Abstract
Genome size is of fundamental biological importance with significance in predicting structural and functional attributes of organisms. Although abundant evidence has shown that the genome size can be largely explained by differential proliferation and removal of non-coding DNA of the genome, the evolutionary and ecological basis of genome size variation remains poorly understood. Nitrogen (N) and phosphorus (P) are essential elements of DNA and protein building blocks, yet often subject to environmental limitation in natural ecosystems. Using phylogenetic comparative methods, we test this hypothesis by determining whether leaf N and P availability affects genome sizes in 99 species of Primulina (Gesneriaceae), a group of soil specialists adapted to limestone karst environment in south China. We find that genome sizes in Primulina are strongly positively correlated with plant N content, but the correlation with plant P content is not significant when phylogeny history was taken into account. This study shows for the first time that N limitation might have been a plausible driver of genome size variation in a group of plants. We propose that competition for nitrogen nutrient between DNA synthesis and cellular functions is a possible mechanism for genome size evolution in Primulina under N-limitation.
Genome size, measured as the haploid nuclear DNA content (1C-value), is of fundamental biological importance with implications for predicting structural and functional attributes of organisms1,2,3. As one of most variable traits of biodiversity, C-value varies over 2400-fold among species of angiosperm plants and has been a longstanding puzzle in evolutionary biology4,5. Although abundant evidence has shown that the C-value enigma can be largely explained by differential proliferation and removal of non-coding DNA of the genome6,7,8, the evolutionary and ecological basis for genome size variation remains poorly understood and highly controversial9,10,11. Nitrogen (N) and phosphorus (P) are essential components for production of nucleic acids and protein, but they are often subjected to ecological limitation in most natural ecosystems. Thus, a biological stoichiometry approach should enable us to unravel the connections between genome and the ecological interactions among organisms and their environments.
Recent biological stoichiometry studies have shown that environmental N-limitation can affect nucleotide composition of an organismal genome12. For example, the genome-wide comparison of nitrogen element between wild and crop plants revealed that the wild Arabidopsis thaliana use less nitrogen in their genomes and proteins than crops, which was attributed to ecological nitrogen limitation in A. thaliana13. Thus, it is conceivable that nutrient limitation may not only influence the molecular composition, but also the size, of the genome (i.e. the amount of DNA) of organisms in resource-limited habitats. No explicit tests of this hypothesis exist, although a few studies support some of the main assumptions. Small genome sizes are frequently reported in carnivorous plants, whose growth is generally both N and P co-limited14. For example, genome sequencing revealed a minute genome size of 82 Mb for the carnivorous plant Utricularia gibba living in P-poor freshwater habitats15. The recently proposed ‘growth rate-genome size-nutrient limitation’ hypothesis (GGNH) predicted that the reallocation of P from DNA to RNA under the selection favoring rapid growth in nutrient-limited environments could lead to genome downsizing in eukaryotes16. This hypothesis received experimental supports from a few studies on zooplanktonic groups such as Caenis spp. and Daphnia spp.17,18,19, but no explicit experimental data from plants available to test the hypothesis. In fact, the relationship between genome size and growth rate in plants has remained controversial20,21. Nevertheless, a long-term fertilization experiment in local plant communities has shown that soil P availability may play a role in selection of plants with different genome sizes22. A recent comparative analysis revealed a tendency of genome size expansion from non-geophytes to their geophytic relatives, for which the storage organs were assumed to be serviced as nutrition reserves and therefore make geophytes relatively independent of the nutrient availability in their environment23. Yet, no direct evidence of nitrogen limitation on genome size variation has been reported in plants.
