Abstract
Microglia regulate the secretion of various immunomediators in central nervous system diseases. Microglial autophagy is the crucial process for cell's survival and cytokine productions. Recent studies have reported that several microRNAs are involved in the autophagy system. miR-Let7A is such a microRNA that plays a role in various inflammation responses, and is magnified as a key modulator particularly in the autophagy system. In present study, we investigated whether miR-Let7A is involved in autophagy in activating microglia. Overexpression of miR-Let7A in LPS-stimulated BV2 microglial cells promoted the induction of the autophagy related factors such as LC3II, Beclin1, and ATG3. Our results suggest a potential role of miR-Let7A in the autophagy process of microglia during CNS inflammation.
Keywords: microRNA-Let7A (miR-Let7A), Microglia, Autophagy, Beclin 1, Microtubule-associated protein light chain 3 (LC3) II, ATG 3
INTRODUCTION
Autophagy is a cardinal cellular mechanism that involves the degradation and digestion of intracellular constituents by lysosomes [1,2,3]. Autophagy controls inflammation through interactions with immune signaling pathways and regulates the secretion of molecular mediators of inflammation [4,5,6]. Autophagy plays a vital role in the physiological conditions of many immune cells including macrophages [7,8,9]. Appropriate autophagy contributes to neuroprotection, whereas inappropriate autophagy could induce cell death [10,11,12]. Autophagy constitutes a lysosome-mediated degradation process [13] and is crucial for the maintenance of metabolic homeostasis by inhibiting the accumulation of misfolded proteins and of damaged cytoplasmic organelles [14,15,16]. Autophagy mitigates neurodegeneration caused by oxidative stress [17], whereas impaired autophagy signaling has been implicated in neurodegenerative disorders such as Parkinson's, Alzheimer's diseases[18,19,20,21], spinal cord injury [22], and stroke [23].
Autophagosome formation is controlled by protein complexes including the mammalian target of rapamycin (mTOR) complex 1 (mTORC1) and the coiled-coil myosin-like BCL2-interacting protein 1 (Beclin 1)-complex [24,25,26,27]. The autophagy process is initiated by the regulation of protein complexes composed of more than 30 autophagy-related (ATG) proteins [28,29,30] and autophagic adaptor light chain 3 protein (LC3) [31]. The phosphatidylethanol-amine-conjugated LC3 (LC3II) is localized at the inner and outer autophagosomal membranes and plays a critical role for the initiation of autophagy process [31]. Autophagy can regulate the production and secretion of diverse cytokines in cells [32,33,34]. Microglia degrades beta-amyloid (Aβ) via autophagy in Alzheimer's disease [35]. The decreased level of Beclin 1 and the blockade of microglial pahgocytosis were observed in Alzheimer's disease patients [36,37].
Several microRNA (miR) are associated with autophagic flux [38]. The overexpression of miR-195[39], miR-14 [40], miRNA-30a [41] and miR-423-5p [42] promote induction of autophagy, whereas suppression of miR-101 induces autophagy in cardiomyocytes [43]. MicroRNA-Let7A (miR-Let7A) is a tumor suppressor miRNA that target transcription-related genes in apoptosis [44]. MiR-Let7A regulates anti-inflammatory responses through repression of specific genes acting in downstream signaling pathways [44] and is involved in the function of microglia in inflammatory injury. Notably, miR-Let7 promotes autophagy by suppression of mTOR signaling [45], and activates the neuronal autophagy in the brain of mice [46,47]. Moreover, the suppression of miR-Let7a in mice leads to the reduction of LC3 II levels [45]. In the present study, we investigated whether miR-Let7a controls autophagy process in microglia activated by lipopolysaccharide (LPS).
MATERIALS AND METHODS
BV2 microglia culture
BV2 microglial cells were obtained from Prof. Eun-hye Joe (Ajou University School of Medicine, Chronic Inflammatory Disease Research Center). BV2 cells cultured in Dulbecco's Modified Eagle's Medium (Gibco, NY, USA) supplemented with 10% fetal bovine serum (Gibco, NY, USA) and 100 µg/ml penicillin-streptomycin (Gibco, NY, USA) at 37℃ in a humidified atmosphere containing 5% CO2. Lipopolysaccharide (LPS, 1 µg/ml; Sigma-Aldrich, St. Louis, MO, USA) was treated in BV2 microglia for 12 h before sampling.
miR-Let7A transfection
Mmu-Let7A-5p (cat#, 4464066) was purchased from Ambion (Ambion, Austin, TX, USA). For the transfection of RNA duplexes, miR-Let7A mimic or miR-Let7A inhibitor was diluted to a 20 nM final concentration in Opti-MEM, and mixed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). After incubation at room temperature for 15 min, the mixture was added to cells. Cells were incubated for 72 h and harvested for total protein or RNA extraction.
