Abstract
AIM: To investigate the distribution of 12 high-pathogenicity island (HPI) genes and the relation between HPI genes and expression of yersiniabactin (Ybt) in enteroaggregative E.coli (EAggEC) isolated from Chinese diarrhea patients.
METHODS: The distribution of 12 HPI genes was investigated by PCR and DNA hybridization in two prototype strains ofEAggEC, EAggEC 17-2, EAggEC O42, and 6 clinical EAggEC isolates from China. The production of siderophore Ybt in HPI-positive strains was detected by reporter gene bioassay to determine the relation between HPI genes and expression of Ybt. Flow cytometry was used to detect fluorescent signal of the reporter strain that could designate production of Ybt.
RESULTS: Seven strains were HPI-positive and one strain was HPI-negative. Six of the seven HPI-positive strains were inserted into asnT-tRNA site. Moreover, seven EAggEC HPI-positive strains revealed enhanced fluorescence signal but the EAggEC HPI-negative strain did not. However, there was a difference in Ybt expression condition and level among these seven EAggEC HPI-positive strains. Although UFT073 strain, the prototype strain of uropathogenic E.coli (UPEC), carried the complete HPI core part, we did not detect the expression of Ybt in it.
CONCLUSION: EAggEC HPI-positive strains can express the Ybt system, but the presence of HPI core part does not mean the functional expression of Ybt.
Keywords: High-pathogenicity island, Enteroaggregative Escherichia coli, Yersiniabactin
INTRODUCTION
High-pathogenicity island (HPI) encoding synthesis and uptake of the siderophore yersiniabactin (Ybt) has been primarily identified in mouse lethal Yersinia: Yersinia pestis, Yersinia pseudotuberculosis serotypes O1-O3, and Yersinia entercolitica 1B biotype[1-5]. The Yersinia HPI consists of a 30.5 kb highly conserved functional core region termed Ybt locus and a 5- to 13-kb AT-rich variable part[1]. The core part comprises genes, irp1-irp5 and irp9 for the biosynthesis of the Ybt, irp6-irp8 and fyuAfor the uptake of Fe-Ybt, the AraC-like regulator ybtA and the integrase intB resides on the 5 end of the HPI right-end extremity next to the asn-tRNA bacterial attachment site[6-9]. Ybt-mediated iron acquisition is the main function of HPI[8,10]. Recently, HPI is subsequently identified in other bacterial genera such as E.coli, Citrobacter diversus, various species of Klebsiella[11-16]. Whether HPI in E.coli has the functional expression of Ybt is the premise to understand that HPI contributes to virulence or ecological fitness. Schubert et al[14] reported that 93% EAggEC, 5% enteropathogenic E.coli, 27% enteroinvasive E.coli and 5% enterotoxigenic E.coli carry HPI by hybridization with an irp2 probe, but none of enterohemorrhagic E.coli harbors it. It is still unclear why the distribution frequency of HPIs is different among various E.coli strains. Considering the high prevalence of HPI in EAggEC, in the present study, we detected all the12 genes of HPI by PCR and DNA hybridization in the EAggEC 17-2, EAggEC O42, and 6 EAggEC clinical isolates from Chinese diarrhea patients. By reporter bioassay, products of Ybt in these strains were determined under iron-rich or -poor conditions to demonstrate the function of Yersiniae HPI in EAggEC strains. The relation between HPI genes and their function was discussed.
MATERIALS AND METHODS
Bacterial strains and growth conditions
EAggEC O42 (O44:H18), 17-2 (O3:H2), CFT 073, JM109, and Y. entercolitica WA (O:8) were provided by Key Laboratory for Molecular Bacteriology in Chinese Center for Disease Control and Prevention. Six EAggEC strains isolated from Chinese diarrhea patients were confirmed by PCR and DNA hybridization with EAEC specific primer[17]. The codes of these strains were EAEC 47/37, EAEC 66, EAEC SS23, EAEC B86-2, EAEC E1073, and EAEC 52/46. The reporter strain WA-CS irp1:KN[14,18,19] was kindly provided by Professor A. Rakin. Yersinia strains were grown in Luria Bertani (LB) broth with shaking at 28 °C for 48 h and other E.coli strains at 37 °C for 24 h. The iron-deficient medium (LBD) was obtained by the addition of 0.2 mmol/L dipyridyl (Sigma) to LB broth. Y. enterocolitica WA (O:8) was used as positive control, while E.coli JM109 as negative control for PCR amplification and DNA blotting and Ybt assay.
