburgh, PA) 60. Since GFP can no longer be detected following this procedure, sections of GFP-labeled animal explants were first individually photographed and then processed for in situ hybridization.
RT-PCR analysis
Total RNAs were extracted form 10–15 animal explants using the RNeasy microRNA isolation kit (Qiagen, Valencia, CA). To avoid contamination from genomic DNA, the RNA samples were digested with RNase-free DNase I. RT-PCR experiments were performed using the One Step RT-PCR kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions using the following primer sets: Six1 (F: CTGGAGAGCCAC CAGTTCTC; R: AGTGGTCTCCCCCTCAGTTT; 30 cycles), Eya1 (F: ATGACACCAAAT GGCACAGA; R: GGGAAAACTGGTGTGCTTGT; 30 cycles) and Odc (F: ACATGGCATTCTCCCTGAAG; R: TGGTCCCAAGGCTAAAGTTG; 25 cycles).
Microarray analysis
RNAs were extracted from animal explants expressing GR, Pax3GR, Zic1GR or a combination of Pax3GR and Zic1GR using the RNeasy microRNA isolation kit. During the extraction procedure, the samples were treated with DNase I to eliminate possible genomic DNA contamination. The amount of isolated RNA was quantified using a spectrophotometer. Extracted RNAs (five replicate for each injection set) were used for the microarray experiment. Probe labelling, hybridization and initial data analysis were performed at the University of Pennsylvania Microarray Facility. Around 0.1–0.2 μg total RNA was converted into first-strand cDNA using Superscript II reverse transcriptase primed by a poly(T) oligomer that incorporates the T7 promoter. Second-strand cDNA synthesis is followed by in vitro transcription for linear amplification of each transcript and incorporation of biotinylated nucleotides. The cRNA products were fragmented to 200 nucleotides or less, heated at 99°C for 5 min and hybridized for 16 hr at 45°C to a GeneChip Xenopus laevis Genome 2.0 Array (Affymetrix, Cleveland, OH). Hybridized arrays were processed by the GeneChip Fluidics system, and scanned in the GeneChip Scanner. Probes intensities were exported in cel files using GCOS software (Affymetrix, Clevland, OH). Subsequent quantification of expression levels and statistical identification of differentially regulated genes was performed using Partek Genomics Suite (Partek, Inc., St Louis, MO). Normalized, log2-transformed probeset intensities were calculated using GC-RMA. ANOVA with multiple testing corrections was used to determine p-values for likelihood of differential expression, based on reproducibility in five replicates to identify cDNAs both activated by Zic1GR and repressed in Zic1GR/Pax3GR samples. Pairwise comparisons yielding fold changes were performed between these conditions. The microarray data have been deposited in the GEO databa se under the accession number GSE68546.
PGD2 signaling inhibitors and treatment
To interfere with PGD2 signaling pathway we used well-characterized pharmacological inhibitors: AT-56 (Tocris Bioscience, Minneapolis, MN) to block LPGDS enzymatic activity, and two antagonists of PGD2 receptors, BWA868C (Cayman Chemical, Ann Arbor, MI) specific for DP1 and ramatroban (Sigma-Aldrich, St Louis, MO) specific for DP2. For treatment, embryos at stage 11 devoid of the vitelline membrane were incubated in the presence of the inhibitors (1 μM, 10 μM and 100 μM in NAM 0.1X), collected at stage 15, and processed for in situ hybridization. DMSO-treated embryos were used as control.
Imaging
Images were captured using an Olympus SZX9 microscope and a QImaging Micro Publisher 3.3 RTV camera. Images from sections of animal explants were obtained on a Nikon SMZ1500 microscope equipped with a Nikon Ds-U3 camera. Composite images were assembled using Adobe Photoshop. In some experiments RFP mRNA was injected as a lineage tracer, in place of GFP mRNA, for consistency the images were then digitally colored (green) using ImageJ, this applies specifically to Fig 5c (RALDH2; lower panels).
