Abstract
Membrane curvature and lipid composition plays critical role in interchanging of matter and energy in cells. Peptide curvature sensors are known to activate signaling pathways, and promote molecular transport across cell membranes. Recently, the 25-mer MARCKS-ED peptide, which is derived from the effector domain of the myristoylated alanine-rich C kinase substrate protein, has been reported to selectively recognize highly curved membrane surfaces. Our previous studies indicated that the naturally occurring L-MARCKS-ED peptide could simultaneously detect both phosphatidylserine (PS) and curvature. Here, we demonstrate that D-MARCKS-ED, composed by unnatural D-amino acids, has the same activities as its enantiomer, LMARCKS-ED, as a curvature and lipid sensor. An atomistic molecular dynamics (MD) simulation suggests that D-MARCKS-ED may change from linear to a boat conformation upon binding to the membrane. Comparable enhancement of fluorescence intensity was observed between D- and L- MARCKS-ED peptides, indicating similar binding affinities. Meanwhile, circular dichroism (CD) spectra of D- and L- MARCKS-ED are almost symmetrical both in the presence and absence of liposomes. These results suggest similar behavior of artificial D- and natural L- MARCKS-ED peptides when binding to curved membranes. Our studies may contribute to further understanding of how MARCKS-ED senses membrane curvature, as well as provide a new direction to develop novel membrane curvature probes.
INTRODUCTION
It has long been thought that the shape of the cell membrane is a passive characteristic of the bilayer. However, recent investigations have uncovered the active role of membrane curvature in controlling cellular organization and activity [1-6]. The shape of the membrane plays an important role in cell signaling and trafficking [7-9] and certain proteins are known to aggregate on curved membranes or generate them [9-15]. Included among proteins that are shown to alter the curvature of membrane bilayers are: the C2B domain of Synaptotagmin-I [16], the endocytosis-associated proteins epsin [17] and dynamin [18], the DP1/Yop1p protein that is associated with the endoplasmic reticulum [19], the Golgi-associated ArfGAP1 lipid packing sensor (ALPS) [20], and the Bin-Amphyphysin-Rvs (BAR) domain of amphiphysin [21]. In addition, highly curved bilayer assemblies have also recently been found to perform cellular-signaling functions that have implications on health and disease [22-25]. These structures, known as exosomes and microvesicles that range in size from 30-1000 nm, have been implicated for applications in cancer [26-31], renal disease [32], HIV [33-35], lipid metabolic diseases [36] and neurodegenerative diseases [37,38]. With this demonstrated importance of membrane curvature, the ability for curvature sensing and understanding the mechanism behind it is essential as this would allow for the targeting of biological processes that are implicated in diseases.
ALPS and the BAR domain are well known as sensors of membrane curvature [39,40] among the proteins mentioned above. In our laboratory, peptides such as the effector domain of myristoylated alanine-rich protein kinase C (MARCKS-ED) and a Synaptotagmin-1-derived cyclic peptide have recently been shown to sense highly curved membrane surfaces [41,42] such as exosomes. These lower-molecular weight peptides have the advantage of being more amenable to large-scale production compared to larger proteins. Mechanisms accounting for curvature-sensing behavior have been proposed [1,10,43] including electrostatic interactions between the concave surfaces of the protein or peptide and membranes enriched with anionic lipids [44,45] or the sensing of membrane surface defects that arise from membrane curvature [20,46-49]. The latter mechanism usually involves the insertion of certain residues into the defects. In the case of MARCKS-ED, our previous work [43] confirms, via computational and experimental means, the essential role of the Phe residues both in the insertion and retention of MARCKS-ED into the curved bilayer. Owing to the hydrophobic character of Phe, molecular dynamics (MD) simulations showed that these aromatic residues descended into the bilayer interface within the first nanosecond of the simulation and, despite the presence of numerous hydrophilic Lys residues, was able to stay buried in the interfacial region and hold the peptide attached to the bilayer. This insertion of the Phe residues was confirmed by electron paramagnetic resonance (EPR) experiments. Such an understanding of the parameters behind curvature sensing can lead to the design of peptides that can bind better to highly curved structures. However, in order to utilize these peptide probes in complex biological environment, modifications are oftentimes required to improve biological ability and stability. One such modification usually employed is the use of D-amino acids to build the sequence of a peptide.