Primers and PCR amplification
PCR was carried out in a 50 mL reaction volume containing template DNA (5 mL of boiled lysate), 1.5 mmol/L MgCl2, 0.4 mmol/L of each of dNTPs, 0.5 mmol/L of each primer, and 25 U of Taq polymerase (TAKARA, Japan) with an automated thermal cycler (GeneAmp 9700, PE Biosystems, USA). The primers were designed according to the HPI sequence of Yersiniafrom GenBank and synthesized by Sangon (Shanghai, China).
Each set of primers and size of the amplified fragments are listed in Table 1. The pre-denaturation (94 °C, 5 min) was followed by 30 cycles of denaturation (94 °C, 1 min), annealing (53-58 °C, 40 s-1 min), and extension (72 °C, 30 s-2 min 30 s) with an additional extension step (72 °C, 7 min). In this study, we amplified all the 12 genes within HPI functional core part. Among them, asnT-tRNA-intB fragment included 3 end partial sequence of asnT-tRNA gene and complete intB gene, while others contained the partial fragment within the internal of the corresponding gene. PCR products were electrophoresed in 1-1.5% agarose gels stained with ethidium bromide and visualized under UV light.
Table 1.
Primers used in this study
| Amplified fragment | Length (bp) | Primers | Reference |
| asnT-tRNA-int | 1 505 | 5’-GCTCTCGACGTCCGTATCGCCTGACAC-3’ | [1], GenBank no.: AJ132668 |
| 5’-GCTCTAGATCAACCTGTTTCGGGTCGG-3’ | |||
| irp9 | 529 | 5’-GCTGTCCTGAAATACGGA-3’ | [1], GenBank no.: AJ132668 |
| 5’-AGTGCGCTCGTTTATGTT-3’ | |||
| irp8 | 1 176 | 5’-TGTCATCAGTTCTCTTGCCG-3’ | [1], GenBank no.: AJ132668 |
| 5’-CTGGTATCGGCGCTGTCTAT-3’ | |||
| irp7 | 1 572 | 5’-ATGGTGGATAAACGGTGAGC-3’ | [1], GenBank no.: AJ132668 |
| 5’-TATCTCCAGCGCCGTTGGCAGTC-3’ | |||
| irp6 | 1 588 | 5’-TCACCATTGAGAGACGATGC-3’ | [1], GenBank no.: AJ132668 |
| 5’-TTTCCTCTCTGGCAGGTGAT-3’ | |||
| ybtA | 555 | 5’-AGTGCGCTCGTTTATGTT-3’ | [1], GenBank no.: AJ132668 |
| 5’-GCAGCAGTTTCTGACGTT-3’ | |||
| irp2 | 267 | 5’-AAGGATTCGCCTGTTACCGGAC-3’ | [19], GenBank no.: L18881 |
| 5’-TCGTCGGGCAGCGTTTCTTCT-3’ | |||
| irp1 | 799 | 5’-CCGGTTATTGTGAAGGTT-3’ | [8], GenBank no.: Y12527 |
| 5’-GTGAATGTTACGGCACTAA-3’ | |||
| irp3 | 1 139 | 5’-GGAATTCCCATGGACCGGAGAACACGTTATG-3’ | [8], GenBank no.: Y12527 |
| 5’-GCTCTAGATCACAGCGCCTCCTTATCATC-3’ | |||
| irp4 | 827 | 5’-GCTCTAGAATGTGCATCCCGCTGCGG-3’ | [8], GenBank no.: Y12527 |
| 5’-AACGCAGGGTACCGTCACCTTTCTGCTGAAGTGC-3’ | |||
| irp5 | 1 636 | 5’-CGCGGATCCGGTACCAGGTGACGCATGAATTCTTC-3’ | [8], GenBank no.: Y12527 |
| 5’-AACTGCAGTCAACCTGTTTCGGGTCGG-3’ | |||
| fyuA | 948 | 5’-GCTTTATCCTCTGGCCTT-3’ | [5], GenBank no.: Z35486 |
| 5’-GGCATAACGATTAACG-3’ | |||
| EAEC | 623 | 5’-CTGGCGAAAGACTGTATCAT-3’ | [17] |
| 5’-CAATGTATAGAAATCCGCTGTT-3’ |
Dot blotting
Dot blotting for the fyuA and irp2 genes was performed on DNA extracted from each strain and spotted on positively charged pyroxylin membranes. Hybridization was performed at 65 °C in 1% SDS/1 moL NaCl/Tris 50 mmol/L HCl, pH 7.5/1% blocking reagent. The membranes were washed in 2 SSC for 15 min at room temperature, then in 2 SSC/0.1% SDS for 30 min at 65°C, finally in 0.1 SSC for 5 min at room temperature. The probes were produced by PCR according to the manufacturer’s instructions (PCR DIG Probe Synthesis kit, Roche), using the primers and amplification procedure described above. Digoxigenin labeling was probed and hybridized with a DNA labeling and detection kit (DIG Luminescent Detection kit for nucleic acids, Roche) according to the manufacturer’s instructions.