The limestone karsts in Southeast Asia harbor a highly diverse and unique biota, and have long been regarded as “natural laboratories” for ecological and evolutionary research addressing natural selection24. Soils in the karst area are typically shallow and characterized by low soil water content, periodic water deficiency, and N limitation25, which exert strong selective forces on plant evolution, resulting in remarkably high species richness and endemism in the region. Primulina, a genus of the African violet family Gesneriaceae, is a monophyletic group comprising more than 140 species of perennials that are widely distributed throughout the karst regions of China and adjacent countries of Southeast Asia26. The distribution of the genus spans a wide latitudinal range (18 °N–31 °N) and most species occur only in calcareous soil associated habitats developing from limestone bedrock (Fig. 1). A recent large-scale analysis of genome size for 101 species revealed variation up to 2.27-fold differences of the 2C DNA content among species although chromosome numbers were constant (2n = 36)27. The rich species diversity and soil-habitat specialization together with a wide range in genome sizes make Primulina an excellent model for studying the impact of nutrient limitation on genome size variation in terrestrial plants.
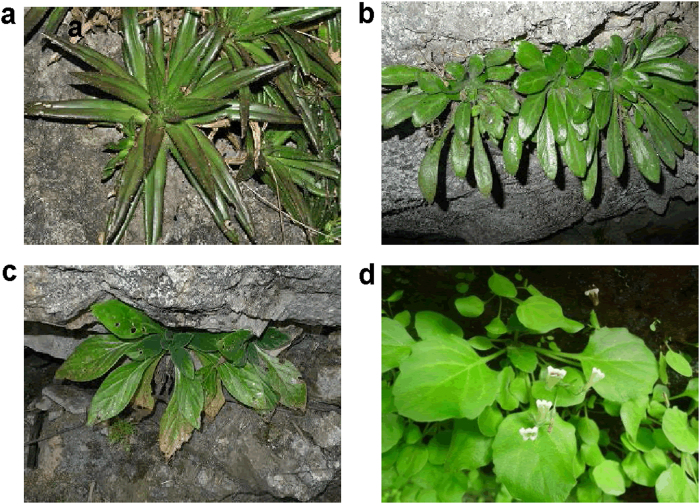
Figure 1. Representative Primulina species growing on limestone karsts with poor soil development.
The order (a–d) shows a gradient of increased genome size: (a) P. spinulosa (2C = 1.47 pg); (b) P. yangchunensis (2C = 1.76 pg); (c) P. eburnea (2C = 2.03 pg) and (d) P. xiuningensis (2C = 2.39 pg).
Phylogeny-based comparative analysis of the genome size in Primulina detected significant positive relationships between genome size and specific leaf area (SLA) as well as between genome size and latitudes, indicating adaptive evolution of genome size in this genus27. We hypothesize that the adaptive genome size in Primulina is likely driven by selection pressure of nutrient limitation towards “efficient” genomes for survival in response to harsh karst habitats. We test this hypothesis by exploring the relationship between genome size and leaf C/N/P content and their respective ratios for 99 species of Primulina with consideration of their phylogenetic history. We find that genome sizes in Primulina are strongly positively correlated with leaf N content. This study represents, to the best of our knowledge, the first test of the hypothesis that nutrient limitation drives genome size evolution in a group of phylogenetically closely related species.
Results
Leaf N and P concentrations and N : P ratio varied greatly across species, with a range of 5.38–38.38 mg g−1 for N, 0.46–5.12 mg g−1 for P and 3.82–41.24 for N : P ratio (Table 1). The average values were 14.65 mg g−1 for N, 1.45 mg g−1 for P and 11.46 mg g−1 for N : P. Leaf N concentration and C : N ratio exhibited a significant amount of phylogenetic signal (λ = 0.759–0.776, P < 0.0001; Table 1), implying strong phylogenetic dependence of these traits. The estimates of λ for leaf P and C : P were only marginally significant (P < 0.048), whereas leaf C and N : P showed no significant phylogenetic signal (P > 0.556).