Reverse transcription PCR
Reverse transcription PCR was performed to measure mRNA levels of Beclin-1, LC3II in BV2 microglia. Briefly, cells were lysed with Trizol reagent (Invitrogen, Carlsbad, CA, USA), and total RNA was extracted according to the manufacturer's protocol. cDNA synthesis from isolated total RNA and sample normalization were performed. PCR was performed using the following thermal cycling conditions: 10 min at 95℃; 35 cycles of denaturing at 95℃ for 15 sec, annealing for 30 sec at 62℃, elongation at 72℃ for 30 sec; final extension for 10 min at 72℃, and held at 4℃. PCR was performed using the following primers (5' to 3'); Beclin-1: 5'-AGC TGC CGT TAT ACT GTT CTG-3', (sense), and 5'-ACT GCC TCC TGT GTC TTC AAT CTT-3', (antisense); LC3II: 5'-GAT GTC CGA CTT ATT CGA GAG C-3', (sense), and 5'-TTG AGC TGT AAG CGC CTT CTA-3' (antisense), GAPDH: GGC ATG GAC TGT GGT CAT GAG (sense), TGC ACC ACC AAC TGC TTA GC (antisense). PCR products were electrophoresed in 1.5% agarose gels and stained with ethidium bromide as described [48].
Western blot analysis
BV2 microglia were washed rapidly with ice-cold PBS, scraped, and collected. Cell pellets were lysed with ice-cold RIPA buffer (Sigma-Aldrich, St. Louis, MO, USA). The lysates were centrifuged at 12,000 rpm for 30 min at 4℃ to produce whole-cell extracts. Protein content was quantified using the BCA method (Pierce, Rockford, IL, USA). Proteins (40 µg) were separated on a 10% SDS-polyacrylamide (PAGE) gel and transferred onto a polyvinylidene difluoride (PVDF) membrane. After blocking with 5% bovine serum albumin, prepared in Tris-buffered saline/Tween (TBS-T; 20 nM Tris [pH 7.2], 150 mM NaCl, and 0.1% Tween 20), for 1 h at room temperature, immunoblots were incubated overnight at 4℃ with primary antibodies that specifically detect Beclin-1 (1 : 1,000, Millipore, Billerica, MA, USA), ATG3 (1 : 1,000, Abcam, Cambridge, MA, USA), LC3II (1 : 1,000, Cell Signaling Technology, Danvers, MA, USA), and β-actin (1 : 2,000, Cell Signaling Technology, Danvers, MA, USA). Next, blots were incubated with HRP-linked anti-rabbit IgG antibodies purchased from Abcam (Abcam, Cambridge, MA, USA) for 1 h 30 min at room temperature. Enhanced chemiluminescence was performed by ECL (Invitrogen, Carlsbad, CA, USA) [49].
Immunocytochemistry
The expression of Beclin1 and ATG3 in BV2 cells was examined by immunocytochemistry. Cells in all experimental groups were washed three times with PBS, fixed with 4% paraformaldehyde for 3 h, and then washed with PBS. BV2 cells were permeabilized with 0.025% Triton X-100 and blocked for 1 h at room temperature with dilution buffer (Invitrogen, Carlsbad, CA, USA). Primary antibodies, anti-rabbit-Beclin1 (1 : 500, Santa Cruz, CA, USA), and anti-rabbit-ATG3 (1 : 500, Santa Cruz, CA, USA) were prepared in dilution buffer, added to samples, and incubated for 3 h at room temperature. Primary antibody was then removed, and cells were washed 3 times for 3 min each with PBS. Next, samples were incubated with Rhodamine-conjugated goat anti-rabbit (1 : 200, Jackson Immunoresearch) for 1 h 30 min at room temperature. Cells were washed again three times for 3 min each with PBS and stained with 1 µg/mL 4',6-diamidino-2-phenylindole (DAPI) (1 : 100, Invitrogen, Carlsbad, CA, USA) for 15 min at room temperature. Fixed samples were imaged using a Zeiss LSM 700 confocal microscope (Carl Zeiss, Thornwood, NY, USA) [49].
Statistical analysis
Statistical analyses were carried out using SPSS 18.0 software (IBM Corp., Armonk, NY, USA). All data are expressed as mean±S. E.M. Significant intergroup differences were determined by one-way analysis of variance (ANOVA) followed by Bonferroni post hoc multiple comparison test. Each experiment included at least 3 replicates per condition. Differences were considered significant at *p<0.05, **p<0.001.