Yersiniabactin assay
A reporter strain system, Y. enterocolitica strain WA-CS irp1:KN (pCJG3.3N), constructed by Schubert et al[18] was used to monitor the expression of Ybt. This strain was deleted in the Ybt biosynthesis gene (irp1) and carried the fyuA-gfp reporter plasmid pCJG3.3N. As the expression of the fyuA gene was unregulated in the presence of the extracellular Ybt, supernatants of bacteria grown under normal (LB medium) or iron-deficient conditions (LBD medium) were applied to this reporter strain, which could lead to increased expression of the fyuA-gfp, if Ybt was present.
We streaked the bacteria on Luria broth agar plates and inoculated about 5 mL LB medium for overnight culture as a start culture. From this preculture we inoculated the main culture on LB and LBD medium, respectively. The strains were incubated in the media for at least 7 d. Supernatants of the strains were added to the reporter strain, and flow cytometry for the reporter strain was done with an Epics flow cytometer (FACSCalibur, Becton Dickinson) and an argon 488-nm laser was used. The scale was logarithmic, and fluorescence data determined in arbitrary units were collected from 50 000 bacteria.
RESULTS
Integration analysis of HPI genes in EAggEC strains
We examined eight EAggEC strains for the presence of all the 12 HPI genes. irp1-irp9, fyuA, and ybtA genes were detected in seven strains of EAggEC by PCR and the amplification products were the same as WA positive control in lengths (Figure 1). One strain (EAEC 47/37) isolated from diarrhea patients did not yield any amplification products of these 12 gene primers.
Figure 1.
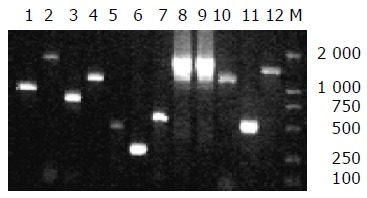
PCR amplification results of 12 genes within HPI core part. Lane 1: fyuA, lane 2: irp5, lane 3: irp4, lane 4: irp3, lane 5: irp1, lane 6: irp2, lane 7: ybtA, lane 8: irp6, lane 9: irp7, lane 10: irp8, lane 11: irp9, lane 12: asnT-tRNA-int, M: DL2000 marker.
In Yersinia, the site of HPI integration, bacterial chromosomes sharing homologies with the bacteriophage P4 attachment site are located within an asn-tRNA locus[4,20]. The asnT-tRNA-int fragment was also identified in six of seven HPI-positive strains, but not in EAEC B86-2, indicating that the strain might have a different insert site from others or the edge of HPI was damaged. We also confirmed the results by DNA-blot hybridization, using irp2 and fyuA genes as probe (data not shown). All the 12 HPI genes were detected also in CFT073 strain, the prototype strain of UPEC, demonstrating that the core region of HPI was present in EAggEC strains with a high frequency. Moreover, HPIs in most of these strains were inserted inasn-tRNA, as in Yersinia.
Detection of yersiniabactin production in EAggEC strains carrying Yersinia HPI
We detected the expressions of Ybt in eight EAggEC strains and UFT073 strain under normal (LB medium) and iron-deficient conditions (LBD medium) using the reporter strain WA-CS irp1:KN. WA stain was used as positive control while WA-CS irp1:KNand HB101 as negative control.
Figure 2 shows that the culture supernatants of these seven HPI-positive EAggEC strains grown on iron-deficient medium exhibited apparently enhanced fluorescent signals of the fyuA-gfp reporter strain. Among these seven strains, EAggEC O42 had the strongest expression of Ybt (90%), which was nearly equal to the positive control strain WA (90.3%), and EAggEC 17-2 had a comparatively weak signal (47.4%), indicating that the seven EAggEC strains could express Ybt under iron-deficient conditions. Interestingly, the supernatants of three isolates, EAEC66, EAEC B86-2 and EAEC E1073, grown on normal media could stimulate the reporter strain too, but the fluorescent signals were weaker than those grown on the iron-deficient medium. On the other hand, in WA strain, EAggEC 17-2, EAggEC O42, and two other HPI-positive isolates, EAEC SS23 and EAEC 52/46, the fluorescence signals were not enhanced when the bacteria were grown on LB medium. In addition, whether bacteria were grown under iron-rich conditions or iron-deficient conditions, the supernatants of two HPI-negative strains, HB101 and EAggEC 47/37, and one HPI-positive strain, CFT 073, did not affect the expression of the fyuA-gfp reporter gene.