The above mentioned study completed in our laboratory used L-MARCKS-ED, where all of the amino acid residues comprising the sequence are of the naturally occurring L-isomer form of amino acids. In many instances, the use of the D-isomer confers certain advantages over the L-form. L-peptides isomers are more prone to proteolytic processes in vivo compared to the D-form [50-53]. D-peptides may also be readily available through oral administration [54,55]. Thus, in the same work, DMARCKS-ED was also studied and was found to have curvature-sensing ability [43]. However, the atomistic picture of such an interaction between the D-form and the lipid bilayer is still lacking. Such information is indispensable if this peptidomimetic form of L-MARCKS-ED is to be subject to further investigation as an effective sensor of curvature in vivo.
In this present study, we performed MD simulations of the D-MARCKS-ED peptide. Such simulations are necessary to observe atomic-level behavior of the peptide with a flat bilayer system. Though we are exploring the mechanisms behind the curvature sensing abilities of MARCKS-ED, our previous work has shown that the interaction of L-MARCKS with a flat membrane in silico was able to predict aspects of the mechanism behind the interactions between L-MARCKS-ED and a curved membrane [43]. We then compared the simulations results of both L- and DMARCKS-ED to see if the two forms behave similarly as they interact with the lipid bilayer. The simulations also allow for the observation of a molecular snapshot of the conformational changes. Based on the simulation results, we further carried out biophysical experiments (e.g. fluorescence enhancement assay, circular dichroism spectroscopy assay etc.) to validate the parallel binding behaviors of L- and DMARCKS-ED.
MATERIALS AND METHODS
General Material Information
The phospholipids 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), and cholesterol were purchased from Avanti Lipids. Ethanol (American Chemical Society reagent, ≥99.5%) was purchased from Sinopharm Chemical Reagent. Milli-Q® water was used (ρ = 18.2 MΩ.cm) (Millipore, Molsheim, France).
Atomistic Membrane Model
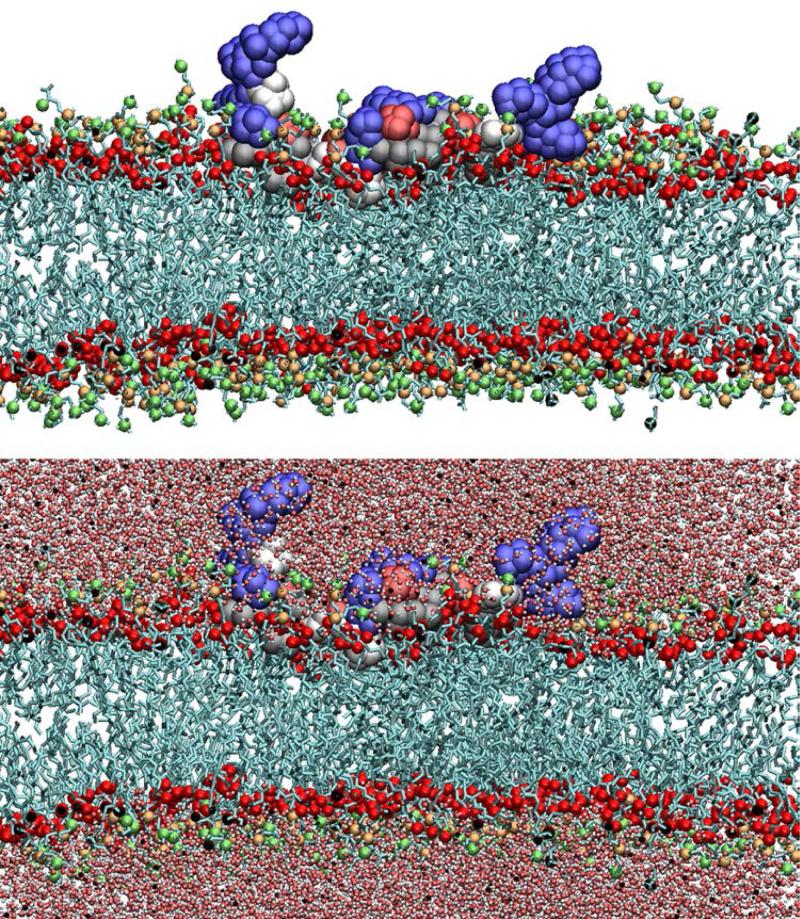
The MARCKS-ED peptide has the amino acid sequence NH2-KKKKKRFSFKKSFKLSGFSFKKNKK-CONH2. MD simulation was employed to model the MARCKS-ED peptide in a fully atomistic biomolecular system composed of lipid bilayers, water molecules, and neutralizing ions for which the Membrane Builder module in CHARMM-GUI [56] was used for construction. The MARCKSED peptides were constructed from internal coordinates found in the CHARMM residue topology files. For the D-MARCKS-ED system, the peptide was placed in the upper leaflet of a flat bilayer composed of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylserine (POPS). The bilayer is composed of 467 POPC and 52 POPS lipid molecules. Of these, 225 POPC and 25 POPS comprise the upper leaflet. Additional