The limited availability of reliable sources of human striatal projection neurons currently hampers understanding of Huntington’s disease mechanisms and hinders the development of novel treatments. In this study, two protocols were developed for the differentiation of neural progenitor cells into striatal medium spiny neurons, and the differentiated neurons were extensively characterized. The data indicate that the three-step differentiation protocol presents a reliable and simplified method for the generation of striatal medium spiny neurons.
Keywords: Neural progenitor cells, Cell differentiation, GABAergic neurons, DARPP-32, CTIP2, Glutamate receptors
Abstract
Huntington’s disease (HD) results from a CAG repeat expansion in the gene encoding the huntingtin protein. This inherited disorder is characterized by progressive neurodegeneration. In particular, HD progression involves the loss of striatal projection neurons. The limited availability of reliable sources of human striatal projection neurons currently hampers our understanding of HD mechanisms and hinders the development of novel HD treatments. In this paper, we described two- and three-step methods for differentiating human neural progenitor cells toward striatal projection neurons. In the two-step differentiation protocol, 90%, 54%, and 6% of MAP2-positive cells were immunopositive for GABA, calbindin (CALB1), and DARPP-32/PPP1R1B, respectively. In the three-step differentiation protocol, 96%, 84%, and 21% of MAP2-positive cells were immunopositive for GABA, calbindin, and DARPP-32/PPP1R1B, respectively. In line with a striatal projection neuron phenotype, cells differentiated with our protocols displayed significantly increased expression of MAP2, CALB1, DARPP-32/PPP1R1B, ARPP21, and CTIP2. Application of glutamate receptor agonists induced calcium influx; accordingly, the cells also expressed various ionotropic glutamate receptor subunits. Differentiated cells also released GABA on stimulation. We suggest that our three-step differentiation protocol presents a reliable and simplified method for the generation of striatal projection neurons, yielding a critical resource for neuronal physiology and neurodegenerative disorder studies.
Significance
The earliest changes in the neurodegenerative disorder Huntington’s disease affect a specific type of brain neurons, the so-called medium spiny neurons of the striatum. In this study, two protocols were developed for the differentiation of neural progenitor cells into striatal medium spiny neurons, and the differentiated neurons were extensively characterized. The data indicate that the three-step differentiation protocol presents a reliable and simplified method for the generation of striatal medium spiny neurons. The generated striatal medium spiny neurons could represent a critical resource for the study of neurodegenerative disorders, a model system for drug discovery, and a step toward cell-based regeneration therapies.
Introduction
Huntington’s disease (HD) is an inherited neurodegenerative disorder caused by a CAG repeat expansion in the gene HTT encoding the huntingtin (HTT) protein [1]. HD patients suffer from progressive motor impairment, cognitive decline, and psychiatric symptoms [2]. The earliest changes in HD affect medium spiny neurons (MSNs), a cell type specific to the striatum [3]. Striatal neurons are predominately MSNs, which account for up to 75%–95% of primate and rodent striatal neuronal populations [4]. The discovery of the HTT gene CAG expansion has been the basis for subsequent HD mechanistic studies. These studies have revealed the multifaceted nature of HD and suggest that this disease affects multiple molecular processes [5]. HD-affected processes include HTT protein misfolding and aggregation [6], ubiquitin-proteasome system dysfunction [7], mitochondrial dysfunction [8], glutamate excitotoxicity [9], loss of brain-derived neurotrophic factor (BDNF) [10], and alterations of the transcriptional profile, which in particular involves neuron-specific genes [11].
The reduced BDNF expression in HD has been attributed to increased binding of the repressor element-1 transcription factor/neuron restrictive silencer factor (REST/NRSF) to a repressor element-1/neuron restrictive silencer element (RE1/NRSE) site within Bdnf promoter II [11]. REST/NRSF binding eventually contributes to neuronal loss in the striatum [10]. In healthy neurons, sequestration of REST/NRSF together with HTT prevents entry of REST/NRSF into the nucleus [11, 12]. Disruption of this interaction in HD allows REST/NRSF to enter the nucleus, where it can bind to RE1/NRSE sites and downregulate BDNF expression [11, 12].
Research on HD pathogenesis and the development of novel treatment strategies would benefit from the availability of human striatal projection neurons. It should be noted, however, that differentiated neurons are postmitotic cells that no longer proliferate; therefore, striatal MSNs cannot be amplified directly in cell culture. In contrast, mitotically active stem cells [13] can be differentiated toward a striatal projection neuron phenotype. Embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) [14, 15], for example, were recently used as starting cells for striatal projection neuron differentiation [16–19].
Current striatal differentiation protocols use a combination of growth factors, morphogens, neurotrophins, and small-molecule inhibitors and analogs [16–19]. A similarity of these protocols is the use of BDNF. BDNF has been shown to be required for striatal neuron survival [10]. Similar to BDNF, the histone deacetylase inhibitor valproic acid (VPA) is also used frequently in differentiation protocols [16, 17] and was shown to inhibit neural progenitor cell (NPC) proliferation and to introduce neuronal differentiation [20]. Neuronal differentiation is also triggered by a dibutyryl-cAMP-induced nuclear accumulation of fibroblast growth factor receptor-1 [21] and by insulin-like growth factor 1 (Igf-1) [22]. Insulin can potentiate the actions of Igf-1 [23]. Moreover, treatment with the ρ-associated protein kinase inhibitor Y-27632 increases neurite outgrowth from neural stem cells (NSCs) [24].
In contrast to factors promoting a general neuronal phenotype, sonic hedgehog (SHH) and Dickkopf 1 (DKK1) support differentiation toward more specific neuronal types [25]. Shh is involved in floor plate and ventral neuron-type induction within the neural tube [26–28]. The production of Shh in ventral parts of the developing central nervous system (CNS) is thought to result in a dorso-ventral concentration gradient [29]. DKK1 blocks WNT signaling by binding to the WNT cell surface receptor LRP6 [30]. Together with SHH, DKK1 favors differentiation of primitive neural cells toward ventral telencephalic neuronal phenotypes [25].
Current differentiation protocols use a mixture of the aforementioned factors. Aubry et al. developed a protocol to induce ESC differentiation toward striatal neurons [16]. Using this protocol, 22% of the cells were immunopositive for the neuronal marker microtubule-associated protein 2 (MAP2); among MAP2-positive cells, 53% were positive for the striatal neuronal marker dopamine and cyclic AMP-regulated phosphoprotein of 32 kDa (DARPP-32; also known as protein phosphatase 1, regulatory inhibitor subunit 1B, PPP1R1B), 10% were positive for calbindin (CALB1), and 36% were positive for GABA [16]. Others have reported higher differentiation efficiencies. Differentiation protocols used by Ma et al. [17] and Delli Carri et al. [19] resulted in 93% of cells expressing the neuronal marker β-III tubulin and 51% of cells expressing MAP2, respectively; moreover, 75% and 10%, respectively, of the total cells were positive for DARPP-32 [17, 19]. Ma et al. [17], however, did not report on the presence of glial cells that are typically observed in similar protocols [16, 18]. The aforementioned differentiation protocols are time-consuming, however, because they require an initial induction of NSC phenotypes prior to more specific differentiation toward striatal projection neurons [16, 17, 19].
An alternative strategy to these differentiation protocols is the use of NPCs as a differentiation protocol starting point [31]. In the present study, we sought to establish a straightforward method to obtain striatal projection neurons for studies of neurodegenerative diseases, such as HD. Specifically, we wished to determine whether NPCs could be used to reliably differentiate functional MSNs that display striatal projection neuronal characteristics including the expression of striatal markers such as DARPP-32, cAMP-regulated phosphoprotein of 21 kDa (ARPP21), and COUP TF1-interacting protein 2 (CTIP2; also known as B-cell leukemia/lymphoma 11B).
Material and Methods
Culture of NPCs
An immortalized human NPC line (ReNcell VM; Millipore, Billerica, MA, http://www.emdmillipore.com) was maintained on laminin-coated plastic ware. NPCs were cultured in NPC medium (Dulbecco’s modified Eagle’s medium nutrient mixture F12 [DMEM/F12] supplemented with the following additives: 20 ng/ml basic fibroblast growth factor [bFGF; Millipore], 20 ng/ml epidermal growth factor [EGF; Millipore], 1× penicillin-streptomycin (Sigma-Aldrich, St. Louis, MO, https://www.sigmaaldrich.com), and 1× B27 supplement minus vitamin A (Gibco; Thermo Fisher Scientific, Waltham, MA, http://www.thermofisher.com). B27 comprises, among others, insulin and triodo-I-thyronine (T3). For subculturing, cells were detached by 3–5 minutes of incubation with Accutase (Millipore). After centrifugation, the supernatant was discarded, the pellet was resuspended in culture medium, and cells were plated on laminin-coated plastic ware (20 μg/ml laminin [Sigma-Aldrich] in DMEM/F12) at an appropriate density. Culture medium was changed every second day, and cells were passaged every 3–5 days. For immunofluorescent staining, cells were cultured on coverslips that had been precoated with poly-l-lysine (100 μg/ml in water; Sigma-Aldrich) for about 24 hours and then with laminin (20μg/ml in DMEM/F12) for another 24 hours. Except for the 21% oxygen experiments (5% CO2/95% air at 37°C with maximum humidity), cells were cultured in an atmosphere of 5% CO2/5% O2/90% N2 at 37°C with maximum humidity.
Spontaneous Differentiation of NPCs
NPCs were dissociated with Accutase and plated on coverslips at an initial cell density of 5 × 104 cells per well and cultured overnight in NPC medium. The following day, the NPC medium was replaced by the spontaneous differentiation medium (Neurobasal medium [Gibco; Thermo Fisher Scientific] supplemented with B-27 Supplement XenoFree CTS [Gibco; Thermo Fisher Scientific], 1× GlutaMAX-I [Gibco; Thermo Fisher Scientific], and 1× penicillin-streptomycin) and cultured further for a total of 14 days (a schematic description of the protocol is given in Fig. 1A). Medium was changed every second day.
