Abstract
We investigate possible improvements in the accuracy of semiempirical quantum chemistry (SQC) methods through the use of machine learning (ML) models for the parameters. For a given class of compounds, ML techniques require sufficiently large training sets to develop ML models that can be used for adapting SQC parameters to reflect changes in molecular composition and geometry. The ML-SQC approach allows the automatic tuning of SQC parameters for individual molecules, thereby improving the accuracy without deteriorating transferability to molecules with molecular descriptors very different from those in the training set. The performance of this approach is demonstrated for the semiempirical OM2 method using a set of 6095 constitutional isomers C7H10O2, for which accurate ab initio atomization enthalpies are available. The ML-OM2 results show improved average accuracy and a much reduced error range compared with those of standard OM2 results, with mean absolute errors in atomization enthalpies dropping from 6.3 to 1.7 kcal/mol. They are also found to be superior to the results from specific OM2 reparameterizations (rOM2) for the same set of isomers. The ML-SQC approach thus holds promise for fast and reasonably accurate high-throughput screening of materials and molecules.
1. Introduction
In the field of de novo in silico materials and drug design, fast and accurate methods are required for high-throughput screening of a wide range of systems.1−6 Density functional theory (DFT) methods are widely used1,3,5,7 because they are robust, often sufficiently accurate, and universally applicable for most of the less exotic materials that can be composed of main group and transition metal elements. Typically, their computational cost is significantly smaller than that of high-level correlated ab initio methods.
Semiempirical quantum chemistry (SQC) and machine learning (ML) methods are much faster than DFT and may thus become viable alternatives to DFT for high-throughput screening. In fact, SQC methods have already been used for such studies, as in the search for the selective kinase inhibitors.8 However, they may often not be accurate enough for this purpose. Their usefulness could be improved significantly by enhancing their average accuracy and transferability and especially by reducing the number of severe outliers in the calculated properties. Parameters in SQC methods are usually fitted in a global way to reproduce available experimental observables or highly accurate quantum chemistry (QC) reference values for a broad range of reference molecules.9,10 While this general-purpose strategy often provides acceptable average accuracy in a statistical sense,9,10 SQC calculations may be quite inaccurate for particular compounds.11,12
SQC parameters are sometimes refitted specifically for some class of compounds (e.g., fullerenes13), for certain reactions (e.g., to study kinetic isotopic effects14) or for intermolecular interactions (e.g., in water15). The resulting special-purpose SQC approaches can achieve high accuracy by closely reproducing experimental or high-level QC data for the target systems. Of course, such special-purpose SQC methods are accurate only for the types of compounds, reactions, and properties for which they have been reparametrized, but generally not for other targets.
ML methods can be used in computational chemistry to infer properties of new molecules through interpolation in chemical compound space.16−18 They employ simple but flexible ad hoc models for interpolation that are trained on a sufficiently large set of compounds and can then be used to predict the properties of related target compounds. Evaluation of an ML model is generally orders of magnitude faster than an SQC calculation, but, due to the lack of rigorous physical approximations, more and larger outliers can be expected.19,20 Obviously, the accuracy and transferability of ML methods depends dramatically on the compound diversity present in the training set.
Here, we explore a novel use of ML methods in SQC. Instead of applying ML methods to directly compute certain properties within a large class of related compounds, we use them to determine optimum SQC parameters for individual molecules within the class of target compounds. In both cases, there is an initial training step for calibrating the ML model on a reference subset of compounds, followed by production runs that yield predictions for the other target compounds. The basic idea is to employ ML techniques to optimize the SQC parameters for individual molecules (within a class of related compounds) such that subsequent SQC calculations are as accurate as possible for predicting the properties of interest. Hence, we introduce a hybrid ML-SQC approach where ML is used as an automatic parametrization technique (APT) to determine on-the-fly optimal individual semiempirical parameters as a function of atomic configuration and composition.
In this article, we first explain the chosen APT approach. Thereafter we present an illustrative application of the ML-SQC method for a set of 6095 constitutional isomers C7H10O2, for which accurate thermochemical reference data from G4MP221 calculations are available.22 These molecules were drawn from the chemical universe database GDB-17 that covers many drug-like molecules and contains 166.4 billion molecules with up to 17 non-hydrogen atoms.23 For the SQC method, we use the semiempirical OM2 (orthogonalization model 2) approach.24,25 In this proof-of-concept study, we evaluate the accuracy that can be achieved by the ML-OM2 method for the chosen target set, and we compare the ML-OM2 results with those obtained using the standard OM2 parameters as well as special-purpose OM2 parameters from reparametrizations for the same target compounds.