Supplementary Material
Acknowledgments
We are grateful to Dr. Yoshihiro Urade, Dr. Ko Fujimori and Dr. Edgar Pera for reagents, to Dr. John Tobias for help with the microarray data and to Dr. Jane McCutcheon for comments on the manuscript. We thank Dr. Juhee Jeong and members of the Jeong laboratory for discussions.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Accession Codes
The microarray data have been deposited in the GEO database under the accession number GSE68546.
Author Contributions
M.B. J. and J-P. S-J. designed the experiments and wrote the manuscript. M.B. J., H. J-G., A. D. and J-P. S-J. performed the experiments and analyzed the data. M.B. J., H. J-G. and J-P. S-J. prepared the figures. All authors have read and approved the final manuscript. Funded by a grant form the National Institutes of Health to J-P. S-J. (R01-DE014212).
References
- 1.Grocott T, Tambalo M, Streit A. The peripheral sensory nervous system in the vertebrate head: a gene regulatory perspective. Dev Biol. 2012;370:3–23. doi: 10.1016/j.ydbio.2012.06.028. [DOI] [PubMed] [Google Scholar]
- 2.Saint-Jeannet JP, Moody SA. Establishing the pre-placodal region and breaking it into placodes with distinct identities. Dev Biol. 2014;389:13–17. doi: 10.1016/j.ydbio.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schlosser G. Making senses development of vertebrate cranial placodes. Int Rev Cell & Mol Biol. 2010;283:129–234. doi: 10.1016/S1937-6448(10)83004-7. [DOI] [PubMed] [Google Scholar]
- 4.Ahrens K, Schlosser G. Tissues and signals involved in the induction of placodal Six1 expression in Xenopus laevis. Dev Biol. 2005;288:40–59. doi: 10.1016/j.ydbio.2005.07.022. [DOI] [PubMed] [Google Scholar]
- 5.Brugmann SA, Pandur PD, Kenyon KL, Pignoni F, Moody SA. Six1 promotes a placodal fate within the lateral neurogenic ectoderm by functioning as both a transcriptional activator and repressor. Development. 2004;131:5871–5881. doi: 10.1242/dev.01516. [DOI] [PubMed] [Google Scholar]
- 6.Litsiou A, Hanson S, Streit A. A balance of FGF, BMP and WNT signalling positions the future placode territory in the head. Development. 2005;132:4051–4062. doi: 10.1242/dev.01964. [DOI] [PubMed] [Google Scholar]
- 7.Hong CS, Saint-Jeannet JP. The activity of Pax3 and Zic1 regulates three distinct cell fates at the neural plate border. Mol Biol Cell. 2007;18:2192–2202. doi: 10.1091/mbc.E06-11-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuo JS, et al. Opl: a zinc finger protein that regulates neural determination and patterning in Xenopus. Development. 1998;125:2867–2882. doi: 10.1242/dev.125.15.2867. [DOI] [PubMed] [Google Scholar]
- 9.Sato T, Sasai N, Sasai Y. Neural crest determination by co-activation of Pax3 and Zic1 genes in Xenopus ectoderm. Development. 2005;132:2355–2363. doi: 10.1242/dev.01823. [DOI] [PubMed] [Google Scholar]
- 10.Monsoro-Burq AH, Wang E, Harland R. Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction. Dev Cell. 2005;8:167–178. doi: 10.1016/j.devcel.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 11.Bae CJ, et al. Identification of Pax3 and Zic1 targets in the developing neural crest. Dev Biol. 2014;386:473–483. doi: 10.1016/j.ydbio.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knecht AK, Good PJ, Dawid IB, Harland RM. Dorsal-ventral patterning and differentiation of noggin-induced neural tissue in the absence of mesoderm. Development. 1995;121:1927–1935. doi: 10.1242/dev.121.6.1927. [DOI] [PubMed] [Google Scholar]