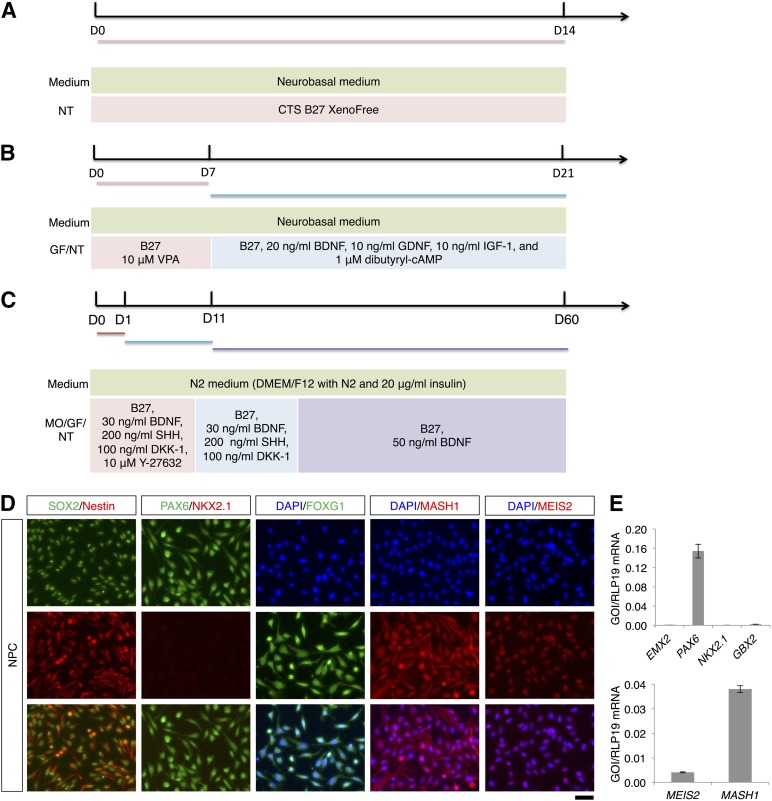
Figure 1.
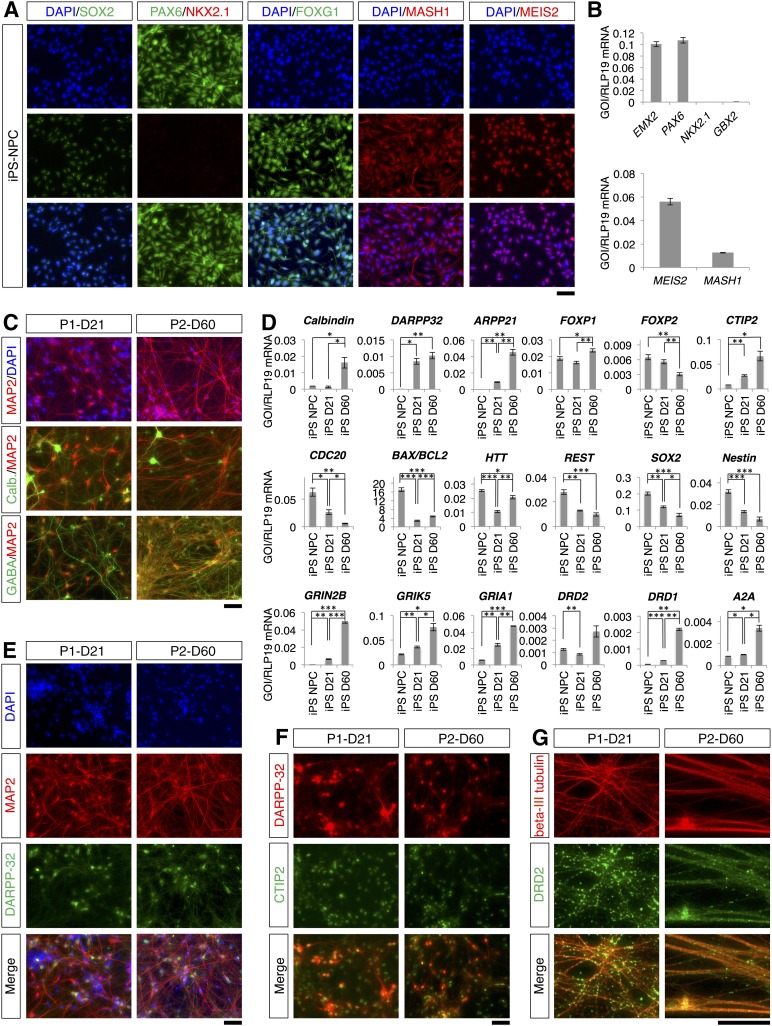
Differentiation of striatal projection neurons from NPCs. (A): Outline of the spontaneous differentiation protocol. (B): Outline of the two-step differentiation protocol. (C): Outline of the three-step differentiation protocol. (D): SOX2, Nestin, PAX6, NKX2.1, FOXG1, MASH1, and MEIS2 immunostaining of NPCs. The scale bar corresponds to 50 μm on the specimen level. (E): Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) analysis of EMX2, PAX6, NKX2.1, GBX2, MEIS2, and MASH1 expression in NPCs. For NKX2.1, no amplification was observed in the qRT-PCR analysis (no Ct value). Abbreviations: B-27, B-27 Supplement XenoFree CTS; D, day; DAPI, 4′,6-diamidino-2-phenylindole; DMEM/F12, Dulbecco’s modified Eagle’s medium nutrient mixture F12; GF, growth factors; IGF-1, insulin-like growth factor 1; MO, morphogens; NPC, neural progenitor cell; NT, neurotrophins; VPA, valproic acid.
Differentiation of Striatal Neurons From NPCs
Modified from previously described methods [17, 19], we used two different protocols to differentiate NPCs toward striatal projection neurons. To allow comparison of our results with these protocols, the duration of relevant differentiation steps were adapted from these protocols. A schematic representation is given in Figure 1B and 1C. NPCs maintained in DMEM/F12 with EGF and bFGF for 2 days served as control cells. Briefly, NPCs in the two-step protocol were initially dissociated with Accutase at 37°C for 5 minutes and then plated on day 0 onto (a) poly-L-lysine/laminin-coated coverslips placed in 24-well dishes (24-WD), (b) laminin-coated 96-WD, or (c) laminin-coated 24-WD. We seeded the NPCs at 5 × 104 cells per well in 24-WD or 7.5 × 103 cells per well in 96-WD. In the first step of the protocol, NPCs were cultured in Neurobasal medium (Gibco; Thermo Fisher Scientific) containing 10 μM VPA (Sigma-Aldrich), 1× B-27 supplement (Gibco; Thermo Fisher Scientific), 1× penicillin-streptomycin, and 1× GlutaMAX-I. Cells were cultured for 7 days in an atmosphere of 5% CO2/5% O2/90% N2 at 37°C with maximum humidity. The medium was changed every second day. In the second step of the protocol (i.e., days 7–21), the above media supplements were replaced with 20 ng/ml BDNF (Peprotech, Rocky Hill, NJ, https://www.peprotech.com), 10 ng/ml glial cell derived neurotrophic factor (GDNF; Peprotech), 10 ng/ml IGF-1 (R&D Systems, Minneapolis, MN, http://www.rndsystems.com), 1 μM dibutyryl-cAMP (Sigma-Aldrich), 1× B-27 supplement, 1× penicillin-streptomycin, and 1× GlutaMAX-I. The medium was replaced every second day. Differentiated cells were harvested on days 14 and 21 for immunostaining, quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR), and calcium imaging and on day 21 for the GABA release assay.
In the three-step protocol, NPCs were dissociated with Accutase at 37°C for 5 minutes and resuspended at final concentration in NBB30 medium. NBB30 medium consists of N2 medium (DMEM/F12 with l-glutamine containing 1× N2 supplement [Gibco; Thermo Fisher Scientific], 20 μg/ml insulin [Gibco; Thermo Fisher Scientific]) supplemented with 1× B-27 supplement, 30 ng/ml BDNF, 200 ng/ml murine SHH, N-terminus (Peprotech), 100 ng/ml DKK1 (Peprotech), and 1× penicillin-streptomycin. On the day of initial plating, NBB30 medium was supplemented with 10 μM Y-27632 (R&D Systems). NPCs were plated at either 5 × 104 cells/25 μl per well in 24-WD or 7.5 × 103 cells/5 μl per well in 96-WD. Cell suspension drops were spotted on plates and incubated in an atmosphere of 5% CO2/5% O2/90% N2 at 37°C with maximum humidity for 20 minutes. After 20 minutes, additional Y-27632-supplemented NBB30 medium was slowly added (225 μl per well for 24-WD and 35 μl per well for 96-WD). Following 24-hour incubation, additional NBB30 medium without Y-27632 was added to the wells (250 μl per well for 24-WD and 40 μl per well for 96-WD). From days 1–10, two-thirds of the NBB30 medium was replaced every 3 days. From days 11–60, NBB30 medium was replaced with NBB50 medium, which consists of DMEM/F12 with l-glutamine containing 1× N2 supplement, 20 μg/ml insulin, 1× B-27 supplement, 50 ng/ml BDNF, and 1× penicillin-streptomycin. Two-thirds of NBB50 medium was replaced every 3 days. Differentiated cells were harvested on days 30, 45, and 60 for immunostaining, qRT-PCR, and calcium imaging and on day 60 for the GABA release assay.
Culture and Differentiation of iPSC-Derived NPCs
NPCs derived from iPSCs (ax0015; Axol Biosciences, Cambridge, U.K., http://www.axolbio.com) were cultured in neural maintenance medium (Axol Biosciences) according to the manufacturer’s protocol. NPCs were cultured on coverslips or plastic culture ware that had been precoated with poly-l-ornithine (100 μg/ml in water; Sigma-Aldrich) for about 24 hours and then with laminin (20 μg/ml in DMEM/F12) for another 24 hours. The differentiation of the iPSC-derived NPCs was essentially performed as described above (see Fig. 1B for the two-step protocol, and Fig. 1C for the three-step protocol), except that we plated at 1 × 105 cells per well in 24-WD. For the spontaneous differentiation of iPSC-derived NPCs, we cultured the cells in DMEM/F12 with l-glutamine supplemented with 1× N2 supplement, 1× B-27 supplement with vitamin A, 20 μg/ml insulin, and 1× penicillin-streptomycin. Culture medium was changed every second day.