Figure 2.
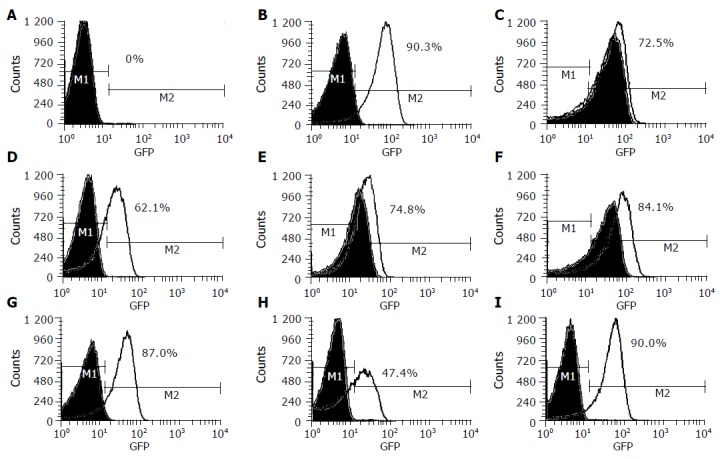
Fluorescence of fyuA-gfp in E.coli grown on LB media (green curve) and LBD media (black curve). A: WA-CS irp1::KN(pCJG3.3N), CFT073(HPI+), HB101(HPI–) and EAEC 47/37(HPI–); B: WA(HPI+); C: EAEC 66(HPI+); D: EAEC SS23(HPI+); E: EAEC B86-2(HPI+); F: EAEC E1073(HPI+); G: EAEC 52/46(HPI+); H: EAEC 17-2 (HPI+); I: EAEC O42 (HPI+); M1: specified range of background of fluorescence signal; M2: specified range of enhanced florescence signal.
The results suggested that EAggEC HPI-positive strains could express the Ybt system, but there was a difference in expression condition and level of Ybt among them.
DISCUSSION
In the genus Yersinia, irp1-irp5 and irp9 genes are essential for Ybt synthesis, and irp6, irp7, fyuA genes for Ybt transport. The pathway of Ybt uptake is blocked if any one of these genes mutates or misses. In this study, we investigated whether EAggEC strains carrying Yersinia HPI genes could express the corresponding products and whether the series of products could synthesize functional Ybt. In order to understand these events, we should know if these EAggEC strains carry the integrated functional core region of HPI.
The presence of HPI in enterobacteriaceae strains has been confirmed only by PCR amplification of irp2/fyuA and DNA hybridization, but the results of irp2/fyuA detection are not always identical[11,14]. Usually, the detection rate of fyuA is lower than that of irp2, which might be explained by the fact that fyuA gene is located in the boundary of HPI functional core region, and during the process of evolution it is liable to be damaged or changed due to the instability of HPI. As the exact mechanisms of transport, excision and synthesis of HPI are still unclear, the detection results of individual genes are not sufficient to confirm the presence of integrated functional core region. To avoid this problem, in this study, all the 12 genes in the functional core region of HPI were amplified by PCR. HPI was positive in seven of eight EAggEC strains, and six of seven HPI-positive EAggEC strains were positive for asnT-tRNA-int gene but not for EAEC B86-2 strain, indicating that the seven HPI-positive EAggEC strains carry the necessary genes for synthesis, transport and regulation of Ybt. HPIs in these EAggEC strains are often inserted into asnT locus on the chromosome, while HPI of EAEC B86-2 strain might be inserted into different loci of other strains or its border sequence is damaged.