Table 1. Statistics summary of leaf nutrient contents (C, N, P) and C : N : P ratio of 99 Primulina species analyzed in this study.
| Mean | SD | CV | Minimum | Maximum | λ | P | |
|---|---|---|---|---|---|---|---|
| C (mg g−1) | 357.97 | 18.39 | 0.05 | 315.43 | 425.66 | 0.137 | 0.556 |
| N (mg g−1) | 14.65 | 5.78 | 0.39 | 5.38 | 38.38 | 0.759 | <0.0001 |
| P (mg g−1) | 1.45 | 0.78 | 0.53 | 0.46 | 5.12 | 0.234 | 0.048 |
| C : N | 28.18 | 11.08 | 0.39 | 8.72 | 66.77 | 0.776 | <0.0001 |
| N : P | 11.46 | 5.09 | 0.44 | 3.82 | 41.24 | 0 | 1 |
| C : P | 308.11 | 142.74 | 0.46 | 74.28 | 772.93 | 0.295 | 0.018 |
Mean, standard deviation (SD), coefficient of variation (CV), minimum and maximum of leaf element concentrations (mg g−1 dry mass), Pagel’s λ and probability (P) testing for phylogenetic signal. Statistically significant values are in bold (P < 0.05).
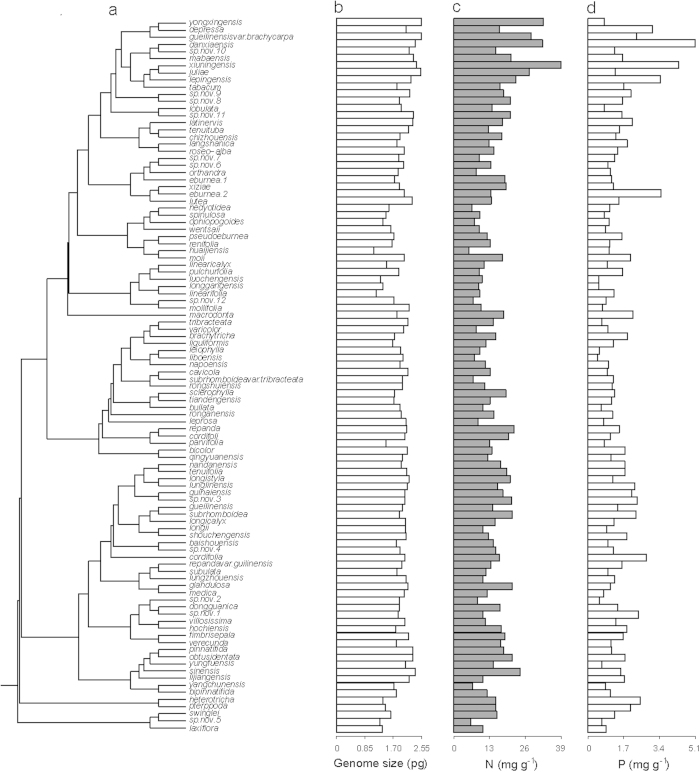
Figure 2 shows the phylogenetic tree of Primulina species with genome size and leaf concentrations of N and P. Genome size was significantly and positively correlated with leaf N concentration and negatively correlated with leaf C : N ratio under both phylogenetic generalized least squares (PGLS) and ordinary least-square (OLS, i.e. nonphylogenetic regression) regressions (Fig. 3, Table S2). Although the PGLS yielded a weaker explanatory power (adjusted R2 = 0.098–0.120) than OLS (adjusted R2 = 0.290–0.299) (Fig. 3, Table S2), the likelihood ratio tests (LRT) showed that the PGLS model fit the data better than the OLS model in both comparisons (Table S2). Because the leaf C content is similar between species (Table 1), the negative correlation of genome size with leaf C : N ratio is likely to be driven by leaf N. Under the PGLS model, leaf N was significantly correlated with latitudinal climate variation, but more tightly with temperature-related than precipitation-related variables (Table S3). Although we found significant relationships between genome size and leaf P and leaf C : P ratios under the OLS model, such relationships were driven by phylogenetic non-independence and disappeared under PGLS model (Table S2).
Figure 2.
A phylogenetic tree of 99 species of Primulina (a) with their genome size (b), leaf N (c) and P (d) concentrations. The phylogenetic tree was adopted from Kang et al. (2014).
Figure 3.