RESULTS
The mRNA levels of LC3II in microglia were increased after miR-Let7A overexpression
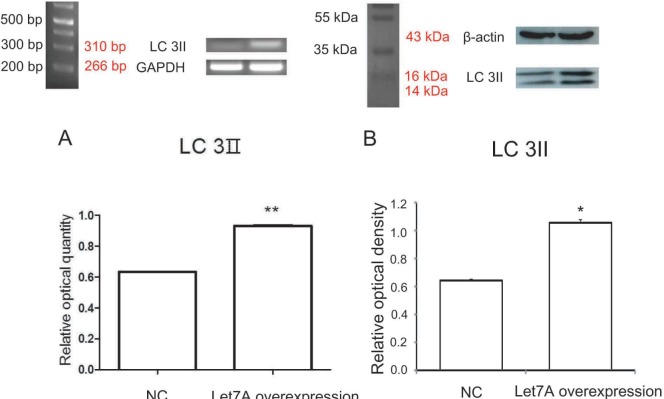
LC3II (the microtubule-associated protein light chain 3) is the key element in the initial isolation membrane nucleation of autophagy process [50]. PCR analysis showed that the mRNA level of LC3II was increased in miR-Let7A-overexpressing BV2 cells (Fig. 1A). Western blot analysis confirmed that the protein level of LC3II in miR-Let7A-overexpressing BV2 cells was increased comparison to the normal group (Fig. 1B).
Fig. 1. miR-Let7A overexpression upregulated LC3II mRNA level in BV2 microglia. (A) PCR data showing enhanced expression of LC3II mRNA level in microglia transfected with miR-Let7A mimic (20 nM). GAPDH was used as a control. (B) Western blotting showing LC3II protein levels in microglia transfected with miR-Let7A mimic. β-actin was used as a control. NC: normal control, Let7A overexpression: miR-Let7A overexpression. Data were expressed as mean±S.E.M, and each experiment included 3 repeats per condition. Differences were considered significant at *p<0.05 and **p<0.001.
The overexpression of miR-Let7A modulated the expression of Beclin1 and ATG3 in inflammation-induced microglia
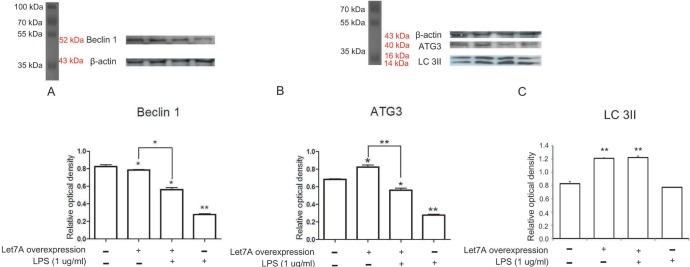
The expression levels of Beclin 1 transcripts (Fig. 2) and Beclin 1 protein (Fig. 3A) were slightly reduced in miR-Let7-Aoverexpressing BV2 cells compared to those in normal BV2 cells. LPS-treated BV2 cells showed more profound reduction of Beclin 1 transcripts and Beclin 1 protein (Fig. 2 and 3A). The miR-Let7A overexpression partially blocked reduced expression of Beclin 1 transcripts and Beclin 1 protein in LPS-treated BV2 cells (Fig. 2 and 3A).
Fig. 2. miR-Let7A overexpression regulated Beclin 1 mRNA level in BV2 cells activated by LPS. The mRNA levels of Beclin 1 in normal BV2 cells, BV2 cells transfected with miR-Let7A, BV2 cells treated with LPS (1 µg/ml), BV2 cells transfected with miR-Let7A and treated with LPS. LPS was treated for 12 h. GAPDH was used as a control. NC: normal control group, Let7A overexpression: miR-Let7A overexpression group, LPS: LPS treatment group. Data were expressed as mean±S.E.M, and each experiment included 3 repeats per condition. Differences were considered significant at *p<0.05, **p < 0.001.