- 13.Lepperdinger G, Engel E, Richter K. A retinoid-binding lipocalin, Xlcpl1, relevant for embryonic pattern formation is expressed in the nervous system of Xenopus laevis. Dev Genes Evol. 1997;207:177–185. doi: 10.1007/s004270050105. [DOI] [PubMed] [Google Scholar]
- 14.Lepperdinger G, et al. The lipocalin Xlcpl1 expressed in the neural plate of Xenopus laevis embryos is a secreted retinaldehyde binding protein. Prot Sci. 1996;5:1250–1260. doi: 10.1002/pro.5560050704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richter K, Grunz H, Dawid IB. Gene expression in the embryonic nervous system of Xenopus laevis. Proc Nat Acad Sci (USA) 1988;85:8086–8090. doi: 10.1073/pnas.85.21.8086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayor R, Morgan R, Sargent MG. Induction of the prospective neural crest of Xenopus. Development. 1995;121:767–777. doi: 10.1242/dev.121.3.767. [DOI] [PubMed] [Google Scholar]
- 17.Pandur PD, Moody SA. Xenopus Six1 gene is expressed in neurogenic cranial placodes and maintained in the differentiating lateral lines. Mech Dev. 2000;96:253–257. doi: 10.1016/s0925-4773(00)00396-8. [DOI] [PubMed] [Google Scholar]
- 18.Schlosser G, Ahrens K. Molecular anatomy of placode development in Xenopus laevis. Dev Biol. 2004;271:439–466. doi: 10.1016/j.ydbio.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 19.Pohl BS, Knochel S, Dillinger K, Knochel W. Sequence and expression of FoxB2 (XFD-5) and FoxI1c (XFD-10) in Xenopus embryogenesis. Mech Dev. 2002;117:283–287. doi: 10.1016/s0925-4773(02)00184-3. [DOI] [PubMed] [Google Scholar]
- 20.Mizuseki K, Kishi M, Matsui M, Nakanishi S, Sasai Y. Xenopus Zic-related-1 and Sox-2, two factors induced by chordin, have distinct activities in the initiation of neural induction. Development. 1998;125:579–587. doi: 10.1242/dev.125.4.579. [DOI] [PubMed] [Google Scholar]
- 21.Huang X, Hong CS, O’Donnell M, Saint-Jeannet JP. The doublesex-related gene, XDmrt4, is required for neurogenesis in the olfactory system. Proc Nat Acad Sci (USA) 2005;102:11349–11354. doi: 10.1073/pnas.0505106102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heller N, Brandli AW. Xenopus Pax-2/5/8 orthologues: novel insights into Pax gene evolution and identification of Pax-8 as the earliest marker for otic and pronephric cell lineages. Dev Gen. 1999;24:208–219. doi: 10.1002/(SICI)1520-6408(1999)24:3/4<208::AID-DVG4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 23.Takabatake Y, Takabatake T, Takeshima K. Conserved and divergent expression of T-box genes Tbx2-Tbx5 in Xenopus. Mech Dev. 2000;91:433–437. doi: 10.1016/s0925-4773(99)00329-9. [DOI] [PubMed] [Google Scholar]
- 24.Urade Y, Hayaishi O. Biochemical, structural, genetic, physiological, and pathophysiological features of lipocalin-type prostaglandin D synthase. Bioch Bioph Acta. 2000;1482:259–271. doi: 10.1016/s0167-4838(00)00161-8. [DOI] [PubMed] [Google Scholar]
- 25.Urade Y, et al. Structural and functional significance of cysteine residues of glutathione-independent prostaglandin D synthase. Identification of Cys65 as an essential thiol. J Biol Chem. 1995;270:1422–1428. doi: 10.1074/jbc.270.3.1422. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka T, et al. Lipocalin-type prostaglandin D synthase (beta-trace) is a newly recognized type of retinoid transporter. J Biol Chem. 1997;272:15789–15795. doi: 10.1074/jbc.272.25.15789. [DOI] [PubMed] [Google Scholar]