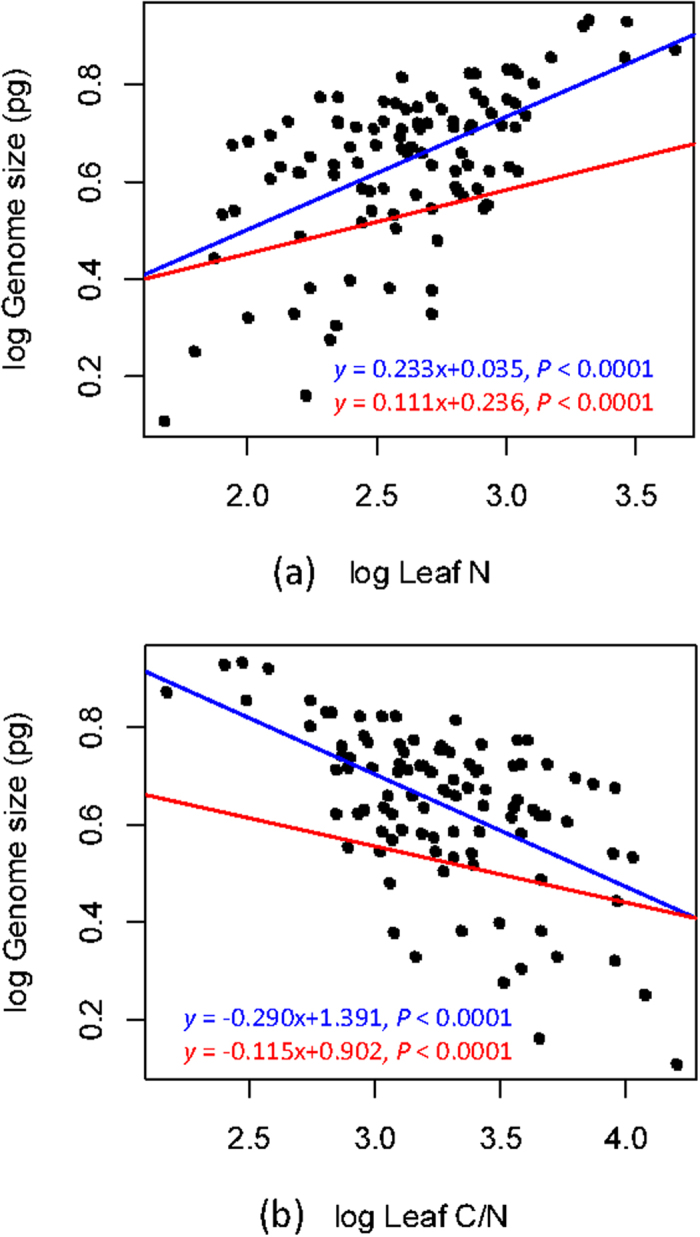
Genome size (2C DNA content) in relation to (a) leaf N concentration and (b) ratio of C:N under regression models of ordinary least squares (OLS; blue) and phylogenetic generalized least squares (PGLS; red).
Discussion
The evolution of genome sizes is probably due to multiple interdependent mechanisms28. Nutrient limitation has been proposed as one such mechanism for plants and invertebrates that need high demands of N and P for growth16. The average leaf N concentration in Primulina (14.65 mg g−1) was much lower than that reported in a study of 1900 plant species across China (22.3 mg g−1)29 and usually well below concentrations that were thought to limit growth30. However, leaf P concentration fell into the general range reported in other plants (1.68 mg g−1)29, and thus resulting in relatively low mean N : P ratio (11.46) (Table 1). According to the criterion of N limitation at N < 20 mg g−1 and N : P < 1430,31, the growth of Primulina appears to be N limited. The strong correlation of genome size with leaf N concentration suggests that N limitation may have been a driving force of genome size variation in Primulina.
To date, the only two experimental studies have found contrasting influence of P availability on plant genome size22,32. A recent study in the Mediterranean region revealed that P availability in soil or plant has not affected genome size stability32, while a long-term (60 years) fertilization experiment shown that soil P availability may play a role in selection of plants with different genome sizes22. It is most likely that variation in sensitivity to P availability among lineages may result in taxon-specific effect on genome size. In our study, we investigated the relationships between genome size and nutrient availability with two different methods: the nonphylogenetic OLS and phylogenetic generalized least-squares (PGLS). Our likelihood ratio tests (LRT) showed that the PGLS model fits the data better than the OLS model. Under PGLS model, there was no significant correlation between plant P concentration and genome size, suggesting P availability is not a determining factor for genome size variation in Primulina.