amounts of POPC and POPS molecules were placed in the lower leaflet to compensate for the area occupied by the peptide in the upper leaflet. This results to a lower leaflet composed of 242 POPC and 27 POPC molecules. For the L-MARCKS ED system, two MARCKS-ED peptides were placed in each leaflet. This was done to explore the effect on sampling if two such peptides were placed in initial identical positions in each leaflet. As the MARCKS-ED peptide is predicted to be constrained in one leaflet [57], the placement of one peptide on each leaflet is not expected to affect the behavior of each peptide. This approach is similar to that employed in potential of mean force (PMF) simulations of amino acids by Maccallum et. al [58]. Each leaflet in the L-MARCKS-ED system was composed of 250 lipids per leaflet (225 POPC and 25 POPS molecules). The investigation of the interactions of the MARCKS-ED peptide with the membrane interfacial region is part of this study. To explore this aspect, the peptide was initially placed in the lipid bilayer in such a way that the peptide lies flat along the interfacial region and thus, the xy-area of the system box includes the contribution of the peptide. This initial conformation of the peptide also orients the side chains to initially lie approximately parallel to the membrane surface. The L-MARCKS-ED system contains ~130,000 atoms, including ~20,000 water molecules. Meanwhile, the D-MARCKS-ED system contain around ~180,000 atoms with ~37000 water molecules. A 0.15-M concentration of KCl was used for both simulations. Rectangular periodic boundary conditions were imposed with a variable box height of ~70 Å and ~100 Å for the L-MARCKS-ED and DMARCKS-ED systems, respectively. The xy-translation lengths were ~135 Å for both systems and was based on 68.3-Å2 and 62-Å2 area/lipid ratios for POPC and POPS, respectively [56]. Fig. 1 shows a snapshot of the D-MARCKS-ED system after 20 ns of simulations.
Fig 1.
Snapshot of the D-MARCKS-ED peptide at t=20 ns of MD simulation.
MD Simulation
The Chemistry at Harvard Macromolecular Mechanics (CHARMM) program [59] was used for this study, making use of the CHARMM36 protein and lipid force fields [60-62]. For water, the TIP3P model [63] was used and bonds to hydrogen atoms were fixed with the SHAKE algorithm [64]. The electrostatics were handled using the particle-mesh Ewald (PME) method [65] using a mesh size of ~1 Å for fast Fourier transformation, κ = 0.34/Å and a sixth-order B-spline interpolation. A 16-Å cutoff distance was used for constructing non-bond pair lists, and the Lennard-Jones potential was smoothly switched off at 10-12 Å using a force-switching function. Following conditions used previously in simulations of biomolecular systems [66-69], simulations were performed under constant temperature (330 K, to ensure bilayer fluidity) and normal pressure (1.0 atm) with a fixed lateral area using Nosé-Hoover methods [70] and the Langevin piston [71].