- 27.Minucci S, et al. Retinoid X receptor-selective ligands produce malformations in Xenopus embryos. Proc Nat Acad Sci (USA) 1996;93:1803–1807. doi: 10.1073/pnas.93.5.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y, Pollet N, Niehrs C, Pieler T. Increased XRALDH2 activity has a posteriorizing effect on the central nervous system of Xenopus embryos. Mech Dev. 2001;101:91–103. doi: 10.1016/s0925-4773(00)00558-x. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Reese DH. A screen for disruptors of the retinol (vitamin A) signaling pathway. Birth Defects Res (Part B) 2013;98:276–282. doi: 10.1002/bdrb.21062. [DOI] [PubMed] [Google Scholar]
- 30.Strate I, Min TH, Iliev D, Pera EM. Retinol dehydrogenase 10 is a feedback regulator of retinoic acid signalling during axis formation and patterning of the central nervous system. Development. 2009;136:461–472. doi: 10.1242/dev.024901. [DOI] [PubMed] [Google Scholar]
- 31.Johnson AT, et al. Synthesis and characterization of a highly potent and effective antagonist of retinoic acid receptors. J Med Chem. 1995;38:4764–4767. doi: 10.1021/jm00024a003. [DOI] [PubMed] [Google Scholar]
- 32.Koide T, Downes M, Chandraratna RAS, Blumberg B, Umesono K. Active repression of RAR signaling is required for head formation. Genes & Dev. 2001;15:2111–2121. doi: 10.1101/gad.908801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hernandez RE, Rikhof HA, Bachmann R, Moens CB. vhnf1 integrates global RA patterning and local FGF signals to direct posterior hindbrain development in zebrafish. Development. 2004;131:4511–4520. doi: 10.1242/dev.01297. [DOI] [PubMed] [Google Scholar]
- 34.Schug TT, Berry DC, Shaw NS, Travis SN, Noy N. Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell. 2007;129:723–733. doi: 10.1016/j.cell.2007.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jetten J. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl Recept Signal. 2009;7:e003. doi: 10.1621/nrs.07003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stehlin-Gaon C, et al. All-trans retinoic acid is a ligand for the orphan nuclear receptor RORb. Natrure Struc Biol. 2003;10:820–825. doi: 10.1038/nsb979. [DOI] [PubMed] [Google Scholar]
- 37.Knecht AK, Harland RM. Mechanisms of dorsal-ventral patterning in noggin-induced neural tissue. Development. 1997;124:2477–2488. doi: 10.1242/dev.124.12.2477. [DOI] [PubMed] [Google Scholar]
- 38.Cornish EJ, Hassan SM, Martin JD, Li S, Merzdorf CS. A microarray screen for direct targets of Zic1 identifies an aquaporin gene, aqp-3b, expressed in the neural folds. Dev Dyn. 2009;238:1179–1194. doi: 10.1002/dvdy.21953. [DOI] [PubMed] [Google Scholar]
- 39.Urade Y, Hayaishi O. Prostaglandin D synthase: structure and function. Vit & Horm. 2000;58:89–120. doi: 10.1016/s0083-6729(00)58022-4. [DOI] [PubMed] [Google Scholar]
- 40.Shiotsugu J, et al. Multiple points of interaction between retinoic acid and FGF signaling during embryonic axis formation. Development. 2004;131:2653–2667. doi: 10.1242/dev.01129. [DOI] [PubMed] [Google Scholar]
- 41.Janesick A, Shiotsugu J, Taketani M, Blumberg B. RIPPLY3 is a retinoic acid-inducible repressor required for setting the borders of the pre-placodal ectoderm. Development. 2012;139:1213–1224. doi: 10.1242/dev.071456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cammas L, et al. Retinoic acid receptor (RAR)-α is not critically required for mediating retinoic acid effects in the developing mouse retina. Inv Oph & Vis Sci. 2010;51:3281–3290. doi: 10.1167/iovs.09-3769. [DOI] [PubMed] [Google Scholar]