It is widely recognized that the elemental demands of N and P for producing nucleic acid are highly costly33. Assuming a 1:1 ratio for purines and pyrimidines, N and P comprise on average approximately 39% and 9% of nucleic acid mass, respectively12. Therefore, the availability of N and P could have a direct influence on nucleic acid synthesis. The ‘growth rate-genome size-nutrient limitation’ hypothesis (GGNH)16 suggests that the reallocation of P (and eventually N) from DNA to RNA synthesis could be under selection in nutrient-limited environments. This hypothesis predicts that species with small genome sizes would have faster growth rates, however this hypothesis is based on a few observations in animals16, so far no experimental test has been attempted in plants. Our field investigations found that Primulina species with small genome sizes usually occur in more harsh southern regions and show slow growth grate. Consistent with the ‘stress resistance syndrome’ (SRS)34, these observations in Primulina may represent an adaptive strategy enabling lineages to persist in the harsh karst environment. Primulina usually grows on shallow, nutrient poor and limestone-rocky soils, which are frequently subject to rapid water loss and drought induced by heat stress, and it is difficult for plants to acquire the necessary resources for fast growth. Therefore, the observed influence of N limitation on genome size in Primulina cannot be explained by the hypothesis of reallocation of N from DNA to RNA synthesis.
We propose an alternative mechanism for genome size evolution in Primulina under N-limitation: competition for nitrogen nutrient between DNA synthesis and cellular functions. Nitrogen is critically important for plants because it is needed in relatively large quantities not only for growth but also for other functions, such as storage, defense, and mechanisms of stress resistance. Recent studies have found that N limitation has caused quantitative and qualitative shifts in the composition of amino acids35,36, providing evidence that plants are able to mobilize N to optimize N metabolism in response to N limitation. Glutamic acid (Glu), which plays a major role in DNA synthesis and as a donor for the synthesis of other amino acids, was found to be significantly reduced under low-N conditions36. Similarly, nitrogen partitioning in oaks revealed both an increase in leaf soluble protein and free amino acid concentrations at the expense of N used for structural components37. It seems that there may be a tradeoff between investment of N in amino acids for plant defense functions and amino acids used for DNA synthesis as an adaptive evolutionary strategy under abiotic stress. N is also an essential component for many classes of metabolites such as alkaloids, cyanogenic glucosides and non-protein amino acids. All of them have been reported to accumulate in plants suffering environmental stresses38,39. In addition, plants normally produce a large variety of phenolic compounds as responses to abiotic and biotic stressors. Increasing stress or decreasing nutrient availability should result in higher production of phenolic compounds, through the competition between protein and phenolic synthesis for their main common precursor, phenylalanine (Phe)40. Thus, N limitation should be a key factor linking the competition between DNA synthesis and amino acids, protein and secondary metabolites, because of N-derived precursors. We believe Primulina, with its smaller genome size in oligotrophic karst habitats, would be subject to N reallocation to amino acids and/or secondary metabolites as an adaptive response. However, the hypothesis should be further tested through quantifying N partitioning among DNA/RNA, protein, amino acids and secondary metabolites.
Methods
Sample collection
The plant sampling has been described in a previous study on genome size and specific leaf area (SLA)27. Briefly, we designated populations as the sampling units, for each population, five adult plants were randomly excavated for element analysis. In total, we used materials of 100 populations of 99 species of Primulina (Fig. S1) collected for a previous analysis of genome size and SLA27. In this study, chemical analyses were performed on dried leaves previously used for SLA calculation. For each species, one population was chosen with the exception for P. eburnea, the most widely distributed species that separated into two clades in the phylogeny, and therefore two populations for this species were used. Since most species are narrow endemics and commonly found in single-site limestone karst habitats, we believe such a sampling design adequately reflects variation within the species.