2. Automatic Parametrization Technique
2.1. Overview
As outlined above, the idea behind APT relies on the use of ML to locally improve upon the global SQC parameter values. To this end, we have implemented the following procedure:
-
(1)
Find optimal corrections to parameter values for each individual molecule in the training set;
-
(2)
Train ML model on the parameter corrections from the previous step;
-
(3)
Use ML model to predict corrections to parameters for target molecules;
-
(4)
Carry out SQC calculations with corrected parameter values for target molecules.
In this procedure, one may, in principle, apply any combination of appropriate parameter optimization and machine learning techniques. In the following, we present the chosen hybrid ML-SQC approach in detail.
2.2. Technical Details
Step 1
Here, we vary only one of the many OM2 parameters at a time. More specifically, we tune a given parameter to minimize the error in the atomization enthalpy for each molecule in the training set using the Levenberg–Marquardt optimization algorithm.26,27 Generally, convergence to complete error depletion was reached after few iterations such that an OM2 calculation with the resulting parameter gives an error-free atomization enthalpy for each molecule (in its standard OM2 geometry) of the training set. Systematic application of this procedure yields a set of changes (ΔPopt) to the standard OM2 parameter values for each molecule in the training set. Failures of this procedure were encountered in a vanishingly small number of cases, which were ignored since they do not affect the overall performance. These minimizations were carried out successively for all OM2 parameters, which are listed in Table 1 in standard notation.24,25
Table 1. Mean Absolute Deviations (MAD) of Parameter Values Optimized in APT Step 1 from the Standard OM2 Values and Mean Absolute Errors (MAEs) in Atomization Enthalpies from ML-OM2//OM2 Calculations at OM2 Geometries for 1000 C7H10O2 Molecules (Drawn at Random) In the Training Set and 5095 C7H10O2 Molecules in the Test Set (Remainder)a.
| hydrogen |
carbon |
oxygen |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| MAE, kcal/mol |
MAE, kcal/mol |
MAE, kcal/mol |
|||||||
| parameter | MAD, % | training | test | MAD, % | training | test | MAD, % | training | test |
| One-Center One-Electron Terms | |||||||||
| Uss | 1.20 | 0.00 | 2.89 | 0.10 | 0.00 | 2.83 | 4.10 | 0.51 | 3.50 |
| Upp | 0.10 | 0.00 | 2.84 | 0.30 | 0.00 | 2.84 | |||
| Orbital Exponent | |||||||||
| ζ | 1.10 | 0.00 | 2.85 | 0.40 | 0.00 | 2.82 | 1.20 | 0.00 | 2.88 |
| Resonance Integrals | |||||||||
| βs | 1.20 | 0.00 | 2.82 | 1.50 | 0.00 | 2.87 | 13.40 | 0.00 | 3.09 |
| βp | 0.90 | 0.00 | 2.84 | 2.50 | 0.00 | 3.04 | |||
| βπ | 3.90 | 0.00 | 3.77 | 9.80 | 0.00 | 3.78 | |||
| βs(X–H) | 2.30 | 0.00 | 2.86 | 117.80 | 0.44 | 6.27 | |||
| βp(X–H) | 1.40 | 0.00 | 2.84 | 35.60 | 0.08 | 6.69 | |||
| αs | 2.50 | 0.00 | 2.82 | 1.30 | 0.00 | 2.84 | 9.40 | 0.00 | 2.99 |
| αp | 0.90 | 0.00 | 2.84 | 2.90 | 0.00 | 3.27 | |||
| απ | 2.50 | 0.00 | 3.49 | 6.60 | 0.00 | 3.33 | |||
| αs(X–H) | 4.40 | 0.00 | 2.88 | 203.20 | 1.37 | 6.01 | |||
| αp(X–H) | 4.70 | 0.00 | 2.99 | 47.40 | 0.24 | 6.28 | |||
| Orthogonalization Factors | |||||||||
| F1 | 4.20 | 0.00 | 2.82 | 0.70 | 0.00 | 2.82 | 1.60 | 0.00 | 2.84 |
| F2 | 5.40 | 0.00 | 2.86 | 8.60 | 0.00 | 2.84 | 4.70 | 0.00 | 2.86 |
| G1 | 40.10 | 0.64 | 3.57 | 17.00 | 0.00 | 3.04 | 215.50 | 0.18 | 5.52 |
| G2 | 26.30 | 0.00 | 2.80 | 11.90 | 0.00 | 2.84 | 223.30 | 0.11 | 4.22 |
| Effective Core Potentials | |||||||||
| ζα | 0.40 | 0.00 | 2.88 | 4.80 | 0.00 | 3.12 | |||
| Fαα | 1.60 | 0.00 | 2.88 | 13.90 | 0.00 | 2.86 | |||
| βα | 6.50 | 0.00 | 2.86 | 116.00 | 0.00 | 3.08 | |||