Immunofluorescence Analysis
Cells were fixed by immersion in 4% paraformaldehyde for 15 minutes at room temperature. Following fixation, cells were incubated overnight at 4°C with primary antibodies in blocking buffer (phosphate-buffered saline [PBS] containing 5% normal goat serum [Invitrogen; Thermo Fisher Scientific] and 0.3% Triton X-100 [Sigma-Aldrich]). After two washes with PBS, cells were incubated with secondary antibodies in blocking buffer for 2 hours at room temperature in the dark. Cells were washed four final times and then mounted on glass slides with Prolong Gold antifade reagent with DAPI (4′,6-diamidino-2-phenylindole, Invitrogen; Thermo Fisher Scientific). Images were acquired using a Nikon Eclipse Ti (Nikon, Tokyo, Japan, http://www.nikon.com) inverted microscope equipped with a Nikon Intensilight C-HGFI unit. Specimen areas were randomly selected by moving the motorized stage of the fluorescence microscope without using the fluorescence signal as a guide. At least five randomly selected areas were imaged, and cells were manually counted by visual evaluation of the images. All primary and secondary antibodies used are listed in supplemental online Table 1.
RNA Isolation and qRT-PCR Analysis
From cells cultured in a single well of a 24-WD, total RNA was isolated using the RNeasy Mini kit (Qiagen, Venlo, The Netherlands, https://www.qiagen.com) and quantified using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific). The Maxima H Minus First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) generated complementary DNA (cDNA) from 0.2 to 2 μg total RNA. We used 1 μl (5–10× diluted) of cDNA product as a qRT-PCR analysis template. Using a Stratagene Mx3005P qPCR system (Agilent Technologies, Santa Clara, CA, http://www.agilent.com) and the Maxima SYBR Green qPCR Master Mix (Thermo Fisher Scientific), we performed qRT-PCR. Three to six biological replicates were used for each group, and three technical replicates were used for each biological replicate. The ribosomal protein gene RPL19 was used for normalization. qRT-PCR primers were chosen from previous publications [19, 32, 33] or from PrimerBank (http://pga.mgh.harvard.edu/primerbank/) or were self-designed and ordered from Sigma-Aldrich. Before use, all primers were tested against a standard curve to check the efficiency of the PCR reaction. Primer sequences used for qRT-PCR data are given in supplemental online Table 2.
Calcium Imaging
Cells were loaded for calcium imaging by 1-hour immersion in a calcium indicator solution (5 μM acetoxymethyl ester form of calcium orange [Invitrogen; Thermo Fisher Scientific] dissolved in Tyrode’s solution). Following loading, cells were gently washed twice with Tyrode’s solution. After 30-minute incubation at 37°C, cells were transferred into a FlexStation3 (Molecular Devices, Sunnyvale, CA, http://www.moleculardevices.com) and intracellular calcium concentration was measured as intensity of emitted light at 576 nm (549 nm excitation wavelength). Values were recorded every 10 seconds. Twenty-five seconds into recording, cells were exposed to Tyrode’s solution containing 50 mM KCl (isosmotic substitution for NaCl), 300 μM GABA, 100 μM N-methyl-D-aspartate (NMDA), 100 μM kainate, and 100 μM l-glutamate/10 μM l-glycine. Each application was performed in quadruplicate, and average fluorescence was calculated.
Analysis of GABA Release
For the measurement of GABA release, the culture medium was removed, and the cells were washed four times with prewarmed Krebs-Ringer solution (20 mM HEPES, pH 7.4, 130 mM NaCl, 3 mM KCl, 2 mM CaCl2, 0.8 mM MgCl2, and 10 mM glucose). Subsequently, the cells were exposed to prewarmed Krebs-Ringer solution supplemented with 50 μM dopamine and 100 μM glutamate for 2 minutes, and the supernatant was collected and stored at −80°C on measurement. The concentration of GABA was determined using a GABA enzyme-linked immunosorbent assay kit, according to the manufacturer’s instructions (Shanghai BlueGene Biotech, Shanghai, People’s Republic of China, http://www.elisakit.cc). Three biological replicates were measured, and two technical replicates were used for each biological replicate. The GABA concentration was determined using a standard curve, according to the manufacturer’s instructions.
Statistical Analysis
NPC and differentiated cell mRNA levels were analyzed with the Student t test (two-sample unequal variance; two-tailed distribution). Similarly, immunofluorescence data in the two- and three-step protocols were analyzed with the Student t test. Data are presented as mean values ±SEM. The level of statistical significance was set at p < .05.
Results
Characterization and Spontaneous Differentiation of the NPCs
We used fetal NPCs that originally were immortalized as bulk NPC culture [34]. To assess the phenotype and the spontaneous differentiation potential of the NPCs, we tested for the expression of different markers by immunofluorescence and qRT-PCR. The NPCs coexpressed SOX2 and nestin in line with their neural identity (Fig. 1D). Moreover, the NPCs expressed PAX6 but were immunonegative for the most ventral marker NKX2.1 (Fig. 1D). NPCs were also immunopositive for FOXG1, MASH1, and MEIS2. We noticed that some of these markers were located in both the nucleus and the cytoplasm (Fig. 1D). These genes are expressed by lateral ganglionic eminescence progenitors, which can differentiate to striatal MSNs [18, 35]. Accordingly, we also confirmed by qRT-PCR that the NPCs express PAX6 but not the medial ganglionic eminence (MGE) marker NKX2.1 or the dorsal pallial marker EMX2 (Fig. 1E). Furthermore, the NPCs expressed very low levels of the caudal marker GBX2. NPCs also expressed MEIS2 and MASH1 (Fig. 1E), in line with the immunofluorescence analysis (Fig. 1D).
Culture of the NPCs in the absence of EGF and bFGF for 14 days (the differentiation protocol is outlined in Fig. 1A) showed that the NPCs could spontaneously differentiate toward neurons and glia (supplemental online Fig. 1A). The majority of the cells differentiated to GFAP-positive astrocytes, and only single cells differentiated toward O1-positive oligodendrocytes. We categorized the neuronal subtypes by immunofluorescence. On spontaneous differentiation of the NPCs, the major neuronal phenotypes are calretinin-positive, DARPP-32-positive, and parvalbumin-positive cells with a minor contribution of TH-positive cells (supplemental online Fig. 1B). DARPP-32-positive cells showed an immature phenotype in part, whereas ∼14% of the DARPP-32-positive cells showed a mature neuronal phenotype (supplemental online Fig. 1B). We concluded that either a longer differentiation period or exposure to additional differentiation factors might be needed for a full maturation of the DARPP-32-positive cells. We thus tested the two- and three-step differentiation protocols, which require differentiation over a period of 21 and 60 days, respectively.
Differentiation of Neurons From NPCs
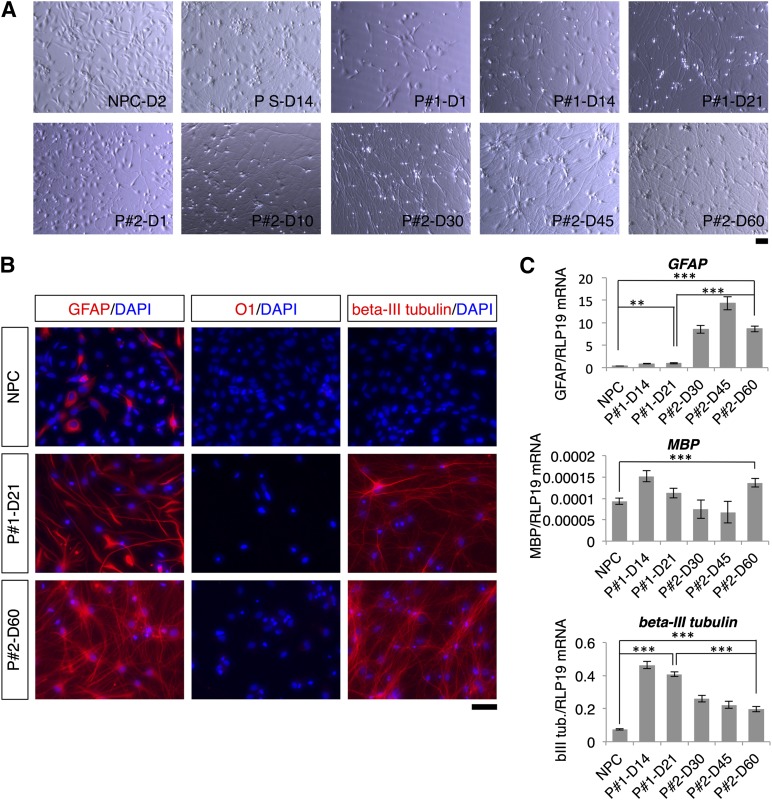
An overview of the two- and three-step differentiation protocols is given in Figure 1B and 1C. NPCs were differentiated under mild hypoxic conditions (i.e., 5% oxygen), which reduced the number of astrocytes in relation to neurons compared with NPCs differentiated at 21% oxygen (supplemental online Fig. 2). After 1 day of differentiation, cells cultured under the three-step protocol displayed somewhat longer neurites than cells cultured under the two-step protocol (Fig. 2A). Neurites had grown very long by the endpoint of differentiation in both protocols (days 21 and 60 for the two- and three-step protocols, respectively) (Fig. 2A). Immunostaining against the markers GFAP, O1, and β-III tubulin at these time points revealed that NPCs had differentiated into neurons and glial cells (Fig. 2B). Although the majority of cells in both protocols differentiated into neurons and astrocytes, only single cells differentiated into oligodendrocytes (<0.1% of the cells) (Fig. 2B). There were more GFAP-positive cells in the three-step protocol than in the two-step protocol. Moreover, qRT-PCR confirmed these results: compared with the two-step protocol, the three-step protocol resulted in significantly higher GFAP expression and significantly lower β-III tubulin expression (Fig. 2C).