Synthetic Lipid Vesicle Preparation
We used ethanol injection method to produce liposomes [75]. PC, Chol, PE, and PS were used to prepare liposomes in mass ratio of 50:15:15:20. Lipids were dissolved in an organic phase (chloroform/methanol, 9:1; v/v), then the organic solvents were removed by evaporation under vacuum to form a dried lipid film. This film was solvated in hot absolute ethanol (500 μL) at 60°C while continuously stirring. This ethanolic solution of lipids was then injected with a syringe through a needle in the middle of a 1-mL-solution of water heated at 70°C contained within a penicillin vial with a flow rate of 3000 μL/min and stirring speed of the aqueous phase of 400 rpm. The stirring of the obtained hydroalcoholic solution lasted 10 minutes. The ethanol was then removed by rotary evaporation under reduced pressure. The final volume of the liposome suspension was 1mL.
Vesicle-size and Zeta Potential Measurements Using Dynamic Light Scattering (DLS)
Size distribution and zeta-potential of the liposome was measured by DLS using the Zetasizer Nano ZS-100 (Horiba Jobin Yvon, Paris, France) [75], with an angle of detection of 90 degrees, measured at 25°C in intensity. Each sample was diluted with the aqueous phase until the appropriate particle concentration was achieved. Each measurement was performed in triplicate and both the average diameter, zeta potential and polydispersity index (PDI) (± the standard error of the mean. The particle size of the liposomes was determined by DLS with the average size of 67 nm. The vesicle diameters distribute ranged from 60 nm to 80 nm is wider than that of extrusive liposomes [76]. In term of heterogeneity, liposomes prepared by ethanol injection method are arguably more comparable to biological extracellular vesicles specimens [77]. Average Zeta potential of liposomes is −34 mV, showing liposome electronegative, caused by the abundance of POPC (20%). Moreover, it is generally acknowledged that the colloid is stable in water if its absolute value of Zeta potential is larger than 30 mV.
Fluorescence Enhancement Assay
The emission spectra of all NBD-labeled peptides were recorded using F-4600 Fluorescence Spectrophotometer (HITACHI, Tokyo, Japan) with λex= 480 nm. D-/LNBD-MARCKS-ED were tested at a concentration of 500 nM in water treated with 500 μM synthetic vesicles. Fluorescence was observed with an emission range of 500−650 nm by 4nm/s.
Circular Dichroism (CD) Spectroscopy
The peptide solutions were prepared at 10 μM in deionized water in the presence/absence of 500 μM liposomes (67 nm pore size with 20% PS) [76]. Circular dichroism spectra were recorded using a Chirascan CD Spectrometer (Applied Photophysics, Surrey, United Kingdom) with a 1 mm path length quartz cuvette at 25 °C using deionized water as a blank. The reading was then converted to molar residue ellipticity (θ). The scans from 190 to 240 nm with data points taken every 1.0 nm were obtained and averaged for each sample.
RESULTS AND DISCUSSION
Distance of Side Chains from the Bilayer Center
Morton et al. [43] established that in its interaction with the lipid bilayer, LMARCKS-ED adopts a “boat conformation” where the Lys-rich ends of the peptides are mostly pointed towards the solvent and at a greater distance from the bilayer center while the central stretch is mainly attached to the lipid bilayer and with the Phe residues found closer to the bilayer center (Fig. 1).