| αα | 4.10 | 0.00 | 2.87 | 250.50 | 1.40 | 25.40 | |||
| One-Center Two-Electron Integrals | |||||||||
| gss | 7.40 | 0.46 | 3.49 | 0.30 | 0.00 | 2.83 | 4.50 | 0.00 | 2.85 |
| gpp | 0.70 | 0.00 | 2.83 | 1.60 | 0.00 | 2.84 | |||
| gsp | 1.50 | 0.10 | 3.18 | 1.30 | 0.00 | 2.89 | |||
| gp2 | 0.20 | 0.00 | 2.83 | 0.60 | 0.00 | 2.84 | |||
| hsp | 11.80 | 0.02 | 3.13 | 11.40 | 0.02 | 3.06 | |||
MADs are given in percent; MAEs are given in kcal/mol. Standard OM2 yields a MAE of 6.30 kcal/mol for these molecules.
Step 2
The corrections {ΔPopt} of the parameter values for each molecule in the training set (obtained at its standard OM2 geometry) are used to train the ML model. We apply an ML approach introduced in 2012,16 which has been described in detail in the literature.16−18 Therefore, we give only a brief outline of the procedure and refer to the original publications for further information.
We employ kernel ridge regression with a Laplacian kernel. In this approach, the default parameter correction ΔP for molecule M is estimated by summing over all Ntrain molecules {Mi} in the training set.
| 1 |
where αi is the regression coefficient for molecule Mi, σ is the length-scale hyperparameter (same value for any pair of molecules M and Mi), and ∥M – Mi∥1 is the 1-norm calculated from the vectorized molecular descriptor X of size Nx by summing the absolute differences between the elements of X(M) and X(Mi)
| 2 |
As molecular representation, we choose the Coulomb matrix C.16−18 It is an atom-by-atom matrix with the elements
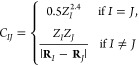 |
3 |
where internuclear distances |RI – RI| are measured between the atomic coordinates R (in Bohr) and all nuclear charges ZI are in e. When one molecule is larger than the other, we extend the Coulomb matrix of the smaller molecule by zeros. The Coulomb matrix is a unique yet nonstereospecific representation of a molecule, and it can thus distinguish diastereomers but not enantiomers. It is translationally and rotationally invariant. In order to also achieve atom-index invariance, we sort all atom indices by the norm of their Coulomb matrix row. Sorted Coulomb matrices are used to calculate the norm ∥M – Mi∥1 according to eq 2, where Xa is an element CIJ of the corresponding Coulomb matrix and the sum runs over all Coulomb matrix elements, with Nx being the square of the number of atoms of the largest molecule.
Training the ML model outlined above requires solving the minimization problem
| 4 |
The analytical solution involves the following matrix transformations16−18
| 5 |
where I is identity matrix, ΔPopt is the vector with corrections to the standard parameter value, and λ is a so-called regularization parameter that ensures the transferability of the model to new compounds.16,17 The elements Kij of the kernel matrix K are defined by
| 6 |
We determined optimal values of hyperparameters by 5-fold cross-validation within the training sets following a previously reported procedure.18 The training set was sorted according to the values of the parameter correction. It was then divided into buckets with only five items each. Thereafter, five splits were created by successively taking out a single item from each bucket. Four of these stratified samples were used to train the ML model, and the fifth out-of-sample split was used to estimate the error of the ML model. All five possible such folds were generated. The error in the out-of-sample split was minimized by varying the hyperparameters σ and λ. Optimal σ and λ values were found for each fold by a simple logarithmic grid search. These hyperparameter values were averaged over five folds to train our final ML model on the entire training set.