Figure 2.
Morphological changes and postdifferentiation distribution of neurons, astrocytes, and oligodendrocytes produced by the two- and three-step protocols. (A): Morphological changes of cells during differentiation in the two- and three-step protocols and in the spontaneous differentiation protocol. NPCs used as starting cells in the two differentiation protocols are shown for comparison. (B): GFAP, O1, and β-III tubulin immunostaining at the beginning and the end of the differentiation protocols. (C): Quantitative reverse transcriptase-polymerase chain reaction analysis of GFAP, MBP, and β-III tubulin expression on different days of the differentiation protocols. Levels of statistical significance were set at ∗∗, p < .01 and ∗∗∗, p < .001. The scale bars correspond to 50 μm on the specimen level. Abbreviations: D, day; DAPI, 4′,6-diamidino-2-phenylindole; NPC, neural progenitor cell; P1, two-step protocol; P2, three-step protocol; PS, spontaneous differentiation protocol.
Throughout differentiation in both protocols, SOX2 and nestin expression were increased (supplemental online Fig. 3A). At later stages of differentiation, however, SOX2 and nestin expression started to decrease (days 14 and 45 in the two- and three-step protocols, respectively). Immunofluorescence analysis revealed that a number of MAP2-positive cells also expressed SOX2 (supplemental online Fig. 3B; spontaneously differentiated cells are described in supplemental online Fig. 1A). Notably, SOX2 nuclear staining was diffuse in the three-step protocol but more speckled in the two-step protocol. Moreover, immunofluorescence staining revealed nestin-positive cells throughout the differentiation process, and there were both β-III tubulin-positive and -negative cells that were immunopositive for nestin (supplemental online Fig. 3C; coimmunofluorescence staining of β-III tubulin and nestin of spontaneously differentiated cells are shown in supplemental online Fig. 1A). Overall, nestin-positive cells appeared to have more immature morphology with larger cell bodies and fewer processes.
Validation of Striatal Projection Neurons
Using immunofluorescence and qRT-PCR analyses, the differentiation process in both protocols was monitored at different time points: days 14 and 21 for the two-step protocol and days 30, 45, and 60 for the three-step protocol. At the early stages of differentiation in both protocols, MAP2-positive neurites were shorter and less mature (Fig. 3A); neurites matured over time. Compared with the two-step protocol, more elaborate dendrites were found after differentiation in the three-step protocol, and a dense network of dendrites was observed.
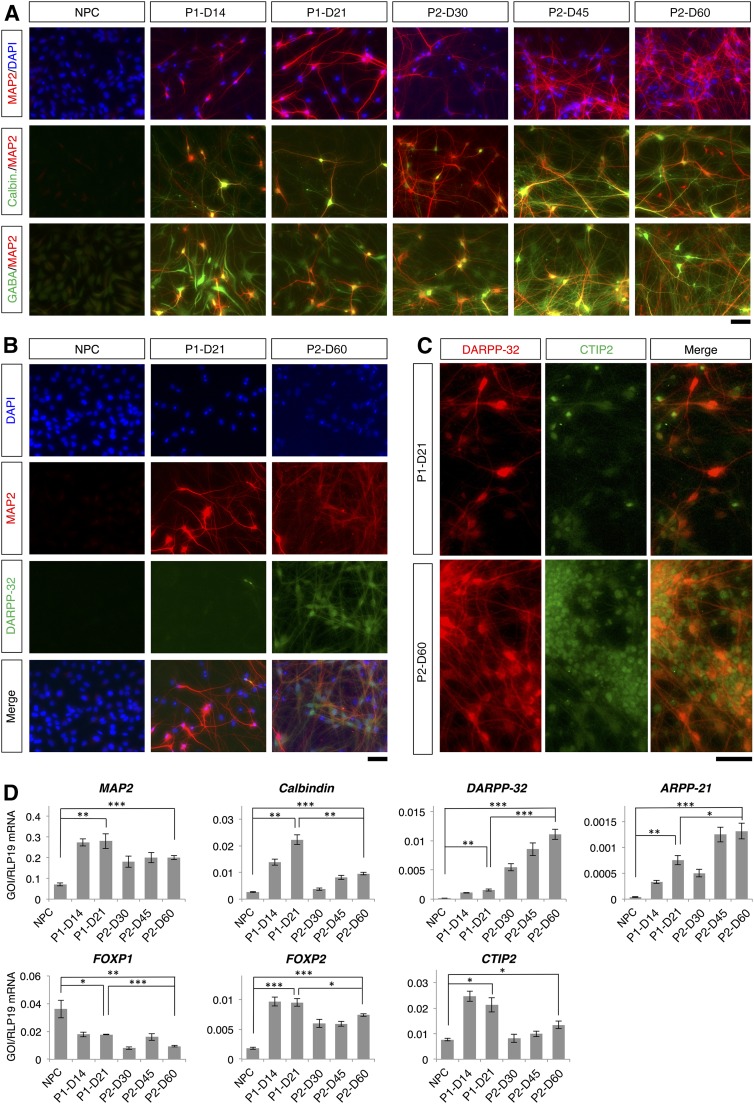
Figure 3.
Validation of striatal neurons. (A): Colocalization of MAP2 with calbindin and with GABA. (B): Colocalization of MAP2 and DARPP-32 at the beginning and the end of the differentiation protocols. (C): Colocalization of DARPP-32 and CTIP2. (D): Quantitative reverse transcriptase-polymerase chain reaction analysis of different MSN markers. Levels of statistical significance were set at ∗, p < .05, ∗∗, p < .01, and ∗∗∗, p < .001. The scale bars correspond to 50 μm on the specimen level. Abbreviations: D, day; DAPI, 4′,6-diamidino-2-phenylindole; NPC, neural progenitor cell; P1, two-step protocol; P2, three-step protocol.
At the end of differentiation, 68% ± 4% and 63% ± 4% of cells were MAP2-positive in the two- and three-step protocols, respectively (supplemental online Table 3). Compared with the two-step protocol, higher percentages of MAP2-positive cells in the three-step protocol were positive for calbindin, GABA, and DARPP-32. Representative immunofluorescence images are given in Figure 3A and 3B, and characterization of the neuronal subpopulations is shown in supplemental online Figure 4A and 4B for the two- and three-step differentiation protocols, respectively. In the two-step protocol, 54% ± 4% of MAP2-positive cells coexpressed calbindin, whereas 84% ± 5% of MAP2-positive cells in the three-step protocol coexpressed calbindin (n = 5; p < .01) (supplemental online Table 3). In the two- and three-step protocols, respectively, 90% ± 3% and 96% ± 1% of MAP2-positive cells were immunoreactive for GABA (n = 5); there was no significant difference between the two differentiation protocols (p > .05). A significantly greater percentage of MAP2-positive cells in the three-step protocol than in the two-step protocol were positive for DARPP-32 (21% ± 1% vs. 6% ± 2%, for the three- and two-step protocols, respectively; p < .001) (Fig. 3B). The vast majority of DARPP-32-positive cells coexpressed CTIP2 in both protocols (Fig. 3C).
In both protocols, qRT-PCR revealed a time-dependent increase in expression of the neuronal marker MAP2 and striatal MSN markers CALB1, DARPP-32, ARPP21, and CTIP2 (Fig. 3D). At the end of differentiation in the three-step protocol, differentiated cells displayed significantly greater DARPP-32 and ARPP21 expression than differentiated cells in the two-step protocol. At the end of differentiation in the two-step protocol, differentiated cells displayed significantly greater CALB1 expression than differentiated cells in the three-step protocol. Using qRT-PCR, expression of forkhead box P1 (FOXP1) and forkhead box P2 FOXP2 were tested as markers of different projection neuron subpopulations [36, 37]. Compared with control cells, both protocols resulted in a significant decrease in FOXP1 expression after differentiation, whereas FOXP2 expression was significantly increased after differentiation. Compared with the three-step protocol, significantly higher expression of FOXP2 was observed in the two-step protocol.
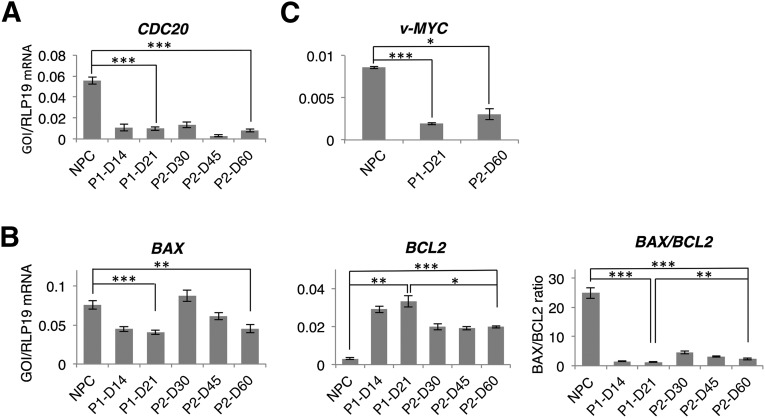
Development of Cell Proliferation and Apoptosis Markers
The expression of CDC20, a cell division cycle regulatory protein [38], was determined at different time points during differentiation to monitor cell proliferation. In both protocols, CDC20 expression was significantly decreased after induction of differentiation, even at early differentiation stages (Fig. 4A). At the endpoint of differentiation, cells produced by either protocol were not significantly different in CDC20 expression. At different time points during differentiation, we also analyzed apoptotic events by using qRT-PCR to determine quantities of both the proapoptotic gene BAX and the antiapoptotic gene BCL2 (Fig. 4B). Both protocols resulted in significantly decreased BAX expression and significantly increased BCL2 expression. Compared with the two-step protocol, however, the three-step protocol resulted in a significantly higher postdifferentiation BAX/BCL2 ratio. Furthermore, we observed a significant decrease of v-MYC expression at the end of both differentiation protocols, and there was no significant difference between the two protocols (Fig. 4C). v-MYC was used to immortalize the NPCs.