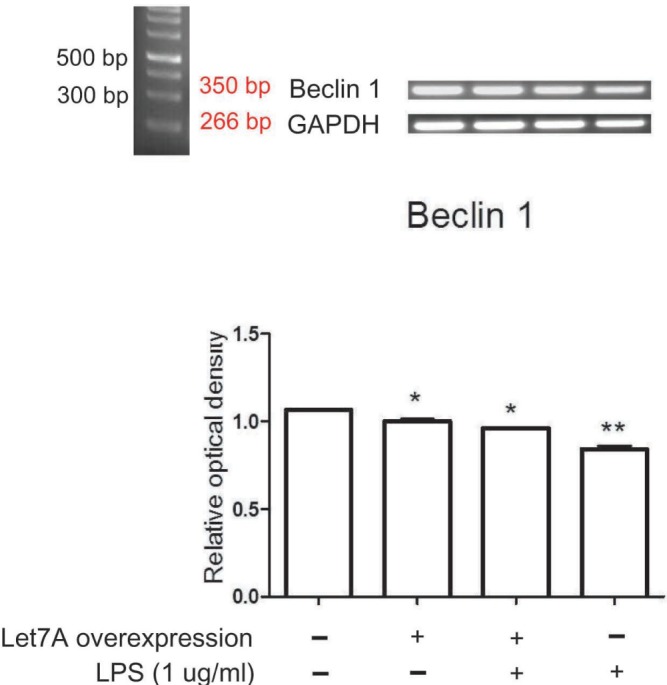
Fig. 3. miR-Let7A overexpression regulated Beclin 1, ATG3, LC3II protein levels in BV2 cells activated by LPS. (A~C) Western blotting showing the expression levels of Beclin 1 (A), ATG3 (B), and LC3II (C) and their quantifications in normal BV2 cells, BV2 cells transfected with miR-Let7A, BV2 cells treated with LPS (1 µg/ml), BV2 cells transfected with miR-Let7A and treated with LPS. miR-Let7A mimic was used at 20 nM and LPS was treated for 12 h. β-actin was used as a control. NC: normal control group, Let7A overexpression: miR-Let7A overexpression group, LPS: LPS treatment group. Data were expressed as mean±S.E.M, and each experiment included 3 repeats per condition. Differences were considered significant at *p<0.05, **p < 0.001.
The ATG3 level was increased in miR-Let7A-overexpressing BV2 cells, whereas the ATG3 level was decreased in LPS-treated BV2 cells compared to that in normal BV2 cells (Fig. 3B). In LPS-treated miR-Let7A-overexpressing BV2 cells, the ATG3 level was lower than that in normal BV2 cells, but it was higher than that in LPS-treated BV2 cells (Fig. 3B).
The LC3II level was increased in miR-Let7A-overexpressing BV2 cells, whereas the LC3II was not significantly changed (Fig. 3C). In LPS-treated miR-Let7A-overexpressing BV2 cells, LC3II level was higher than that in normal BV2 cells (Fig. 3C).
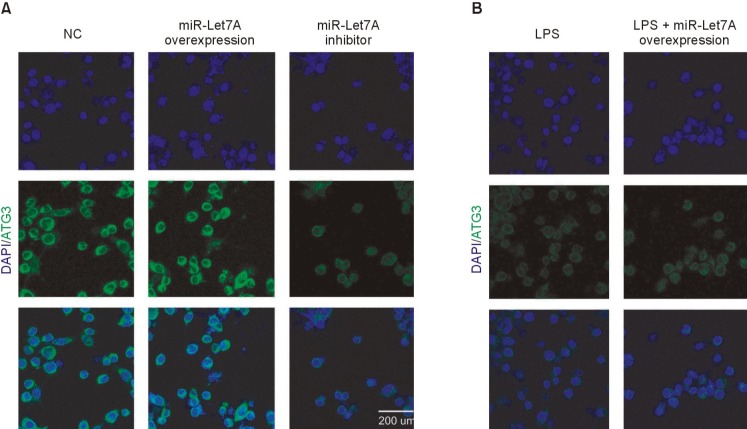
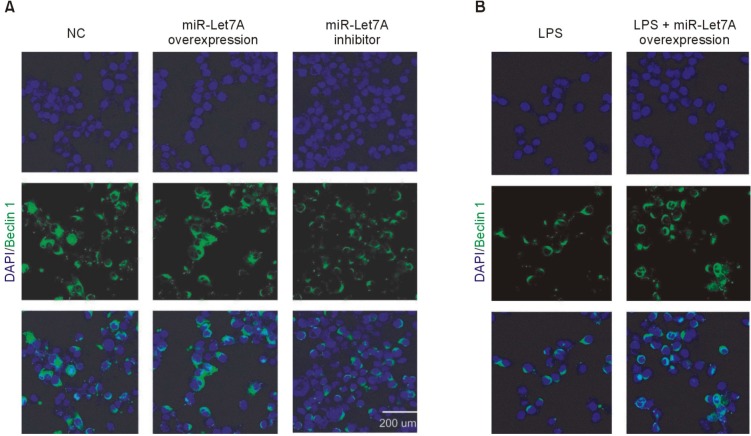
Immunocytochemical analyses were performed to visualize the miR-Let7A-dependent regulation of ATG3 (Fig. 4) and Beclin 1 (Fig. 5) expressions in a cellular level. The miR-Let7A overexpression in BV2 cells increased the expression of ATG3, whereas LPS treatment suppressed ATG3 in BV2 cells (Fig. 4A). Overexpression of miR-Let7A partially recovered LPS-induced reduced expression of ATG3 (Fig. 4B). The overexpression of miR-Let7A in LPS-stimulated BV2 cells also partially recovered LPS-induced reduced expression of Beclin 1 (Fig. 5B).