Step 3
The ML model trained in the previous step was employed to predict corrections to the OM2 parameters for other molecules (outside the training set) according to eq 1, using geometries optimized with default OM2 parameters.
Step 4
The corrections to the OM2 parameters predicted in the previous step were added to their OM2 default values, and the resulting parameters were used in a subsequent OM2 calculation of the atomization enthalpy.
3. Computational Details
All OM2 calculations with default and modified parameters were carried out with our locally modified MNDO2005(28) program. The SCF energy convergence criterion was set to 10–8 eV. In addition, the diagonal elements of the density matrix were converged to less than 10–8. Geometry optimizations were considered converged when the Cartesian gradient norm dropped below 0.1 kcal/(mol·Å). No cutoffs were applied for the three-center orthogonalization corrections in the OM2 calculations.
4. Results and Discussion
The G4MP2 atomization enthalpies at T = 298 K of the 6095 constitutional isomers C7H10O222 that can be extracted from GDB-1723 (see above) served as reference data.
4.1. Application of APT
In an initial screening of all OM2 parameters, we estimated the potential improvements in accuracy that one might expect from a hybrid ML-SQC approach. ML-OM2 calculations were performed at OM2 geometries (denoted ML-OM2//OM2). In the screening of the 61 OM2 parameters for hydrogen, carbon, and oxygen, we used 1000 randomly taken molecules for training and the remaining 5095 molecules for testing (APT Step 1). The resulting mean absolute errors (MAEs) in the atomization enthalpies for the training and test sets are given in Table 1. In addition, mean absolute deviations (MADs) of the individually optimized parameters (ATP Step 1) from the standard OM2 parameters are also listed.
The MAEs in the atomization enthalpies calculated with ML-OM2//OM2 for the test set typically improve from 6.30 kcal/mol (standard OM2 for these molecules) down to 2.80–3.00 kcal/mol in 38 out of 61 cases when a single OM2 parameter is adjusted individually through ML. Thus, in principle, any of these 38 parameters could be used for APT. Here, we have chosen to develop ML models for corrections to the ζ parameter (orbital exponent) of carbon. This choice is motivated by the fact that tuning ζ apparently leads to minimal changes in parameter value combined with maximal changes in the computed property (Table 1). More specifically, optimizing the ζ parameter of carbon for the individual molecules in the training set leads to a mean absolute change of only 0.4% in the parameter value, while the MAE for the atomization enthalpy is reduced from 6.30 to 2.82 kcal/mol in the test set. Such small changes of parameters can be considered to be fine-tuning rather than drastic reparametrization to populate significantly different regions of parameter space. In addition, it seems natural to use an OM2 parameter of carbon for fine-tuning since our present application deals exclusively with organic molecules. The OM2 parameters for the effective core potential might offer a promising alternative for fine-tuning because they yield MAEs of less than 3 kcal/mol for parameter changes in the single-digit percentage range.
Next, we studied the effect of the size of the chosen training set on the ML-OM2 results (using, again, the ζ parameter of carbon for ML). We considered Ntrain = 10, 100, 1000, 2000, 3000, 4000, and 5000, with molecules being drawn at random from the full set of 6095 C7H10O2 isomers. After applying the ML model to the training set, the remaining fitting errors were vanishingly small in all cases (i.e., for all Ntrain values). Not surprisingly, the accuracy for the out-of-sample test set improved systematically with increasing Ntrain. The results are summarized in Table 2 and shown in Figure 1. The 5k ML-OM2//OM2 model (Ntrain = 5000, MAE = 1.72 kcal/mol) has a substantially improved accuracy when compared to that of standard OM2 (MAE = 6.30 kcal/mol) and even approaches the highly coveted target of chemical accuracy (1 kcal/mol) at an overall computational cost of about 6 CPU hours. We also note that a 2 kcal/mol accuracy for atomization enthalpies is on par with (if not better than) many of the more advanced DFT methods.29
Table 2. Mean Absolute Errors (MAEs) in the Predicted Atomization Enthalpies of the Constitutional Isomers of C7H10O2 from OM2 (Ntrain = 0) and ML-OM2//OM2 Calculations at OM2 Geometriesa.
| Ntrain | training set | test set |
|---|---|---|
| 0 | 6.30 | |
| 10 | 0.00 | 6.31 |
| 100 | 0.00 | 5.46 |
| 1000 | 0.00 | 2.88 |
| 2000 | 0.00 | 2.29 |
| 3000 | 0.00 | 1.96 |
| 4000 | 0.00 | 1.81 |
| 5000 | 0.00 | 1.72 |
See the text for details. MAEs are given in kcal/mol for Ntrain molecules in the training sets and for 6095 – Ntrain molecules in the test sets.