To demonstrate the production of Ybt, we used a detection system based on Ybt-induced upregulation of a Ybt reporter gene fyuA-gfp. Schubert et al[18] found that 49 of 60 HPI-positive enterobacteriaceae isolated from blood cultures and urine samples can produce Ybt using this system. Our study showed that Ybt could be synthesized in the seven HPI-positive EAggEC strains, including the strain of EAEC B86-2 with an incomplete asn-tRNA-int gene. However, not all HPI-positive strains could express Ybt. Based on the HPI transfer mechanism and bacterial evolution, we can make one intriguing hypothesis that under certain selective pressure, bacteria can obtain HPI, and then mutations occur during the process of evolution, thus resulting in the loss of function. However, bacteria could survive by changing themselves to adapt to the environment in the evolutionary process. Another hypothesis is that various bacteria can obtain HPI independently, a kind of random event rather than necessary for living, and the non-functional HPI remains in bacteria. The results suggest that the presence of irp2/fyuAdoes not necessarily mean that the integrated ‘functional core region’ of HPI is complete, and the presence of integrated functional core region of HPI does not necessarily mean that functional Ybt is synthesized.
Under iron-deficient condition (LBD medium), the seven HPI-positive EAggEC strains of Ybt were differently expressed, and some strains were different from WA strain in characteristics of Ybt expression. Only under iron-deficient condition, the Ybt system of WA strain could be activated. On the contrary, in our study expression of Ybt under normal condition could also be detected in three HPI-positive EAggEC strains. The reason why Ybt systems in these strains are easy to be activated remains to be clarified. A possible explanation might be that the difference in genetic structure of bacteria can result in the different need of iron. Though most enterobacteriaceae carry genes encoding high-affinity iron uptake systems, such as enterobactin system and aerobactin system, the fact that most E.coli HPI-positive strains have the ability to synthesize Ybt while others do not, may mean that the expression of Ybt possess more specific functions during different infection phases. It was reported that Yen-HPI core genes are insufficient to confer a positive chrome azurol S (CAS) phenotype and mouse virulence to the siderophore-deficient strain Y. enterocolitica NF-O (BG 1A), but sufficient to confer MRS40 (serotype O:9, BG 2; phenotype, CAS negative)[21]. Therefore, it is conceivable that the expression of Ybt may also be regulated by genes outside the HPI to adjust bacteria to make a living in iron-limited environment. More studies are needed to clarify the contribution of Yersinia HPI to the virulence of E.coli.
ACKNOWLEDGMENTS
The authors thank Rakin A (Max von Pettenkofer Institute, Germany) for kindly providing WA-CS irp1:KN (pCJG3.3N) reporter strain system and Schubert S (Max von Pettenkofer-Institute, Germany) for technical assistance and helpful discussion.
Footnotes
Science Editor Wang XL and Guo SY Language Editor Elsevier HK
Supported by the National Basic Research Program of Ministry of Science and Technology of China, No. G1999054101
Co-first-author: Jing Hu
Co-correspondent: Shou-Yi Yu
References
- 1.Rakin A, Noelting C, Schubert S, Heesemann J. Common and specific characteristics of the high-pathogenicity island of Yersinia enterocolitica. Infect Immun. 1999;67:5265–5274. doi: 10.1128/iai.67.10.5265-5274.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchrieser C, Rusniok C, Frangeul L, Couve E, Billault A, Kunst F, Carniel E, Glaser P. The 102-kilobase pgm locus of Yersinia pestis: sequence analysis and comparison of selected regions among different Yersinia pestis and Yersinia pseudotuberculosis strains. Infect Immun. 1999;67:4851–4861. doi: 10.1128/iai.67.9.4851-4861.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bach S, Buchrieser C, Prentice M, Guiyoule A, Msadek T, Carniel E. The high-pathogenicity island of Yersinia enterocolitica Ye8081 undergoes low-frequency deletion but not precise excision, suggesting recent stabilization in the genome. Infect Immun. 1999;67:5091–5099. doi: 10.1128/iai.67.10.5091-5099.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchrieser C, Brosch R, Bach S, Guiyoule A, Carniel E. The high-pathogenicity island of Yersinia pseudotuberculosis can be inserted into any of the three chromosomal asn tRNA genes. Mol Microbiol. 1998;30:965–978. doi: 10.1046/j.1365-2958.1998.01124.x. [DOI] [PubMed] [Google Scholar]