Fig. 4. Immunocytochemical analysis of ATG3 expression in miR-Let7A-transfected BV2 microglia activated by LPS. (A, B) Immunocytochemical images showing ATG3 immunoreactivity (green) in normal BV2 cells, BV2 cells transfected with miR-Let7A, BV2 cells treated with miR-Let7A inhibitor, BV2 cells treated with LPS (1 µg/ml), BV2 cells transfected with miR-Let7A and treated with LPS. miR-Let7A mimic was used at 20 nM and LPS was treated for 12 h. NC: normal control group, miR-Let7A overexpression: Let7A overexpression group, LPS: LPS treatment group, LPS+miR-Let7A overexpression: LPS treatment plus miR-Let7A overexpression group, miR-Let7A inhibitor: miR-Let7A suppression group. Scale bar: 200 µm, 4', 6-diamidino-2-phenylindole (DAPI): blue, ATG3: green.
Fig. 5. Immunocytochemical analysis of Beclin 1 expression in miR-Let7A-transfected BV2 microglia activated by LPS. (A, B) Immunocytochemical images showing Beclin 1 immunoreactivity (green) in normal BV2 cells, BV2 cells transfected with miR-Let7A, BV2 cells treated with miR-Let7A inhibitor, BV2 cells treated with LPS (1 µg/ml), BV2 cells transfected with miR-Let7A and treated with LPS. miR-Let7A mimic was used at 20 nM and LPS was treated for 12 h. NC: normal control group, miR-Let7A overexpression: Let7A overexpression group, LPS: LPS treatment group, LPS+miR-Let7A overexpression: LPS treatment plus miR-Let7A overexpression group, miR-Let7A inhibitor: miR-Let7A suppression group. Scale bar: 200 µm, 4', 6-diamidino-2-phenylindole (DAPI): blue, Beclin 1: green.
Considering that Beclin1 is crucial in the initiation and activation of autophagy [51,52] and ATG3 protein is essential in the formation of autophagic vesicles [53], these results suggest that miR-Let7A promotes the initiation and activation of autophagy in inflammation induced BV2 cells by increasing the level of Beclin 1 and ATG3.
DISCUSSION
Autophagy is an essential mechanism in degrading dysfunctional proteins and damaged mitochondria in cells [29,53,54,55,56,57]. Autophagy regulates inflammatory responses [58,59]. On the other hand, impairment in autophagy is harmful; it aggravates endoplasmic reticulum (ER) stress in cells [14,57,60] and is associated with neurodegeneration such as Alzheimer's disease [61,62,63]. Microtubule-associated protein LC3II is located on the internal surface of autophagosomes [14]. Several studies suggest that upregulation of LC3II increases the autophagic flux and early autophagosome formation, which requires also Beclin 1 [64,65]. LC3II levels are reduced in the brain of anti-miR-Let7 treated Huntington's disease mice [45]. These results, together with miR-Let7-dependent induction of LC3II in BV2 cells (Fig. 3), support the notion that miR-Let7A plays a role in autophagy induction in microglia.
Numerous studies have reported that Beclin1 is a key factor in the activation of the autophagy [51,52] and the initiation of autophagy by enhancing vesicle nucleation [66]. A pivotal role of Beclin1 in regulating autophagy in microglia has been described [67]. Considering that Beclin 1 is essential in autophagy [51,52], we speculate that miR-Let7A may boost the initiation and activation of microglial autophagy by up-regulating Beclin 1 against inflammatory stress. ATG3 is a component of autophagy-related ubiquitination-like systems [24] and functions as a regulator of cell survival [68] by generating the autophagic vesicle [53,69,70]. miR-Let7A partially increased ATG3 expression in LPS-treated BV2 cells (Fig. 3B). This result suggests that miR-Let7A promotes autophagy in microglia via up-regulating ATG3.
Disruption of autophagy flux contributes to neuronal cell death in Alzherimer's disease and Huntington disease [71,72]. Considering that several autophagic factors including LC3II, Beclin1, and ATG3 were regulated by miR-Let7A in activating microglia, miR-Let7A-depdent regulation of microglial autophagy needs be studied in more detail in the context and neuroinflammation and neurodegeneration.
ACKNOWLEDGEMENTS
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (NRF-2014R1A2A2A01006556).