Chemical analyses
Leaves of the five individuals were washed with double-distilled water and dried at 60°C to constant weight, then ground to a fine powder (2-mm-mesh pore size) and stored in plastic vials for later analyses. Concentrations of leaf C and N were analyzed using a PDZ Europa ANCA-GSL elemental analyzer interfaced to a PDZ Europa 20-20 isotope ratio mass spectrometer (Sercon Ltd., Cheshire, UK). All the analyses were carried out at the University of California (Davis) stable isotope facility. The total P concentrations in leaves were determined via inductively coupled plasma optical emission spectrometry (ICP-OES) following digestion in Teflon beakers with a mixture of ultrapure concentrated HNO3 + HClO4 (4:1 v/v). The solutions were evaporated to dryness, redissolved in HNO3, diluted with 50 mL of MilliQ water and then filtered through a 0.45-mm cellulose acetate membrane (Millipore, Billerica, MA, USA) for analysis. The precision and accuracy of the measurements were controlled through repeated analysis of National Standard Reference materials consisting of poplar leaves (GBW 07604) and agricultural soils (GBW E 070045), respectively.
Data analysis
The descriptive statistics of leaf elements and their ratios were conducted using JMP pro 10 (SAS Institute Inc., Cary, NC). Based on the uniformity in chromosome numbers in the genus, we interpreted 2C DNA content as genome size throughout the article. Throughout this study, genome size data (2C DNA content) of Primulina species were taken from a previous study27. The analysis of phylogenetic signal of leaf traits was based on a previously published phylogenetic tree of 104 species of Primulina27. In this study, we treated all variables as continuous traits. Pagel’s λ41 measures the phylogenetic signals in relation to leaf traits. The parameter λ assesses the contribution of phylogeny to the covariance among species for a given trait. If there is no phylogenetic signal, λ will be close to zero, indicating that trait evolution has proceeded independent of the phylogeny. If λ = 1, then the trait has exactly the amount of signal expected for the phylogenetic tree employed and a model of evolution based on a random walk (Brownian motion). Intermediate values of 0 < λ < 1 indicate different degrees of phylogenetic signals. The analyses were carried out using species means with the R package ‘caper’42. For each analysis, the significance of the phylogenetic signal was assessed by estimating the likelihood under λ = 0 and λ = 1 via likelihood ratio tests (LRT)43.
We examined the relationships between genome size and leaf nutrient concentrations using a phylogenetic generalized least-squares (PGLS) approach44, in which phylogenetic regression was performed with a phylogenetic tree whose internal branches were all multiplied by λ, leaving the tip branches at their original length. In this approach, when λ is forced to 0, it is equivalent to ordinary (nonphylogenetic) least-squares regression (OLS), which assumes a star –shaped phylogeny in which residual variation is independent among species. In this study, the R package ‘caper’42 was used to compute both types of regression models with λ forced to equal 0 and estimated values, with genome size as dependent variables and leaf nutrient concentrations as independent variable. Likelihood ratio tests (LRT) were used to assess which model had the best fit.
To explore the influence of environmental conditions on leaf nutrients, PGLS was used to assess the relationship between leaf concentrations of N and P and a suit of factors of climatic variables. We extracted 19 bioclimatic variables for average georeferenced records per species from the WorldClim climate database (at 2.5 min scale; http://worldclim.org/bioclim)45. These bioclimatic variables represent summaries of temperature and precipitation dimensions of environment.
Additional Information
How to cite this article: Kang, M. et al. Nitrogen limitation as a driver of genome size evolution in a group of karst plants. Sci. Rep. 5, 11636; doi: 10.1038/srep11636 (2015).
Supplementary Material
Acknowledgments
We thank Y.M. Buckley and J. Xiang for comments on the manuscript drafts. This work was supported by Natural Science Foundation of China (31270427; 31370366).
Footnotes
Author Contributions M.K. and H.W.H. designed the work and wrote the main manuscript text. M.K. collected the sample. J.W. conducted laboratory experiments. M.K. analyzed the data. All the authors have reviewed and approved the manuscript.