Figure 1.
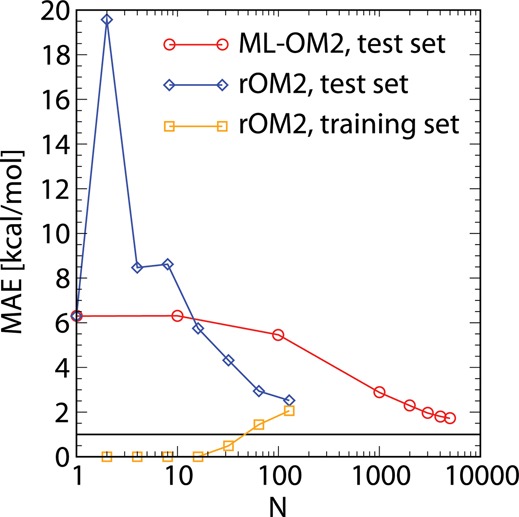
Mean absolute errors (MAEs) in the predicted atomization enthalpies for the out-of-sample test set of molecules with C7H10O2 stoichiometry for ML-OM2//OM2 and rOM2 (see text). MAEs for the training set are shown only for rOM2 (vanishingly small for ML-OM2//OM2). The MAEs are plotted as a function of the training set size (Ntrain, logarithmic scale). The horizontal line at 1.0 kcal/mol indicates the onset of chemical accuracy.
4.2. Special-Purpose Reparametrization of OM2
For the sake of comparison, we performed a conventional reparametrization of all OM2 parameters for the same set of 6095 C7H10O2 isomers using the same reference atomization enthalpies. The first 2n (n = 1–7) molecules from the randomly ordered set served as training sets for the reparametrization. All OM2 parameters were reoptimized using a modified implementation of the Subplex method30 based on the NLopt library,31 without imposing any limits or constraints. The standard OM2 parameters were taken as starting values. The accuracy of the resulting series of reparametrized OM2 (rOM2) methods was evaluated on the corresponding test sets consisting of the remaining 6095 – 2n molecules. For a fair comparison with the APT approach, the rOM2 calculations on the test molecules were done at geometries optimized at the standard OM2 level (designated rOM2//OM2).
For small training sets (Ntrain = 2–8), the reparameterization results in overfitting, as indicated by MAEs for the test set that are larger than the MAE of standard OM2 (Table 3). For larger training sets, the MAE for the training set grows monotonically, whereas the MAE for test set decreases monotonically. With increasing size of the training set size, the MAEs for both sets converge to the same range, reaching values of 2.06–2.52 kcal/mol for Ntrain = 128 (Figure 1).
Table 3. Mean Absolute Errors (MAEs) in Atomization Enthalpies from OM2 (Ntrain = 0) and rOM2//OM2 Calculations at OM2 Geometries for Ntrain Molecules in the Training Sets and 6095 – Ntrain in the Test Setsa.
| Ntrain | training set | test set |
|---|---|---|
| 0 | 6.30 | |
| 2 | 0.00 | 19.57 |
| 4 | 0.00 | 8.47 |
| 8 | 0.00 | 8.62 |
| 16 | 0.00 | 5.75 |
| 32 | 0.49 | 4.32 |
| 64 | 1.44 | 2.94 |
| 128 | 2.06 | 2.52 |
MAEs are given in kcal/mol.
Obviously, the MAE for the test set is generally higher than that for the training set. Since the MAE for the training set continually increases with the size of the set, it is safe to assume that the MAEs for both sets will be slightly larger than 2.06 kcal/mol for larger Ntrain values.