References
- 1.Jiang T, Yu JT, Zhu XC, Wang HF, Tan MS, Cao L, Zhang QQ, Gao L, Shi JQ, Zhang YD, Tan L. Acute metformin preconditioning confers neuroprotection against focal cerebral ischaemia by pre-activation of AMPK-dependent autophagy. Br J Pharmacol. 2014;171:3146–3157. doi: 10.1111/bph.12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson MP, Shacka JJ. Autophagy modulation in disease therapy: where do we stand? Curr Pathobiol Rep. 2013;1:239–245. doi: 10.1007/s40139-013-0032-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Urbanek T, Kuczmik W, Basta-Kaim A, Gabryel B. Rapamycin induces of protective autophagy in vascular endothelial cells exposed to oxygen-glucose deprivation. Brain Res. 2014;1553:1–11. doi: 10.1016/j.brainres.2014.01.017. [DOI] [PubMed] [Google Scholar]
- 4.Oka T, Hikoso S, Yamaguchi O, Taneike M, Takeda T, Tamai T, Oyabu J, Murakawa T, Nakayama H, Nishida K, Akira S, Yamamoto A, Komuro I, Otsu K. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485:251–255. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green DR, Galluzzi L, Kroemer G. Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science. 2011;333:1109–1112. doi: 10.1126/science.1201940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korkaya H, Wicha MS. Inflammation and autophagy conspire to promote tumor growth. Cell Cycle. 2011;10:2623–2624. doi: 10.4161/cc.10.16.16414. [DOI] [PubMed] [Google Scholar]
- 7.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 8.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, Tanaka K, Kawai T, Tsujimura T, Takeuchi O, Yoshimori T, Akira S. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 9.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, Fitzgerald KA, Ryter SW, Choi AM. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puyal J, Clarke PG. Targeting autophagy to prevent neonatal stroke damage. Autophagy. 2009;5:1060–1061. doi: 10.4161/auto.5.7.9728. [DOI] [PubMed] [Google Scholar]
- 11.Nixon RA, Yang DS. Autophagy failure in Alzheimer's disease--locating the primary defect. Neurobiol Dis. 2011;43:38–45. doi: 10.1016/j.nbd.2011.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosello A, Warnes G, Meier UC. Cell death pathways and autophagy in the central nervous system and its involvement in neurodegeneration, immunity and central nervous system infection: to die or not to die--that is the question. Clin Exp Immunol. 2012;168:52–57. doi: 10.1111/j.1365-2249.2011.04544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meng Y, Yong Y, Yang G, Ding H, Fan Z, Tang Y, Luo J, Ke ZJ. Autophagy alleviates neurodegeneration caused by mild impairment of oxidative metabolism. J Neurochem. 2013;126:805–818. doi: 10.1111/jnc.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, Massey DC, Menzies FM, Moreau K, Narayanan U, Renna M, Siddiqi FH, Underwood BR, Winslow AR, Rubinsztein DC. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 15.Zhang T, Wang H, Li Q, Huang J, Sun X. Modulating autophagy affects neuroamyloidogenesis in an in vitro ischemic stroke model. Neuroscience. 2014;263:130–137. doi: 10.1016/j.neuroscience.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 16.Luo T, Park Y, Sun X, Liu C, Hu B. Protein misfolding, aggregation, and autophagy after brain ischemia. Transl Stroke Res. 2013;4:581–588. doi: 10.1007/s12975-013-0299-5. [DOI] [PubMed] [Google Scholar]
- 17.Xing S, Zhang J, Dang C, Liu G, Zhang Y, Li J, Fan Y, Pei Z, Zeng J. Cerebrolysin reduces amyloid-β deposits, apoptosis and autophagy in the thalamus and improves functional recovery after cortical infarction. J Neurol Sci. 2014;337:104–111. doi: 10.1016/j.jns.2013.11.028. [DOI] [PubMed] [Google Scholar]
- 18.Klionsky DJ. Neurodegeneration: good riddance to bad rubbish. Nature. 2006;441:819–820. doi: 10.1038/441819a. [DOI] [PubMed] [Google Scholar]