Figure 4.
Cell proliferation and apoptosis during differentiation in the two- and the three-step protocols. (A): Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) analysis of the cell proliferation marker CDC20. (B): qRT-PCR analysis of the apoptotic markers: BAX is proapoptotic and BCL2 is antiapoptotic. (C): qRT-PCR analysis of v-MYC. Levels of statistical significance were set at ∗, p < .05, ∗∗, p < .01, and ∗∗∗, p < .001. Abbreviations: D, day; NPC, neural progenitor cell; P1, two-step protocol; P2, three-step protocol.
HD-Related Gene Expression
Throughout the differentiation process, HTT and REST/NRSF gene expression was analyzed to determine the suitability of neurons differentiated with the two protocols for HD studies. Both protocols resulted in a significant increase in HTT gene expression (Fig. 5). At the end of differentiation, HTT gene expression was not significantly different between the two protocols. In contrast to HTT gene expression, REST/NRSF expression differed between the two protocols. Whereas REST/NRSF expression after differentiation was unaltered in the three-step protocol, the two-step protocol resulted in a significantly decreased REST/NRSF expression relative to NPCs.
Figure 5.
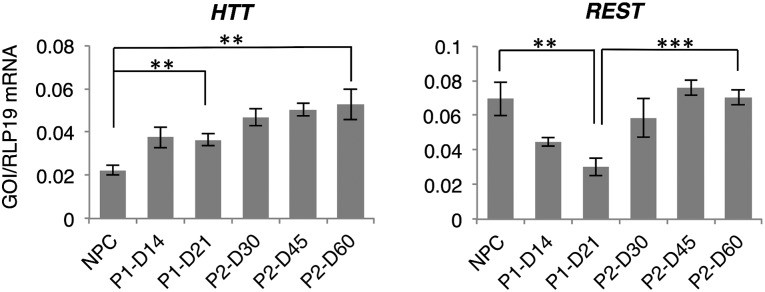
Gene expression of Huntington’s disease-related genes. Quantitative reverse transcriptase-polymerase chain reaction analysis was used to measure expression of HTT and REST/NRSF during differentiation in the two- and three-step protocols. Levels of statistical significance were set at ∗∗, p < .01 and ∗∗∗, p < .001. Abbreviations: D, day; NPC, neural progenitor cell; P1, two-step protocol; P2, three-step protocol.
Calcium Response of Differentiated Cells After Stimulation
Differentiated cell function was determined based on stimulation-induced calcium response. In both protocols, extracellular application of KCl, GABA, l-glutamate/l-glycine, NMDA, or kainate increased intracellular calcium levels (Fig. 6A). In addition, we used qRT-PCR in differentiated cells to validate gene expression of ionotropic glutamate receptor subtypes (i.e., GRIN2B, GRIK5, and GRIA1). Both protocols resulted in significantly increased expression of all three glutamate receptor genes (Fig. 6B). At the end of differentiation, the two protocols did not produce significant differences in GRIN2B expression; however, GRIK5 and GRIA1 expression were significantly higher in the three-step protocol than in the two-step protocol.
Figure 6.
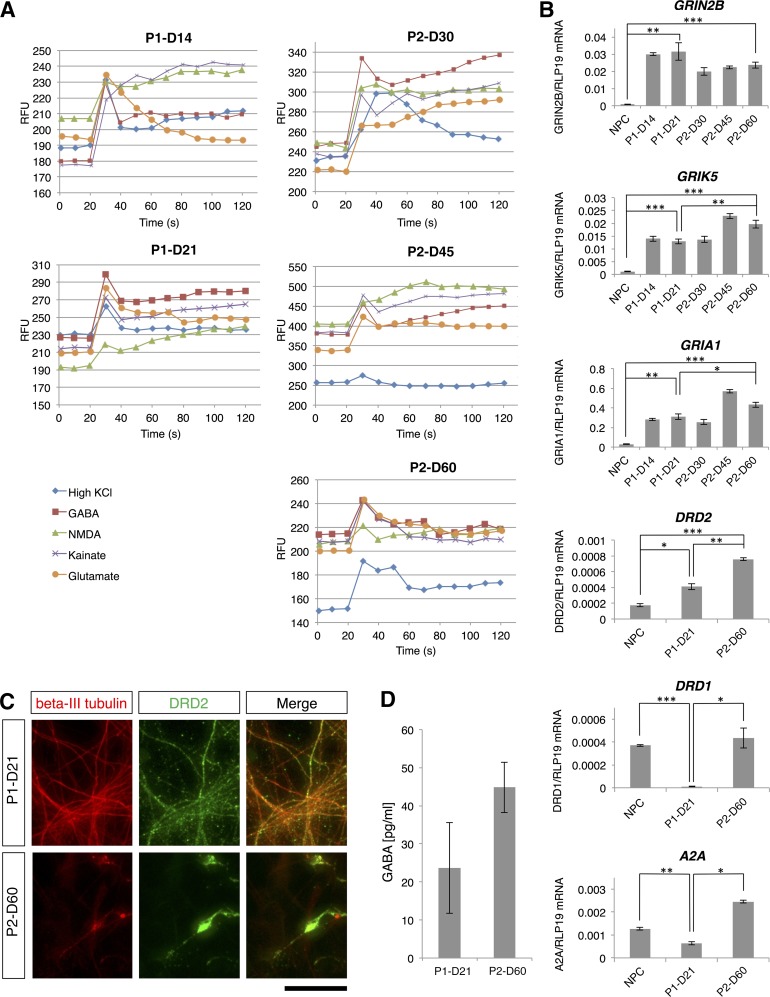
Differentiated cell functional characterization. (A): Intracellular calcium measurement in cells differentiated according to the two-step (left) and three-step (right) protocols. In both protocols, intracellular calcium was measured following extracellular application of 50 mM KCl (isosmotic substitution for NaCl), 300 μM GABA, 100 μM NMDA, 100 μM kainate, and 100 μM l-glutamate/10 μM l-glycine. The days of the protocols when measurements were performed are indicated above each graph. (B): Quantitative reverse transcriptase-polymerase chain reaction analysis of expression of three ionotropic glutamate receptor subtypes, GRIN2B, GRIK5 and GRIA1, the dopamine receptors DRD1 and DRD2, and the adenosine receptor A2A. Levels of statistical significance were set at ∗, p < .05, ∗∗, p < .01, and ∗∗∗, p < .001. (C): Colocalization of β-III tubulin and DRD2. The scale bar corresponds to 50 μm on the specimen level. (D) GABA release assay on stimulation of the differentiated cells. Abbreviations: D, day; NPC, neural progenitor cell; P1, two-step protocol; P2, three-step protocol; RFU, relative fluorescence units; s, seconds.
Expression of Dopamine and Adenosine Receptors and GABA Release
Striatal medium spiny neurons differ in the expression of dopamine and adenosine receptors and thus can be classified into two different subpopulations: striatonigral neurons predominantly express dopamine receptor D1 (DRD1), whereas striatopallidal neurons predominantly express dopamine receptor D2 (DRD2) [39, 40]. We confirmed by immunofluorescence that the differentiated neurons expressed DRD2 in both differentiation protocols (Fig. 6C), and these results were corroborated by qRT-PCR (Fig. 6B). Both protocols resulted in a significant increase in gene expression of DRD2, and expression was significantly higher in the in the three-step protocol than in the two-step protocol. DRD1 was expressed in the samples of the three-step differentiation protocol but was significantly reduced in the samples of the two-step protocol (Fig. 6B). Moreover, in both protocols, expression of the adenosine A2A receptor ADORA2A (A2A) was found, but its expression was significantly higher in the three-step protocol than in the two-step protocol (Fig. 6B). Furthermore, we tested whether the differentiated cells released GABA on stimulation with dopamine and glutamate. Neurons differentiated in both protocols released GABA (Fig. 6D), and there was a trend toward increased GABA release in the three-step protocol (p = .07).
Differentiation of iPSC-Derived NPCs
Finally, we used the two- and three-step differentiation protocols to induce differentiation of iPSC-derived NPCs toward striatal neurons. The iPSC-derived NPCs were immunopositive for the same markers as the fetal NPCs (Fig. 7A). In particular, the SOX2-positive NPCs were immunonegative for NKX2.1 but expressed PAX6. Moreover, the NPCs also expressed FOXG1, MASH1, and MEIS2. Of note, some of these transcription factors were predominantly located in the cytoplasm as observed for the fetal NPCs (compare Fig. 1D, 7A). Quantitative RT-PCR corroborated the expression of the markers (Fig. 7B). The iPSC-derived NPCs expressed PAX6, MEIS2, and MASH1 but did not express either NKX2.1 or GBX2. In contrast to the fetal NPCs, the iPSC-derived NPCs expressed significant levels of the dorsal pallial marker EMX2 (p < .001). On spontaneous differentiation of the NPCs in the absence of EGF and bFGF for 14 days, some DARPP-32-positive neurons were observed, and DARPP-32-positive neurons coexpressed CTIP2 (supplemental online Fig. 5). This observation suggested that the iPSC-derived NPCs can differentiate toward neurons coexpressing DARPP-32 and CTIP2 on spontaneous differentiation.
Figure 7.
Differentiation of striatal projection neurons from iPS-derived NPCs. (A): iPS-derived NPCs immunostained for SOX2, PAX6, NKX2.1, FOXG1, MASH1, and MEIS2. (B): Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) analysis of EMX2, PAX6, NKX2.1, GBX2, MEIS2, and MASH1 expression. (C): Colocalization of MAP2 with calbindin and with GABA at the endpoint of differentiation. (D): qRT-PCR analysis of marker expression in iPS-derived NPCs and differentiated cells. Levels of statistical significance were set at ∗, p < .05, ∗∗, p < .01, and ∗∗∗, p < .001. (E): Colocalization of MAP2 and DARPP-32 at the end of the differentiation protocols. (F): Colocalization of DARPP-32 and CTIP2 at the end of the differentiation protocols. (G): Colocalization of β-III tubulin and DRD2. The scale bars correspond to 50 μm on the specimen level. Abbreviations: D, day; DAPI, 4′,6-diamidino-2-phenylindole; IPS, induced pluripotent stem cell; NPC, neural progenitor cell; P1, two-step protocol; P2, three-step protocol.