- 5.Rakin A, Saken E, Harmsen D, Heesemann J. The pesticin receptor of Yersinia enterocolitica: a novel virulence factor with dual function. Mol Microbiol. 1994;13:253–263. doi: 10.1111/j.1365-2958.1994.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 6.Bearden SW, Fetherston JD, Perry RD. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect Immun. 1997;65:1659–1668. doi: 10.1128/iai.65.5.1659-1668.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brem D, Pelludat C, Rakin A, Jacobi CA, Heesemann J. Functional analysis of yersiniabactin transport genes of Yersinia enterocolitica. Microbiology. 2001;147:1115–1127. doi: 10.1099/00221287-147-5-1115. [DOI] [PubMed] [Google Scholar]
- 8.Pelludat C, Rakin A, Jacobi CA, Schubert S, Heesemann J. The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation. J Bacteriol. 1998;180:538–546. doi: 10.1128/jb.180.3.538-546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perry RD, Balbo PB, Jones HA, Fetherston JD, DeMoll E. Yersiniabactin from Yersinia pestis: biochemical characterization of the siderophore and its role in iron transport and regulation. Microbiology. 1999;145(Pt5):1181–1190. doi: 10.1099/13500872-145-5-1181. [DOI] [PubMed] [Google Scholar]
- 10.de Almeida AM, Guiyoule A, Guilvout I, Iteman I, Baranton G, Carniel E. Chromosomal irp2 gene in Yersinia: distribution, expression, deletion and impact on virulence. Microb Pathog. 1993;14:9–21. doi: 10.1006/mpat.1993.1002. [DOI] [PubMed] [Google Scholar]
- 11.Bach S, de Almeida A, Carniel E. The Yersinia high-pathogenicity island is present in different members of the family Enterobacteriaceae. FEMS Microbiol Lett. 2000;183:289–294. doi: 10.1111/j.1574-6968.2000.tb08973.x. [DOI] [PubMed] [Google Scholar]
- 12.Clermont O, Bonacorsi S, Bingen E. The Yersinia high-pathogenicity island is highly predominant in virulence-associated phylogenetic groups of Escherichia coli. FEMS Microbiol Lett. 2001;196:153–157. doi: 10.1111/j.1574-6968.2001.tb10557.x. [DOI] [PubMed] [Google Scholar]
- 13.Karch H, Schubert S, Zhang D, Zhang W, Schmidt H, Olschläger T, Hacker J. A genomic island, termed high-pathogenicity island, is present in certain non-O157 Shiga toxin-producing Escherichia coli clonal lineages. Infect Immun. 1999;67:5994–6001. doi: 10.1128/iai.67.11.5994-6001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schubert S, Rakin A, Karch H, Carniel E, Heesemann J. Prevalence of the "high-pathogenicity island" of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect Immun. 1998;66:480–485. doi: 10.1128/iai.66.2.480-485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gophna U, Oelschlaeger TA, Hacker J, Ron EZ. Yersinia HPI in septicemic Escherichia coli strains isolated from diverse hosts. FEMS Microbiol Lett. 2001;196:57–60. doi: 10.1111/j.1574-6968.2001.tb10540.x. [DOI] [PubMed] [Google Scholar]
- 16.Xu JG, Cheng B, Wen X, Cui S, Ye C. High-pathogenicity island of Yersinia spp. in Escherichia coli strains isolated from diarrhea patients in China. J Clin Microbiol. 2000;38:4672–4675. doi: 10.1128/jcm.38.12.4672-4675.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schubert S, Cuenca S, Fischer D, Heesemann J. High-pathogenicity island of Yersinia pestis in enterobacteriaceae isolated from blood cultures and urine samples: prevalence and functional expression. J Infect Dis. 2000;182:1268–1271. doi: 10.1086/315831. [DOI] [PubMed] [Google Scholar]
- 19.Guilvout I, Mercereau-Puijalon O, Bonnefoy S, Pugsley AP, Carniel E. High-molecular-weight protein 2 of Yersinia enterocolitica is homologous to AngR of Vibrio anguillarum and belongs to a family of proteins involved in nonribosomal peptide synthesis. J Bacteriol. 1993;175:5488–5504. doi: 10.1128/jb.175.17.5488-5504.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schubert S, Rakin A, Fischer D, Sorsa J, Heesemann J. Characterization of the integration site of Yersinia high-pathogenicity island in Escherichia coli. FEMS Microbiol Lett. 1999;179:409–414. doi: 10.1111/j.1574-6968.1999.tb08756.x. [DOI] [PubMed] [Google Scholar]
- 21.Pelludat C, Hogardt M, Heesemann J. Transfer of the core region genes of the Yersinia enterocolitica WA-C serotype O: 8 high-pathogenicity island to Y. enterocolitica MRS40, a strain with low levels of pathogenicity, confers a yersiniabactin biosynthesis phenotype and enhanced mouse virulence. Infect Immun. 2002;70:1832–1841. doi: 10.1128/IAI.70.4.1832-1841.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]