- 19.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 21.Nixon RA. Autophagy in neurodegenerative disease: friend, foe or turncoat. Trends Neurosci. 2006;29:528–535. doi: 10.1016/j.tins.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 22.Liu S, Sarkar C, Dinizo M, Faden AI, Koh EY, Lipinski MM, Wu J. Disrupted autophagy after spinal cord injury is associated with ER stress and neuronal cell death. Cell Death Dis. 2015;6:e1582. doi: 10.1038/cddis.2014.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Z, Zhong L, Zhong S, Xian R, Yuan B. Hypoxia induces microglia autophagy and neural inflammation injury in focal cerebral ischemia model. Exp Mol Pathol. 2015;98:219–224. doi: 10.1016/j.yexmp.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 25.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, Gruss P, Piacentini M, Chowdhury K, Cecconi F. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 26.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 29.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Satoh J, Motohashi N, Kino Y, Ishida T, Yagishita S, Jinnai K, Arai N, Nakamagoe K, Tamaoka A, Saito Y, Arima K. LC3, an autophagosome marker, is expressed on oligodendrocytes in Nasu-Hakola disease brains. Orphanet J Rare Dis. 2014;9:68. doi: 10.1186/1750-1172-9-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris J. Autophagy and cytokines. Cytokine. 2011;56:140–144. doi: 10.1016/j.cyto.2011.08.022. [DOI] [PubMed] [Google Scholar]
- 33.Harris J. Autophagy and IL-1 family cytokines. Front Immunol. 2013;4:83. doi: 10.3389/fimmu.2013.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal. 2014;20:460–473. doi: 10.1089/ars.2013.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho MH, Cho K, Kang HJ, Jeon EY, Kim HS, Kwon HJ, Kim HM, Kim DH, Yoon SY. Autophagy in microglia degrades extracellular β-amyloid fibrils and regulates the NLRP3 inflammasome. Autophagy. 2014;10:1761–1775. doi: 10.4161/auto.29647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, Small S, Spencer B, Rockenstein E, Levine B, Wyss-Coray T. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118:2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lucin KM, O'Brien CE, Bieri G, Czirr E, Mosher KI, Abbey RJ, Mastroeni DF, Rogers J, Spencer B, Masliah E, Wyss-Coray T. Microglial beclin 1 regulates retromer trafficking and phagocytosis and is impaired in Alzheimer's disease. Neuron. 2013;79:873–886. doi: 10.1016/j.neuron.2013.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gan J, Qu Y, Li J, Zhao F, Mu D. An evaluation of the links between microRNA, autophagy, and epilepsy. Rev Neurosci. 2015;26:225–237. doi: 10.1515/revneuro-2014-0062. [DOI] [PubMed] [Google Scholar]
- 39.Shi G, Shi J, Liu K, Liu N, Wang Y, Fu Z, Ding J, Jia L, Yuan W. Increased miR-195 aggravates neuropathic pain by inhibiting autophagy following peripheral nerve injury. Glia. 2013;61:504–512. doi: 10.1002/glia.22451. [DOI] [PubMed] [Google Scholar]
- 40.Song L, Su M, Wang S, Zou Y, Wang X, Wang Y, Cui H, Zhao P, Hui R, Wang J. MiR-451 is decreased in hypertrophic cardiomyopathy and regulates autophagy by targeting TSC1. J Cell Mol Med. 2014;18:2266–2274. doi: 10.1111/jcmm.12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar S, Gupta P, Khanal S, Shahi A, Kumar P, Sarin SK, Venugopal SK. Overexpression of microRNA-30a inhibits hepatitis B virus X protein-induced autophagosome formation in hepatic cells. FEBS J. 2015;282:1152–1163. doi: 10.1111/febs.13209. [DOI] [PubMed] [Google Scholar]
- 42.Stiuso P, Potenza N, Lombardi A, Ferrandino I, Monaco A, Zappavigna S, Vanacore D, Mosca N, Castiello F, Porto S, Addeo R, Prete SD, De Vita F, Russo A, Caraglia M. MicroRNA-423-5p promotes autophagy in cancer cells and is increased in serum from hepatocarcinoma patients treated with sorafenib. Mol Ther Nucleic Acids. 2015;4:e233. doi: 10.1038/mtna.2015.8. [DOI] [PubMed] [Google Scholar]
- 43.Wu D, Jiang H, Chen S, Zhang H. Inhibition of microRNA-101 attenuates hypoxia/reoxygenationinduced apoptosis through induction of autophagy in H9c2 cardiomyocytes. Mol Med Rep. 2015;11:3988–3994. doi: 10.3892/mmr.2015.3215. [DOI] [PubMed] [Google Scholar]
- 44.Chen XM, Splinter PL, O'Hara SP, LaRusso NF. A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J Biol Chem. 2007;282:28929–28938. doi: 10.1074/jbc.M702633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dubinsky AN, Dastidar SG, Hsu CL, Zahra R, Djakovic SN, Duarte S, Esau CC, Spencer B, Ashe TD, Fischer KM, MacKenna DA, Sopher BL, Masliah E, Gaasterland T, Chau BN, Pereira de, Morrison BE, La Spada AR. Let-7 coordinately suppresses components of the amino acid sensing pathway to repress mTORC1 and induce autophagy. Cell Metab. 2014;20:626–638. doi: 10.1016/j.cmet.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jewell JL, Russell RC, Guan KL. Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol. 2013;14:133–139. doi: 10.1038/nrm3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding Z, Wang X, Schnackenberg L, Khaidakov M, Liu S, Singla S, Dai Y, Mehta JL. Regulation of autophagy and apoptosis in response to ox-LDL in vascular smooth muscle cells, and the modulatory effects of the microRNA hsa-let-7 g. Int J Cardiol. 2013;168:1378–1385. doi: 10.1016/j.ijcard.2012.12.045. [DOI] [PubMed] [Google Scholar]