References
- Grime J. P. & Mowforth M. A. Variation in genome size–an ecological interpretation. Nature 299, 151–153 (1982). [Google Scholar]
- Jasienski M. & Bazzaz F. A. Genome size and high CO2. Nature 376, 559–560 (1995). [Google Scholar]
- MacGillivray C. W. & Grime J. P. Genome size predicts frost resistance in British herbaceous plants: implications for rates of vegetation response to global warming. Funct. Ecol. 9, 320–325 (1995). [Google Scholar]
- Bennett M. D. & Leitch I. J. Plant genome size research: A field in focus. Ann. Bot. 95, 1–6 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M. D. & Leitch I. J. Nuclear DNA amounts in angiosperms: targets, trends and tomorrow. Ann. Bot. 107, 467–590 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M. T. Transposable element number in mixed mating populations. Genet. Res. 77, 261–275 (2001). [DOI] [PubMed] [Google Scholar]
- Wendel J. F., Cronn R. C., Johnston J. S. & Price H. J. Feast and famine in plant genomes. Genetica 115, 37–47 (2002). [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Ma J. X. & Devos K. Mechanisms of recent genome size variation in flowering plants. Ann. Bot. 95, 127–132 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov D. A. Mutational equilibrium model of genome size evolution. Theor. Pop. Bio. 61, 531–544 (2002). [DOI] [PubMed] [Google Scholar]
- Lynch M. & Conery J. S. The origins of genome complexity. Science 302, 1401–1404 (2003). [DOI] [PubMed] [Google Scholar]
- Vinogradov A. E. Compactness of human housekeeping genes: selection for economy or genomic design? Trends Genet. 20, 248–253 (2004). [DOI] [PubMed] [Google Scholar]
- Elser J. J., Acquisti C. & Kumar S. Stoichiogenomics: the evolutionary ecology of macromolecular elemental composition. Trends Ecol. Evol. 26, 38–44 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acquisti C., Elser J. J. & Kumar S. Ecological nitrogen limitation shapes the DNA composition of plant genomes. Mol. Bio. Evol. 26, 953–956 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison A. M. Nutrient limitation and stoichiometry of carnivorous plants. Plant Biol. 8, 740–747 (2006). [DOI] [PubMed] [Google Scholar]
- Ibarra-Laclette E. et al. Architecture and evolution of a minute plant genome. Nature 498, 94–98 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessen D. O., Jeyasingh P. D., Neiman M. & Weider L. J. Genome streamlining and the elemental costs of growth. Trends Ecol. Evol. 25, 75–80 (2010). [DOI] [PubMed] [Google Scholar]
- Frost P. C. & Elser J. J. Growth responses of littoral mayflies to the phosphorus content of their food. Ecol Lett. 5, 232–240 (2002). [Google Scholar]
- Weider L. J., Glenn K. L., Kyle M. & Elser J. J. Associations among ribo-somal (r) DNA intergenic spacer length, growth rate, and C: N: P stoichiometry in the genus Daphnia. Limnol. Oceanogr. 49, 1417–1423 (2004). [Google Scholar]
- Hessen D. O., Ventura M. & Elser J. J. Do phosphorus requirements for RNA limit genome size in crustacean zooplankton? Genome 51, 685–691 (2008). [DOI] [PubMed] [Google Scholar]
- Knight C. A., Molinari N. A. & Petrov D. A. The large genome constraint hypothesis: Evolution, ecology and phenotype. Ann. Bot. 95, 177–190 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Símová I. & Herben T. Geometrical constraints in the scaling relationships between genome size, cell size and cell cycle length in herbaceous plants. P. Roy. Soc. B: Bio. Sci. 279, 867–875 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šmarda P. et al. Effect of phosphorus availability on the selection of species with different ploidy levels and genome sizes in a long-term grassland fertilization experiment. New Phytol. 200, 911–921 (2013). [DOI] [PubMed] [Google Scholar]