- 43.Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134:921–931. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kam RK, Deng Y, Chen Y, Zhao H. Retinoic acid synthesis and functions in early embryonic development. Cell & Biosci. 2012;2:11. doi: 10.1186/2045-3701-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rhinn M, Dolle P. Retinoic acid signalling during development. Development. 2012;139:843–858. doi: 10.1242/dev.065938. [DOI] [PubMed] [Google Scholar]
- 46.Wu J, Yang J, Klein PS. Neural crest induction by the canonical Wnt pathway can be dissociated from anterior-posterior neural patterning in Xenopus. Dev Biol. 2005;279:220–232. doi: 10.1016/j.ydbio.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 47.Mandal A, et al. Transgenic retinoic acid sensor lines in zebrafish indicate regions of available embryonic retinoic acid. Dev Dyn. 2013;242:989–1000. doi: 10.1002/dvdy.23987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ribes V, Wang Z, Dolle P, Niederreither K. Retinaldehyde dehydrogenase 2 (RALDH2)-mediated retinoic acid synthesis regulates early mouse embryonic forebrain development by controlling FGF and sonic hedgehog signaling. Development. 2006;133:351–361. doi: 10.1242/dev.02204. [DOI] [PubMed] [Google Scholar]
- 49.Blentic A, Gale E, Maden M. Retinoic acid signalling centres in the avian embryo identified by sites of expression of synthesising and catabolising enzymes. Dev Dyn. 2003;227:114–127. doi: 10.1002/dvdy.10292. [DOI] [PubMed] [Google Scholar]
- 50.Maden M, Gale E, Kostetskii I, Zile M. Vitamin A-deficient quail embryos have half a hindbrain and other neural defects. Curr Biol. 1996;6:417–426. doi: 10.1016/s0960-9822(02)00509-2. [DOI] [PubMed] [Google Scholar]
- 51.Maden M, et al. Retinoic acid is required for specification of the ventral eye field and for Rathke’s pouch in the avian embryo. Int J Dev Biol. 2007;51:191–200. doi: 10.1387/ijdb.062175mm. [DOI] [PubMed] [Google Scholar]
- 52.Lee LM, et al. A paradoxical teratogenic mechanism for retinoic acid. Proc Nat Acad Sci (USA) 2012;109:13668–13673. doi: 10.1073/pnas.1200872109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanibe M, et al. Retinoic acid metabolizing factor xCyp26c is specifically expressed in neuroectoderm and regulates anterior neural patterning in Xenopus laevis. Int J Dev Biol. 2008;52:893–901. doi: 10.1387/ijdb.082683mt. [DOI] [PubMed] [Google Scholar]
- 54.Drummond DL, et al. The role of Zic transcription factors in regulating hindbrain retinoic acid signaling. BMC Dev Biol. 2013;13:13–31. doi: 10.1186/1471-213X-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maurus D, Harris WA. Zic-associated holoprosencephaly: zebrafish Zic1 controls midline formation and forebrain patterning by regulating Nodal, Hedgehog, and retinoic acid signaling. Genes & Dev. 2009;23:1461–1473. doi: 10.1101/gad.517009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kolm PJ, Sive HL. Efficient hormone-inducible protein function in Xenopus laevis. Dev Biol. 1995;171:267–272. doi: 10.1006/dbio.1995.1279. [DOI] [PubMed] [Google Scholar]
- 57.Nieuwkoop PD, Faber J. Normal table of Xenopus leavis. North Holland Publishing Company; Amsterdam: 1967. [Google Scholar]
- 58.Slack JM, Forman D. An interaction between dorsal and ventral regions of the marginal zone in early amphibian embryos. J Embr Exp Morph. 1980;56:283–299. [PubMed] [Google Scholar]
- 59.Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- 60.Henry GL, Brivanlou IH, Kessler DS, Hemmati-Brivanlou A, Melton DA. TGF-beta signals and a pattern in Xenopus laevis endodermal development. Development. 1996;122:1007–1015. doi: 10.1242/dev.122.3.1007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.