We differentiated the iPSC-derived NPCs according to the two- and three-step differentiation protocols. At the end of two- and three-step differentiation protocols, we found both GFAP-positive astrocytes and β-III tubulin-positive neurons, which formed a dense network (supplemental online Fig. 6). Accordingly, MAP2-positive neurons were observed by immunofluorescence staining (Fig. 7C). In the two-step protocol, 60% ± 2% and 77% ± 4% of the MAP2-positive cells were immunopositive for calbindin and GABA, respectively, whereas in the three-step protocol, ∼64% ± 4% and 96% ±3% of the MAP2-positive cells coexpressed calbindin and GABA, respectively (Fig. 7C; supplemental online Table 4). Immunopositivity for GABA was significantly greater in the three-step differentiation protocol (p < .001).
In both protocols, differentiated cells showed an increase of the MSN markers. At the end of the two- and three-step differentiation protocol, qRT-PCR revealed a significant increase in the expression of DARPP-32, ARPP21 and CTIP2 (Fig. 7D). ARPP21 expression was significantly higher in the three-step protocol than in the two-step protocol. We found a significant increase of the CALB1 and FOXP1 expression in the three-step protocol. Expression of FOXP2 did not change significantly in the two-step protocol but decreased significantly in the three-step protocol. At the end of both differentiation protocols, the expression of the proliferation marker CDC20 decreased significantly, and the BAX/BCL2 ratio also decreased concurrently and significantly. Likewise, the expression of HTT and REST decreased significantly at the end of the protocols, as did the expression of SOX2 and nestin. In line with the qRT-PCR results, a significantly greater percentage of MAP2-positive cells were immunopositive for DARPP-32 in the three-step protocol: ∼51% ± 7% and 86% ± 3% of the MAP2-positive cells coexpressed DARPP-32 in the two- and three-step differentiation protocols, respectively (p < .001) (Fig. 7E; supplemental online Table 4). Basically, all DARPP-32-positive cells coexpressed CTIP2 (Fig. 7F).
The expression of glutamate, dopamine, and adenosine receptors typical for striatal projection neurons was confirmed by qRT-PCR (Fig. 7D). We found a significant increase in the expression of GRIN2B, GRIK5, and GRIA1 in both protocols, and the expression of all three genes was significantly higher in the three-step protocol than in the two-step protocol. In both protocols, the cells also expressed the DRD1, DRD2, and A2A receptors. Compared with the two-step protocol, significantly higher expression of DRD1 and A2A was observed in the three-step protocol. There was no significant change in the expression of DRD2 in the three-step protocol. By using immunofluorescence, we confirmed that β-III tubulin-positive neurons were immunopositive for the DRD2 receptor (Fig. 7G).
Discussion
In this study, we established two protocols for the differentiation of NPCs toward MSNs: a two-step 21-day protocol and a three-step 60-day protocol. Both protocols were performed under hypoxic conditions (5% O2). We confirmed that differentiated cells expressed the general neuronal markers β-III tubulin and MAP2 and the striatal markers DARPP-32, ARPP21, and CTIP2. The differentiated cells also expressed functional glutamate and GABA receptors in addition to dopamine receptors. On stimulation, the differentiated cells released GABA. Finally, we provided insight into the expression of HD-related genes and followed the expression of traditional NSC markers.
Precise regulation is required for extracellular signal-induced differentiation of NPCs to striatal projection neurons. Both of our differentiation protocols successfully produced DARPP-32-positive striatal neurons from NPCs. In this paper, we described important differences between the two protocols including basic media, morphogens, growth factors, neurotrophins, and differentiation time; these protocol alterations resulted in different differentiation efficiencies. The only growth factor common to both protocols was BDNF. Along with important striatal neuronal maintenance functions, higher BDNF concentrations in the three-step protocol may have contributed to this protocol’s improved specific differentiation efficiency.
In the two-step protocol, we used VPA in the first step. During the second step, VPA was replaced with BDNF, GDNF, IGF-1, and dibutyryl-cAMP. The hormones insulin and T3, which are known to support neuronal differentiation [23, 41], also were included in the media supplement. Combined, these aforementioned factors promoted differentiation toward a more general neuronal phenotype. For neurogenesis in the striatum, several genes that encode morphogens (e.g., WNT and SHH) are involved in neuronal differentiation. In contrast to the factors used in the two-step protocol that promoted general neuronal differentiation, the three-step protocol used a more specific differentiation strategy. Specifically, the three-step differentiation protocol was based on a combination of SHH and DKK-1 treatment; simultaneous SHH and DKK-1 exposure has been suggested as a trigger for specific differentiation toward ventral telencephalic progenitors [25].
The differentiation protocols used in our study were modified from two previous reports: the two-step protocol was based on work by Ma et al. [17] and the three-step protocol was based on a study by Delli Carri et al. [19]; however, we modified these protocols to optimize NPC differentiation. In contrast to Ma et al. [17], for example, we found that omitting the B-27 supplement and l-glutamine in the two-step protocol resulted in severe cell death at very early stages of differentiation; therefore, we included these two supplements in our media. In contrast to Delli Carri et al. [19], both of our protocols used hypoxic culturing conditions throughout differentiation. Hypoxia has been shown to support both ESC and NSC proliferation [42] and to improve neuronal differentiation [43].
Immunofluorescence and qRT-PCR analysis demonstrated that neurons differentiated according to both of our protocols expressed striatal markers. As a major marker of MSNs [44], Darpp-32 is expressed in the vast majority of striatal MSNs [45]. Darpp-32 is a phosphorylation-dependent inhibitor of protein phosphatase-1 [46]. Whereas dopamine binding to dopamine D1-like receptors results in Darpp-32 phosphorylation and activation, glutamate binding to NMDA receptors dephosphorylates and inactivates Darpp-32 [47]. DARPP-32 has thus been suggested as an integrator of neurotransmission [48]. ARPP21 is another cyclic AMP-regulated phosphoprotein in MSNs. Similar to Darpp-32, Arpp21 is dopamine regulated [49] and highly expressed in the striatum [50]. Consequently, we selected these two genes as valid markers of striatal projection neurons. Importantly, our three-step protocol significantly increased expression of both genes.
The FOXP1 and FOXP2 genes encode transcription factors important for brain development; however, these genes are also expressed in the adult brain [51–53]. Although the exact function of FOXP1 is unclear, FOXP2 contributes to plasticity and neurite outgrowth during development [54, 55]. Foxp1 and Foxp2 are expressed in subsets of neurons in various CNS regions [36, 37, 51]. In mouse cerebral cortex, for example, Foxp1 is expressed in neocortical neurons from layers III to VIa, whereas Foxp2 is expressed in Darpp-32-positive projection neurons from layer VI [37]. In mouse striatum, ∼65% of Map2-positive neurons and 72% of Darpp-32-positive neurons express Foxp1 [51]. Because interneurons are Foxp1 negative, Foxp1 expression appears to be restricted to projection neurons [51]. In both rodents and primates, FOXP2 is expressed in the developing and postnatal striatum [52, 56]. Acting via the striatal Shh pathway, recent studies indicated that Foxp2 supports MSN development [57]. Together, these findings suggest that FOXP1 and FOXP2 might distinguish subpopulations of striatal neurons. Accordingly, we suggest that the FOXP1 and FOXP2 expression profiles observed in our study reflect preferential differentiation toward one subpopulation of striatal neuron subpopulations.
In both the developing and adult striatum, the transcription factor Ctip2 is highly expressed and has been shown to be essential for MSN full differentiation [58]. Within the brain, CTIP2 has been described as an integral part of the ATP-dependent chromatin remodeling complex SWI/SNF [59]. CTIP2-induced gene regulation contributes to the specific gene expression profiles of striatal MSNs [60]. Consequently, the increased CTIP2 expression observed in differentiated cells in our study supports the identification of these cells as MSNs.
In addition to confirming that our differentiated cells expressed striatal markers, we tested whether the differentiated cells would respond to ionotropic glutamate receptor stimulation. Ionotropic glutamate receptor stimulation results in cellular calcium influx and increases intracellular calcium concentrations [61]. The striatum contains increased levels of different ionotropic glutamate receptor subtypes. The NMDA receptor subunits NR1/GRIN1 and NR2B/GRIN2B, for example, are highly expressed in all striatal neuronal subtypes; however, striatal projection neurons have lower NR2A/GRIN2A expression [62, 63]. Striatal MSNs also express the AMPA receptor subunits GluR1/GRIA1 and GluR2/GRIA2 [64] and the kainate receptor subunits GluR6/GRIK2 and KA2/GRIK5 [65, 66].
To characterize differentiated neuronal function in our study, we tested the calcium response to various agonists. In neurons differentiated according to either protocol, treatment with extracellular KCl, NMDA, kainate, l-glutamate/l-glycine, or GABA elevated intracellular calcium levels. This functional characterization was confirmed by expression analysis. Differentiated cells expressed subunits of all three types of ionotropic glutamate receptors: the NMDA receptor GRIN2B, the AMPA receptor GRIA1, and the kainate receptor GRIK5. Although there was not a difference between the two protocols in terms of GRIN2B expression, the three-step protocol resulted in higher expression of GRIK5 and GRIA1.