4.3. Comparison of ML-OM2 and rOM2 Results
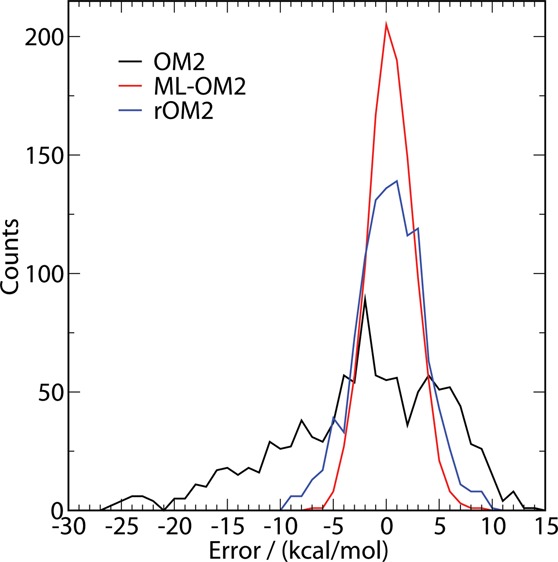
We now analyze the distribution of errors for both reparametrization approaches. For a set of 1095 randomly drawn constitutional isomers of C7H10O2, Figure 2 displays the error distributions of their atomization enthalpies obtained from the 5k ML-OM2//OM2 model as well as rOM2 (Ntrain = 128) and standard OM2 calculations. In the error distribution of standard OM2, there is a systematic shift (i.e., an underestimation of atomization enthalpies) and a substantial skew. The rOM2 reparametrization overcomes both these problems, yielding a more normal distribution centered at zero. However, in the case of 5k ML-OM2//OM2, the error distribution is more narrow, suggesting a higher degree of fidelity and a lower number of outliers. The 5k ML-OM2 model has the lowest MAE (1.72 kcal/mol). The worst outlier has an error of more than 26 kcal/mol in OM2, which is reduced to 9.8 and 8.2 kcal/mol in rOM2 and ML-OM2//OM2, respectively.
Figure 2.
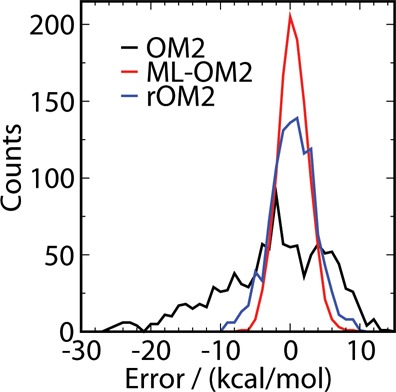
Error histogram for OM2, 5k ML-OM2//OM2, and rOM2//OM2 for a test set of 1095 molecules (see text).
We have already noted that the conventional reparametrization of OM2 appears to have a lower bound for MAEs for the test set, in our case 2.06 kcal/mol (see above), presumably due to the fixed functional form dictated by the use of OM2. The ML-based APT approach, on the other hand, is highly flexible because of the use of an expansion in nonlinear terms, which can be systematically improved by adding more examples to the training set. However, conventional reparametrization schemes do have the advantage of providing rapid improvements even for small training sets, whereas APT typically requires thousands of reference data points. Therefore, APT is particularly suited to problems involving big data sets. Another advantage of APT over conventional reparametrizations is due to the fact that its kernel inversion is convenient and computationally less demanding, whereas in the case of conventional reparametrizations, complex multidimensional optimization problems must be solved.
Yet another important issue arises when it comes to transferability. More specifically, one might wonder what happens when we attempt to compute properties of molecules that differ substantially from those present in the training set. Such molecules will normally not be well represented by the modified parameters, for obvious reasons, and one may thus expect huge errors. In the case of APT, by contrast, the ML model will predict vanishing corrections to the individual parameters for molecules that are very different, and, consequently, the results will be close to those obtained with the standard parameters. In this sense, the APT-ML model is well-tempered and transferable. It can only improve (and will never deteriorate) performance, regardless whether we consider molecules structurally similar to or different from the species in the training set.
For a more quantitative study of this aspect, we performed a comparative error analysis of the OM2, 5k ML-OM2//OM2, and rOM2//OM2 results on a validation set of 100 molecules drawn at random from the big database of ca. 134 000 molecules,22 which covers all organic molecules with up to nine heavy atoms (not counting hydrogens). We compared the three computed sets of atomization enthalpies to the reference G4MP2 values.22 Many molecules in the validation set have N or F atoms, not present in C7H10O2 isomers, which poses a severe challenge. In the case of rOM2, we combined the modified parameters for H, C, and O (see above) with the standard OM2 parameters for N and F, whereas the trained 5k ML-OM2 model employed only one modified parameter (ζ for C) together with the standard values of all other OM2 parameters in a given molecule. Consequently, it is not too surprising that rOM2//OM2 yields dramatic errors that may exceed 400 kcal/mol (Table 4). In the case of ML-OM2//OM2, the MAE is drastically reduced to ∼20 kcal/mol, with a maximum outlier of ∼50 kcal/mol. As expected, these ML-OM2//OM2 results are not too far off from the corresponding standard OM2 results (MAE ∼10 kcal/mol, maximum error ∼40 kcal/mol). This confirms that the ML-OM2//OM2 approach is fairly robust even in difficult cases.