- Veselý P., Bureš P. & Šmarda P. Nutrient reserves may allow for genome size increase: evidence from comparison of geophytes and their sister non-geophytic relatives. Ann. Bot. 112, 1193–1200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements R., Sodhi N. S., Schilthuizen M. & Ng P. K. L. Limestone Karsts of southeast Asia: Imperiled arks of biodiversity. Bioscience 56, 733–742 (2006). [Google Scholar]
- Yuan D. X. Karst of China, Geological Publishing House, Beijing (1991). [Google Scholar]
- Weber A., Clark J. L. & Möller M. A new formal classification of Gesneriaceae, Selbyana 31, 68–94 (2013). [Google Scholar]
- Kang M. et al. Adaptive and nonadaptive genome size evolution in Karst endemic flora of China. New Phytol. 202, 1371–1381 (2014). [DOI] [PubMed] [Google Scholar]
- Petrov D. A. Evolution of genome size: new approaches to an old problem. Trends Genet. 17, 23–28 (2001). [DOI] [PubMed] [Google Scholar]
- Han W. X., Fang J. Y., Reich P. B., Woodward F. I. & Wang Z. H. Biogeography and variability of eleven mineral elements in plant leaves across gradients of climate, soil and plant functional type in China. Ecol. Lett. 14, 788–796 (2011). [DOI] [PubMed] [Google Scholar]
- Aerts R. & Chapin F. S. The mineral nutrition of wild plants revisited: a re-evaluation of processes and patterns. Adv. Ecol. Res. 30, 1–67 (2000). [Google Scholar]
- Koerselman W. & Meuleman A. F. W. The vegetation N: P ratio: a new tool to detect the nature of nutrient limitation. J. Appl. Ecol. 33, 1441–1450 (1996). [Google Scholar]
- Pellicer J., Fay M. F. & Leitch I. J. The largest eukaryotic genome of them all? Bot. J. Linn. Soc. 164, 10–15 (2010). [Google Scholar]
- Elser J. J., Dobberfuhl D. R., MacKay N. A. & Schampel J. H. Organism size, life history, and N: P stoichiometry: towards a unified view of cellular and ecosystem processes. BioScience 46, 674–684 (1996). [Google Scholar]
- Chapin F. S., Autumn K. & Pugnaire F. Evolution of suites of traits in response to environmental stress. Am. Nat. 142, S78–S92 (1993). [Google Scholar]
- Ikram S., Bedu M., Daniel-Vedele F., Chaillou S. & Chardon F. Natural variation of Arabidopsis response to nitrogen availability. J Exp. Bot. 63, 91–105 (2012). [DOI] [PubMed] [Google Scholar]
- Schlüter U. et al. Maize source leaf adaptation to nitrogen deficiency affects not only nitrogen and carbon metabolism but also control of phosphate homeostasis. Plant Physiol. 160, 1384–1406 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B. et al. Nitrigen partitioning in oak leaves depends on species, provenance, climate conditions and soil type. Plant Bio. 15, 198–209 (2013). [DOI] [PubMed] [Google Scholar]
- Ahuja I., de Vos R. C., Bones A. M. & Hall R. D. Plant molecular stress responses face climate change. Trends Plant Sci. 15, 664–674 (2010). [DOI] [PubMed] [Google Scholar]
- Selmar D. & Kleinwächter M. Stress enhances the synthesis of secondary plant products: the impact of stress-related over-reduction on the accumulation of natural products. Plant Cell Physiol. 54, 817–826 (2013). [DOI] [PubMed] [Google Scholar]
- Jones C. G. & Hartley S. E. A protein competition model of phenolic allocation. Oikos 86, 27–44 (1999). [Google Scholar]
- Pagel M. Inferring the historical patterns of biological evolution. Nature 401, 877–884 (1999). [DOI] [PubMed] [Google Scholar]
- Orme D. et al. Caper: Comparative analyses of phylogenetics and evolution in R. Methods Ecol. Evol. 3, 145–151 (2012). [Google Scholar]
- Huelsenbeck J. P. & Rannala B. Phylogenetic methods come of age: testing hypotheses in an evolutionary context. Science 276, 227–232 (1997). [DOI] [PubMed] [Google Scholar]
- Revell L. J. Phylogenetic signal and linear regression on species data. Methods Ecol. Evol. 1, 319–329 (2010). [Google Scholar]
- Hijmans R. J., Cameron S. E., Parra J. L., Jones P. G. & Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978 (2005). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.