Differentiated cells also expressed dopamine and adenosine receptors and released GABA on stimulation. There are two subpopulations of MSNs in the striatum, and they differ by their expression of receptors: striatonigral neurons primarily express the dopamine receptor subtype Drd1, and striatopallidal neurons primarily express the dopamine receptor subtype Drd2 [39, 40]. The vast majority of the latter subpopulation coexpresses the adenosine receptor A2a, which is typically not expressed by the former subpopulation [40]. Of note, both differentiation protocols used in this study resulted in differentiated cells expressing DRD2 and A2A receptors, whereas expression of DRD1 decreased on differentiation of the fetal NPCs using the two-step protocol. In contrast, the iPSC-derived NPCs expressed significant levels of DRD1 in both differentiation protocols.
Limited self-renewal capacity and the tendency toward acquisition of karyotypic aberrations are regarded as potential limitations of nonimmortalized NPCs for basic science and translational medicine studies [67, 68]. In the present study, we used NPCs that had been immortalized by transduction with v-MYC [34]. Consequently, the NPCs can be propagated efficiently without visible karyotypic instability [34]. MYC was originally described as a proto-oncogene, and a number of malignancies overexpress MYC [69]; however, MYC overexpression alone does not appear to be sufficient to induce tumorigenic transformation in normal human cells [69]. More recently, increasing evidence suggests that MYC is a stemness gene [70]. Taken together, the neoplastic potential of MYC-immortalized NPCs is suggested to be rather low. Further support for this conclusion comes from transplantation experiments in which the expression of v-MYC in immortalized human NSCs was shown to be spontaneously downregulated within the first days following engraftment [71]. Along these lines, we observed a significant decrease in the expression of v-MYC at the end of the neuronal differentiation process.
Conclusion
Using a multistep differentiation protocol, previous studies have started striatal neuronal differentiation using ESCs and iPSCs [16–19]. The stepwise differentiation process of ESCs and iPSCs to striatal neurons can take up to 80 days. This study reports the successful differentiation of fetal and iPSC-derived NPCs into striatal projection neurons that express MSN markers and functional glutamate receptors. Our three-step protocol provides a simple and reliable method to differentiate striatal projection neurons for use in striatal neuron physiology and neurodegenerative disease studies.
Supplementary Material
Acknowledgments
We thank Dr. Tue Banke for access to experimental facilities and helpful discussions and Dr. Jens Nyengaard for general support. We thank Susanne Stubbe for expert technical assistance. We also acknowledge the use of experimental facilities at the DNC House. This study was supported by the Sapere Aude Program of the Danish Council for Independent Research and the Lundbeck Foundation’s Fellowship Program to M.M.G. The Center for Stochastic Geometry and Advanced Bioimaging is supported by the Villum Foundation.
Author Contributions
L.L. and B.S.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; J.Y.: collection and/or assembly of data, data analysis and interpretation, final approval of manuscript; M.M.G.: conception and design, financial support, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicated no potential conflicts of interest.
References
- 1.A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Walker FO. Huntington’s disease. Lancet. 2007;369:218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- 3.Graveland GA, Williams RS, DiFiglia M. Evidence for degenerative and regenerative changes in neostriatal spiny neurons in Huntington’s disease. Science. 1985;227:770–773. doi: 10.1126/science.3155875. [DOI] [PubMed] [Google Scholar]
- 4.Graveland GA, DiFiglia M. The frequency and distribution of medium-sized neurons with indented nuclei in the primate and rodent neostriatum. Brain Res. 1985;327:307–311. doi: 10.1016/0006-8993(85)91524-0. [DOI] [PubMed] [Google Scholar]
- 5.Labbadia J, Morimoto RI. Huntington’s disease: Underlying molecular mechanisms and emerging concepts. Trends Biochem Sci. 2013;38:378–385. doi: 10.1016/j.tibs.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiFiglia M, Sapp E, Chase KO, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 7.Bennett EJ, Shaler TA, Woodman B, et al. Global changes to the ubiquitin system in Huntington’s disease. Nature. 2007;448:704–708. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- 8.Panov AV, Gutekunst CA, Leavitt BR, et al. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 9.Taylor-Robinson SD, Weeks RA, Sargentoni J, et al. Evidence for glutamate excitotoxicity in Huntington’s disease with proton magnetic resonance spectroscopy. Lancet. 1994;343:1170. doi: 10.1016/s0140-6736(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 10.Zuccato C, Ciammola A, Rigamonti D, et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington’s disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 11.Zuccato C, Tartari M, Crotti A, et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- 12.Shimojo M. Huntingtin regulates RE1-silencing transcription factor/neuron-restrictive silencer factor (REST/NRSF) nuclear trafficking indirectly through a complex with REST/NRSF-interacting LIM domain protein (RILP) and dynactin p150 Glued. J Biol Chem. 2008;283:34880–34886. doi: 10.1074/jbc.M804183200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Filippis L, Binda E. Concise review: Self-renewal in the central nervous system: Neural stem cells from embryo to adult. Stem Cells Translational Medicine. 2012;1:298–308. doi: 10.5966/sctm.2011-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puri MC, Nagy A. Concise review: Embryonic stem cells versus induced pluripotent stem cells: The game is on. Stem Cells. 2012;30:10–14. doi: 10.1002/stem.788. [DOI] [PubMed] [Google Scholar]
- 15.Bilic J, Izpisua Belmonte JC. Concise review: Induced pluripotent stem cells versus embryonic stem cells: Close enough or yet too far apart? Stem Cells. 2012;30:33–41. doi: 10.1002/stem.700. [DOI] [PubMed] [Google Scholar]
- 16.Aubry L, Bugi A, Lefort N, et al. Striatal progenitors derived from human ES cells mature into DARPP32 neurons in vitro and in quinolinic acid-lesioned rats. Proc Natl Acad Sci USA. 2008;105:16707–16712. doi: 10.1073/pnas.0808488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma L, Hu B, Liu Y, et al. Human embryonic stem cell-derived GABA neurons correct locomotion deficits in quinolinic acid-lesioned mice. Cell Stem Cell. 2012;10:455–464. doi: 10.1016/j.stem.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delli Carri A, Onorati M, Lelos MJ, et al. Developmentally coordinated extrinsic signals drive human pluripotent stem cell differentiation toward authentic DARPP-32+ medium-sized spiny neurons. Development. 2013;140:301–312. doi: 10.1242/dev.084608. [DOI] [PubMed] [Google Scholar]
- 19.Delli Carri A, Onorati M, Castiglioni V, et al. Human pluripotent stem cell differentiation into authentic striatal projection neurons. Stem Cell Rev. 2013;9:461–474. doi: 10.1007/s12015-013-9441-8. [DOI] [PubMed] [Google Scholar]
- 20.Yu IT, Park JY, Kim SH, et al. Valproic acid promotes neuronal differentiation by induction of proneural factors in association with H4 acetylation. Neuropharmacology. 2009;56:473–480. doi: 10.1016/j.neuropharm.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 21.Stachowiak EK, Fang X, Myers J, et al. cAMP-induced differentiation of human neuronal progenitor cells is mediated by nuclear fibroblast growth factor receptor-1 (FGFR1) J Neurochem. 2003;84:1296–1312. doi: 10.1046/j.1471-4159.2003.01624.x. [DOI] [PubMed] [Google Scholar]
- 22.Brooker GJ, Kalloniatis M, Russo VC, et al. Endogenous IGF-1 regulates the neuronal differentiation of adult stem cells. J Neurosci Res. 2000;59:332–341. doi: 10.1002/(sici)1097-4547(20000201)59:3<332::aid-jnr6>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 23.Arsenijevic Y, Weiss S. Insulin-like growth factor-I is a differentiation factor for postmitotic CNS stem cell-derived neuronal precursors: Distinct actions from those of brain-derived neurotrophic factor. J Neurosci. 1998;18:2118–2128. doi: 10.1523/JNEUROSCI.18-06-02118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu H, Yu SP, Gutekunst CA, et al. Inhibition of the Rho signaling pathway improves neurite outgrowth and neuronal differentiation of mouse neural stem cells. Int J Physiol Pathophysiol Pharmacol. 2013;5:11–20. [PMC free article] [PubMed] [Google Scholar]
- 25.Li XJ, Zhang X, Johnson MA, et al. Coordination of sonic hedgehog and Wnt signaling determines ventral and dorsal telencephalic neuron types from human embryonic stem cells. Development. 2009;136:4055–4063. doi: 10.1242/dev.036624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ericson J, Muhr J, Placzek M, et al. Sonic hedgehog induces the differentiation of ventral forebrain neurons: A common signal for ventral patterning within the neural tube. Cell. 1995;81:747–756. doi: 10.1016/0092-8674(95)90536-7. [DOI] [PubMed] [Google Scholar]
- 27.Krauss S, Concordet JP, Ingham PW. A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell. 1993;75:1431–1444. doi: 10.1016/0092-8674(93)90628-4. [DOI] [PubMed] [Google Scholar]
- 28.Echelard Y, Epstein DJ, St-Jacques B, et al. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75:1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- 29.Chamberlain CE, Jeong J, Guo C, et al. Notochord-derived Shh concentrates in close association with the apically positioned basal body in neural target cells and forms a dynamic gradient during neural patterning. Development. 2008;135:1097–1106. doi: 10.1242/dev.013086. [DOI] [PubMed] [Google Scholar]