Table 4. Mean Absolute Errors (MAEs) of Atomization Enthalpies of 100 Molecules Drawn at Random from GDB-1723a.
| method | MAE, kcal/mol | range of errors, kcal/mol |
|---|---|---|
| OM2 | 10.94 | –39.57 to 9.6 |
| ML-OM2//OM2 | 21.58 | –52.02 to 0.87 |
| rOM2//OM2 | 145.39 | –414.15 to 484.33 |
Results are given for OM2 with default parameters, rOM2//OM2 was reparametrized using 128 C7H10O2 isomers, and the ML-OM2//OM2 model was trained on 5k C7H10O2 isomers. MAEs are given in kcal/mol.
5. Conclusions
We have introduced an automatic parametrization technique that augments semiempirical parameters for any new molecule. It is based on machine learning models of parameters as a function of molecular structure (requiring as input only the identities of the constituent atoms and their coordinates). After training the model on sufficiently large training sets (yielding precalculated corrections to parameters), it can be applied to other new molecules for predicting molecule-specific corrections to the parameters that allow semiempirical quantum chemical calculations with improved accuracy.
For numerical demonstration, we chose the OM2 method, which has a mean absolute error of 6.3 kcal/mol in atomization enthalpy for the 6095 constitutional isomers of C7H10O2 stoichiometry, in calculations with standard OM2 parameters. After individually adjusting the parameters in the ML-OM2 approach for the largest training set of 5000 isomers, the mean absolute error for the remaining 1095 isomers in the test set can be reduced from 6.3 kcal/mol (standard OM2, same value as for the full set) to 1.7 kcal/mol, and the largest error can be lowered from 26.3 to 8.2 kcal/mol, respectively. Furthermore, ML-OM2 has a narrower error distribution than OM2, or than that of a conventionally reparametrized variant of OM2 (rOM2), and it is found to be quite robust even when screening structures that differ substantially from those present in the training set.
To summarize, we have presented numerical evidence that the ML-APT approach can significantly improve the predictive accuracy of well-established semiempirical quantum-chemical methods for large sets of molecules, without increasing the computational burden beyond the need of having a reference database at disposal. We emphasize that, due to its general nature, the APT idea may be useful for any method of fixed functional form that depends on parameters, e.g., in DFT or in the learning-on-the-fly approach to ab initio molecular dynamics.32
Acknowledgments
The authors thank Xin Wu for kindly providing the parametrization program. This research used resources of the Argonne Leadership Computing Facility at Argonne National Laboratory, which is supported by the Office of Science of the U.S. DOE under contract no. DE-AC02-06CH11357.
W.T. is grateful to the European Research Council (ERC) for financial support through an ERC Advanced Grant. O.A.v.L. acknowledges support from the Swiss National Science foundation (no. PP00P2_138932).
The authors declare no competing financial interest.