- 48.Ding ZB, Shi YH, Zhou J, Qiu SJ, Xu Y, Dai Z, Shi GM, Wang XY, Ke AW, Wu B, Fan J. Association of autophagy defect with a malignant phenotype and poor prognosis of hepatocellular carcinoma. Cancer Res. 2008;68:9167–9175. doi: 10.1158/0008-5472.CAN-08-1573. [DOI] [PubMed] [Google Scholar]
- 49.Song J, Cheon SY, Jung W, Lee WT, Lee JE. Resveratrol induces the expression of interleukin-10 and brain-derived neurotrophic factor in BV2 microglia under hypoxia. Int J Mol Sci. 2014;15:15512–15529. doi: 10.3390/ijms150915512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scarlatti F, Maffei R, Beau I, Codogno P, Ghidoni R. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ. 2008;15:1318–1329. doi: 10.1038/cdd.2008.51. [DOI] [PubMed] [Google Scholar]
- 52.Tian S, Lin J, Jun Zhou J, Wang X, Li Y, Ren X, Yu W, Zhong W, Xiao J, Sheng F, Chen Y, Jin C, Li S, Zheng Z, Xia B. Beclin 1-independent autophagy induced by a Bcl-XL/Bcl-2 targeting compound, Z18. Autophagy. 2010;6:1032–1041. doi: 10.4161/auto.6.8.13336. [DOI] [PubMed] [Google Scholar]
- 53.Mizushima N. Autophagy in protein and organelle turnover. Cold Spring Harb Symp Quant Biol. 2011;76:397–402. doi: 10.1101/sqb.2011.76.011023. [DOI] [PubMed] [Google Scholar]
- 54.Hung TH, Hsieh TT, Chen SF, Li MJ, Yeh YL. Autophagy in the human placenta throughout gestation. PLoS One. 2013;8:e83475. doi: 10.1371/journal.pone.0083475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu Y, Yuan J, Lipinski MM. Live imaging and single-cell analysis reveal differential dynamics of autophagy and apoptosis. Autophagy. 2013;9:1418–1430. doi: 10.4161/auto.25080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 57.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 58.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13:722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 60.Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, Shiosaka S, Hammarback JA, Urano F, Imaizumi K. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Héraud C, Griffiths A, Pandruvada SN, Kilimann MW, Pata M, Vacher J. Severe neurodegeneration with impaired autophagy mechanism triggered by ostm1 deficiency. J Biol Chem. 2014;289:13912–13925. doi: 10.1074/jbc.M113.537233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 63.Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 64.Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, Green DR. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 65.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 66.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.O'Brien CE, Wyss-Coray T. Sorting through the roles of beclin 1 in microglia and neurodegeneration. J Neuroimmune Pharmacol. 2014;9:285–292. doi: 10.1007/s11481-013-9519-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Criollo A, Chereau F, Malik SA, Niso-Santano M, Mariño G, Galluzzi L, Maiuri MC, Baud V, Kroemer G. Autophagy is required for the activation of NFκB. Cell Cycle. 2012;11:194–199. doi: 10.4161/cc.11.1.18669. [DOI] [PubMed] [Google Scholar]
- 69.Ngu M, Hirata E, Suzuki K. Visualization of Atg3 during autophagosome formation in Saccharomyces cerevisiae. J Biol Chem. 2015;290:8146–8153. doi: 10.1074/jbc.M114.626952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oral O, Oz-Arslan D, Itah Z, Naghavi A, Deveci R, Karacali S, Gozuacik D. Cleavage of Atg3 protein by caspase-8 regulates autophagy during receptor-activated cell death. Apoptosis. 2012;17:810–820. doi: 10.1007/s10495-012-0735-0. [DOI] [PubMed] [Google Scholar]
- 71.Schaeffer V, Lavenir I, Ozcelik S, Tolnay M, Winkler DT, Goedert M. Stimulation of autophagy reduces neurodegeneration in a mouse model of human tauopathy. Brain. 2012;135:2169–2177. doi: 10.1093/brain/aws143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rusten TE, Stenmark H. p62, an autophagy hero or culprit? Nat Cell Biol. 2010;12:207–209. doi: 10.1038/ncb0310-207. [DOI] [PubMed] [Google Scholar]