- 30.Bafico A, Liu G, Yaniv A, et al. Novel mechanism of Wnt signalling inhibition mediated by Dickkopf-1 interaction with LRP6/Arrow. Nat Cell Biol. 2001;3:683–686. doi: 10.1038/35083081. [DOI] [PubMed] [Google Scholar]
- 31.El-Akabawy G, Medina LM, Jeffries A, et al. Purmorphamine increases DARPP-32 differentiation in human striatal neural stem cells through the Hedgehog pathway. Stem Cells Dev. 2011;20:1873–1887. doi: 10.1089/scd.2010.0282. [DOI] [PubMed] [Google Scholar]
- 32.Kang J, Perry JK, Pandey V, et al. Artemin is oncogenic for human mammary carcinoma cells. Oncogene. 2009;28:2034–2045. doi: 10.1038/onc.2009.66. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Huang CT, Chen J, et al. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell. 2010;7:90–100. doi: 10.1016/j.stem.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donato R, Miljan EA, Hines SJ, et al. Differential development of neuronal physiological responsiveness in two human neural stem cell lines. BMC Neurosci. 2007;8:36. doi: 10.1186/1471-2202-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toresson H, Mata de Urquiza A, Fagerström C, et al. Retinoids are produced by glia in the lateral ganglionic eminence and regulate striatal neuron differentiation. Development. 1999;126:1317–1326. doi: 10.1242/dev.126.6.1317. [DOI] [PubMed] [Google Scholar]
- 36.Ferland RJ, Cherry TJ, Preware PO, et al. Characterization of Foxp2 and Foxp1 mRNA and protein in the developing and mature brain. J Comp Neurol. 2003;460:266–279. doi: 10.1002/cne.10654. [DOI] [PubMed] [Google Scholar]
- 37.Hisaoka T, Nakamura Y, Senba E, et al. The forkhead transcription factors, Foxp1 and Foxp2, identify different subpopulations of projection neurons in the mouse cerebral cortex. Neuroscience. 2010;166:551–563. doi: 10.1016/j.neuroscience.2009.12.055. [DOI] [PubMed] [Google Scholar]
- 38.Gieffers C, Peters BH, Kramer ER, et al. Expression of the CDH1-associated form of the anaphase-promoting complex in postmitotic neurons. Proc Natl Acad Sci USA. 1999;96:11317–11322. doi: 10.1073/pnas.96.20.11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerfen CR, Engber TM, Mahan LC, et al. D1 and D2 dopamine receptor-regulated gene expression of striatonigral and striatopallidal neurons. Science. 1990;250:1429–1432. doi: 10.1126/science.2147780. [DOI] [PubMed] [Google Scholar]
- 40.Oude Ophuis RJ, Boender AJ, van Rozen AJ, et al. Cannabinoid, melanocortin and opioid receptor expression on DRD1 and DRD2 subpopulations in rat striatum. Front Neuroanat. 2014;8:14. doi: 10.3389/fnana.2014.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen C, Zhou Z, Zhong M, et al. Thyroid hormone promotes neuronal differentiation of embryonic neural stem cells by inhibiting STAT3 signaling through TRα1. Stem Cells Dev. 2012;21:2667–2681. doi: 10.1089/scd.2012.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santilli G, Lamorte G, Carlessi L, et al. Mild hypoxia enhances proliferation and multipotency of human neural stem cells. PLoS One. 2010;5:e8575. doi: 10.1371/journal.pone.0008575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Studer L, Csete M, Lee SH, et al. Enhanced proliferation, survival, and dopaminergic differentiation of CNS precursors in lowered oxygen. J Neurosci. 2000;20:7377–7383. doi: 10.1523/JNEUROSCI.20-19-07377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walaas SI, Aswad DW, Greengard P. A dopamine- and cyclic AMP-regulated phosphoprotein enriched in dopamine-innervated brain regions. Nature. 1983;301:69–71. doi: 10.1038/301069a0. [DOI] [PubMed] [Google Scholar]
- 45.Ouimet CC, Langley-Gullion KC, Greengard P. Quantitative immunocytochemistry of DARPP-32-expressing neurons in the rat caudatoputamen. Brain Res. 1998;808:8–12. doi: 10.1016/s0006-8993(98)00724-0. [DOI] [PubMed] [Google Scholar]
- 46.Hemmings HC, Jr, Greengard P, Tung HY, et al. DARPP-32, a dopamine-regulated neuronal phosphoprotein, is a potent inhibitor of protein phosphatase-1. Nature. 1984;310:503–505. doi: 10.1038/310503a0. [DOI] [PubMed] [Google Scholar]
- 47.Halpain S, Girault JA, Greengard P. Activation of NMDA receptors induces dephosphorylation of DARPP-32 in rat striatal slices. Nature. 1990;343:369–372. doi: 10.1038/343369a0. [DOI] [PubMed] [Google Scholar]
- 48.Svenningsson P, Nishi A, Fisone G, et al. DARPP-32: An integrator of neurotransmission. Annu Rev Pharmacol Toxicol. 2004;44:269–296. doi: 10.1146/annurev.pharmtox.44.101802.121415. [DOI] [PubMed] [Google Scholar]
- 49.Tsou K, Girault JA, Greengard P. Dopamine D1 agonist SKF 38393 increases the state of phosphorylation of ARPP-21 in substantia nigra. J Neurochem. 1993;60:1043–1046. doi: 10.1111/j.1471-4159.1993.tb03252.x. [DOI] [PubMed] [Google Scholar]
- 50.Ouimet CC, Hemmings HC, Jr, Greengard P. ARPP-21, a cyclic AMP-regulated phosphoprotein enriched in dopamine-innervated brain regions. II. Immunocytochemical localization in rat brain. J Neurosci. 1989;9:865–875. doi: 10.1523/JNEUROSCI.09-03-00865.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamura S, Morikawa Y, Iwanishi H, et al. Foxp1 gene expression in projection neurons of the mouse striatum. Neuroscience. 2004;124:261–267. doi: 10.1016/j.neuroscience.2003.11.036. [DOI] [PubMed] [Google Scholar]
- 52.Takahashi K, Liu FC, Hirokawa K, et al. Expression of Foxp2, a gene involved in speech and language, in the developing and adult striatum. J Neurosci Res. 2003;73:61–72. doi: 10.1002/jnr.10638. [DOI] [PubMed] [Google Scholar]
- 53.Tamura S, Morikawa Y, Iwanishi H, et al. Expression pattern of the winged-helix/forkhead transcription factor Foxp1 in the developing central nervous system. Gene Expr Patterns. 2003;3:193–197. doi: 10.1016/s1567-133x(03)00003-6. [DOI] [PubMed] [Google Scholar]
- 54.Vernes SC, Oliver PL, Spiteri E, et al. Foxp2 regulates gene networks implicated in neurite outgrowth in the developing brain. PLoS Genet. 2011;7:e1002145. doi: 10.1371/journal.pgen.1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spiteri E, Konopka G, Coppola G, et al. Identification of the transcriptional targets of FOXP2, a gene linked to speech and language, in developing human brain. Am J Hum Genet. 2007;81:1144–1157. doi: 10.1086/522237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takahashi K, Liu FC, Oishi T, et al. Expression of FOXP2 in the developing monkey forebrain: Comparison with the expression of the genes FOXP1, PBX3, and MEIS2. J Comp Neurol. 2008;509:180–189. doi: 10.1002/cne.21740. [DOI] [PubMed] [Google Scholar]
- 57.Chiu YC, Li MY, Liu YH, et al. Foxp2 regulates neuronal differentiation and neuronal subtype specification. Dev Neurobiol. 2014;74:723–738. doi: 10.1002/dneu.22166. [DOI] [PubMed] [Google Scholar]
- 58.Arlotta P, Molyneaux BJ, Jabaudon D, et al. Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. J Neurosci. 2008;28:622–632. doi: 10.1523/JNEUROSCI.2986-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kadoch C, Hargreaves DC, Hodges C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Desplats PA, Lambert JR, Thomas EA. Functional roles for the striatal-enriched transcription factor, Bcl11b, in the control of striatal gene expression and transcriptional dysregulation in Huntington’s disease. Neurobiol Dis. 2008;31:298–308. doi: 10.1016/j.nbd.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.MacDermott AB, Mayer ML, Westbrook GL, et al. NMDA-receptor activation increases cytoplasmic calcium concentration in cultured spinal cord neurones. Nature. 1986;321:519–522. doi: 10.1038/321519a0. [DOI] [PubMed] [Google Scholar]
- 62.Küppenbender KD, Standaert DG, Feuerstein TJ, et al. Expression of NMDA receptor subunit mRNAs in neurochemically identified projection and interneurons in the human striatum. J Comp Neurol. 2000;419:407–421. doi: 10.1002/(sici)1096-9861(20000417)419:4<407::aid-cne1>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 63.Landwehrmeyer GB, Standaert DG, Testa CM, et al. NMDA receptor subunit mRNA expression by projection neurons and interneurons in rat striatum. J Neurosci. 1995;15:5297–5307. doi: 10.1523/JNEUROSCI.15-07-05297.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stefani A, Chen Q, Flores-Hernandez J, et al. Physiological and molecular properties of AMPA/Kainate receptors expressed by striatal medium spiny neurons. Dev Neurosci. 1998;20:242–252. doi: 10.1159/000017318. [DOI] [PubMed] [Google Scholar]
- 65.Chergui K, Bouron A, Normand E, et al. Functional GluR6 kainate receptors in the striatum: Indirect downregulation of synaptic transmission. J Neurosci. 2000;20:2175–2182. doi: 10.1523/JNEUROSCI.20-06-02175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bischoff S, Barhanin J, Bettler B, et al. Spatial distribution of kainate receptor subunit mRNA in the mouse basal ganglia and ventral mesencephalon. J Comp Neurol. 1997;379:541–562. doi: 10.1002/(sici)1096-9861(19970324)379:4<541::aid-cne6>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 67.Park TI, Monzo H, Mee EW, et al. Adult human brain neural progenitor cells (NPCs) and fibroblast-like cells have similar properties in vitro but only NPCs differentiate into neurons. PLoS One. 2012;7:e37742. doi: 10.1371/journal.pone.0037742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang AH, Kaushal D, Rehen SK, et al. Chromosome segregation defects contribute to aneuploidy in normal neural progenitor cells. J Neurosci. 2003;23:10454–10462. doi: 10.1523/JNEUROSCI.23-32-10454.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gabay M, Li Y, Felsher DW. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med. 2014;4 doi: 10.1101/cshperspect.a014241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Laurenti E, Wilson A, Trumpp A. Myc’s other life: Stem cells and beyond. Curr Opin Cell Biol. 2009;21:844–854. doi: 10.1016/j.ceb.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 71.Flax JD, Aurora S, Yang C, et al. Engraftable human neural stem cells respond to developmental cues, replace neurons, and express foreign genes. Nat Biotechnol. 1998;16:1033–1039. doi: 10.1038/3473. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.