References
- Hachmann J.; Olivares-Amaya R.; Atahan-Evrenk S.; Amador-Bedolla C.; Sánchez-Carrera R. S.; Gold-Parker A.; Vogt L.; Brockway A. M.; Aspuru-Guzik A. J. Phys. Chem. Lett. 2011, 2, 2241–2251. [Google Scholar]
- Yang L.; Ceder G. Phys. Rev. B 2013, 88, 224107. [Google Scholar]
- Jain A.; Ong S. P.; Hautier G.; Chen W.; Richards W. D.; Dacek S.; Cholia S.; Gunter D.; Skinner D.; Ceder G.; Persson K. A. APL Mater. 2013, 1, 011002. [Google Scholar]
- Jorgensen W. L. Science 2004, 303, 1813–1818. [DOI] [PubMed] [Google Scholar]
- Nørskov J. K.; Bligaard T.; Rossmeisl J.; Christensen C. H. Nat. Chem. 2009, 1, 37–46. [DOI] [PubMed] [Google Scholar]
- Schneider G. Nat. Rev. Drug Discovery 2010, 9, 273–276. [DOI] [PubMed] [Google Scholar]
- Hautier G.; Miglio A.; Ceder G.; Rignanese G.-M.; Gonze X. Nat. Commun. 2013, 4, 2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T.; Caflisch A. ChemMedChem 2010, 5, 1007–1014. [DOI] [PubMed] [Google Scholar]
- Thiel W. Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2014, 4, 145–157and references therein. [Google Scholar]
- Clark T.; Stewart J. J. P.. MNDO-like semiempirical molecular orbital theory and its application to large systems. In Computational Methods for Large Systems: Electronic Structure Approaches for Biotechnology and Nanotechnology; Reimers J. R., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, 2011; pp 259–286. [Google Scholar]
- Jensen F.Introduction to Computational Chemistry; John Wiley & Sons Ltd: Chichester, England, 1999; pp 81–97. [Google Scholar]
- Korth M.; Thiel W. J. Chem. Theory Comput. 2011, 7, 2929–2936. [DOI] [PubMed] [Google Scholar]
- Tseng S.-P.; Shen M.-Y.; Yu C.-H. Theor. Chim. Acta 1995, 92, 269–280. [Google Scholar]
- Gonzalez-Lafont A.; Truong T. N.; Truhlar D. G. J. Phys. Chem. 1991, 95, 4618–4627. [Google Scholar]
- Wu X.; Thiel W.; Pezeshki S.; Lin H. J. Chem. Theory Comput. 2013, 9, 2672–2686. [DOI] [PubMed] [Google Scholar]
- Rupp M.; Tkatchenko A.; Müller K.-R.; von Lilienfeld O. A. Phys. Rev. Lett. 2012, 108, 058301. [DOI] [PubMed] [Google Scholar]
- von Lilienfeld O. A. Int. J. Quantum Chem. 2013, 113, 1676–1689. [Google Scholar]
- Hansen K.; Montavon G.; Biegler F.; Fazli S.; Rupp M.; Scheffler M.; von Lilienfeld O. A.; Tkatchenko A.; Müller K.-R. J. Chem. Theory Comput. 2013, 9, 3404–3419. [DOI] [PubMed] [Google Scholar]
- Moussa J. E. Phys. Rev. Lett. 2012, 109, 059801. [DOI] [PubMed] [Google Scholar]
- Rupp M.; Tkatchenko A.; Müller K.-R.; von Lilienfeld O. A. Phys. Rev. Lett. 2012, 109, 059802. [DOI] [PubMed] [Google Scholar]
- Curtiss L. A.; Redfern P. C.; Raghavachari K. J. Chem. Phys. 2007, 127, 124105. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan R.; Dral P. O.; Rupp M.; von Lilienfeld O. A. Sci. Data 2014, 1, 140022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruddigkeit L.; van Deursen R.; Blum L.; Reymond J.-L. J. Chem. Inf. Model. 2012, 52, 2684. [DOI] [PubMed] [Google Scholar]
- Weber W.Ein neues semiempirisches NDDO-Verfahren mit Orthogonalisierungskorrekturen: Entwicklung des Modells, Parametrisierung und Anwendungen. Ph.D. Thesis, Universität Zürich, 1996. [Google Scholar]
- Weber W.; Thiel W. Theor. Chem. Acc. 2000, 103, 495–506. [Google Scholar]
- Levenberg K. Q. Appl. Math. 1944, 2, 164–168. [Google Scholar]
- Marquardt D. W. SIAM J. Appl. Math. 1963, 11, 431–441. [Google Scholar]
- Thiel W.MNDO2005, version 7.0; Max-Planck-Institut für Kohlenforschung: Mülheim an der Ruhr, Germany, 2005.
- Cohen A. J.; Mori-Sánchez P.; Yang W. Chem. Rev. 2012, 112, 289–320. [DOI] [PubMed] [Google Scholar]
- Rowan T.Functional Stability Analysis of Numerical Algorithms. Ph.D. Thesis, University of Texas at Austin, 1990. [Google Scholar]
- Johnson S. G.NLopt: Nonlinear-Optimization Package. http://ab-initio.mit.edu/nlopt. [Google Scholar]
- Csányi G.; Albaret T.; Payne M. C.; Vita A. D. Phys. Rev. Lett. 2004, 93, 175503. [DOI] [PubMed] [Google Scholar]