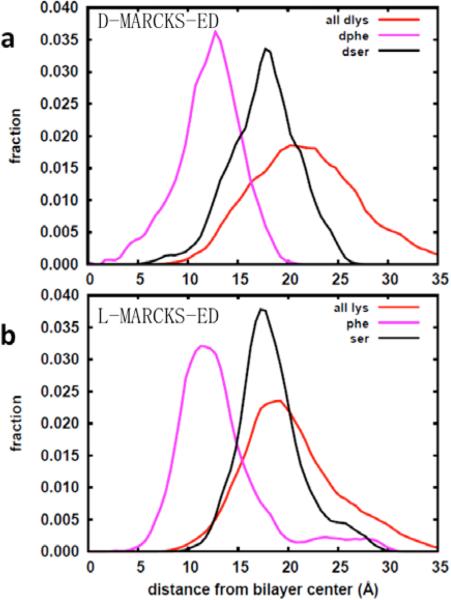
Fig. 2 shows the average insertion depth of side chains of the D-MARCKS-ED and L-MARCKS-ED residues (Fig. 2a and 2b, respectively). The values presented were obtained from the average z-coordinate of the center of mass of the side chains ranging from the 10-ns to the 30-ns time point of simulation. It shows that indeed, the lysine residues at the ends of the peptides (blue bars) are found interacting more strongly with the bulk water while the Phe residues are found more buried and closer to the bilayer center. Though there are also Lys residues found in the central stretch, they are found closer to the bilayer center by around 4-5 Å compared to the lysine residues at the ends of the peptides. This behavior points to how strongly the central stretch is held by the membrane. Meanwhile, comparing the side chain depths of the L-MARCKS-ED and D-MARCKS-ED residues, it can also be observed that the average distances of these residues from the bilayer center are fairly similar to about 1-3 Å indicating that the two enantiomeric peptides interact similarly with the bilayer.
Fig 2.
Average insertion depth of the centers-of-mass of the side chains of D-MARCKS and L-MARCKS.
To gain a better overall picture of how the three main residues of MARCKS-ED (Lys, Phe and Ser) behave during the course of the simulation, histograms of the side-chain insertion depths were constructed. Fig. 3 shows the distribution of these insertion depths for the Lys, Phe and Ser residues comprising L-MARCKS-ED and D-MARCKS-ED (Fig. 3a and 3b, respectively). For both systems, the average distances of the lipid head group components from the bilayer center are as follows: choline N= ~±20 Å, phosphate P= ~18 Å, and carbonyl C= ~± 13 Å. As with Fig. 2, it can clearly be observed from Fig. 3 that the average position of the Phe alanine residues at ~10-12 Å is closer to the bilayer center by around 5-7 Å compared to the hydrophilic Ser and Lys residues. The Lys residues (red curve) are found as far as past the choline region. There is a slight difference of the positioning of the middle of the Lys peaks of the L- and D-MARCKS-ED peptides made evident by the broadness of the Lys peak for the D-MARKCS-ED peptide. This broadness is due to the fact that the curve for the L-MARCKS-ED peptide was constructed from twice the amount of data than that of D-MARCKS-ED. This arises from the presence of two peptides, one in each leaflet, for the L-MARCKS-ED system. Calculating the averages of the Lys side chain depths shows average values of 20.7 ± 1.2 and 21.4 ± 1.2 Å for the L- and D-MARCKS-ED peptides, respectively. Ser residues (black curve) are also found closer to the bulk solvent owing to the hydrogen-bond donor present in the side chain that can interact with water and the head groups. The preferred position of Phe from Fig. 3 (~12 Å from the bilayer center) agrees with previous simulations of just the side-chain analog reported by MacCallum and coworkers [58]. However, the same cited work also reports that the Ser side chain analog is closer to the bilayer center by ~4 Å compared to our results. This discrepancy can be attributed to the fact that the presence of a number of Lys residues has the effect of pulling the Ser residues farther away from the bilayer center. Zhang, Yethiraj and Cui [72] also report this trend in implicit membrane simulations of the MARCKS-ED peptide where the Phe residues are found closer to the dielectric boundary while the charged and polar residues are found in the high dielectric constant region. Once more, this shows how the aromatic Phe residues are able to hold the whole peptide into the bilayer interface despite the fact that there are more hydrophilic residues that are interacting with the bulk solvent.
Fig 3.
Distribution of the Z-coordinates of the side chains of Lys, Ser, and Phe of a) D-MARCKS-ED and b) L-MARCKS-ED.
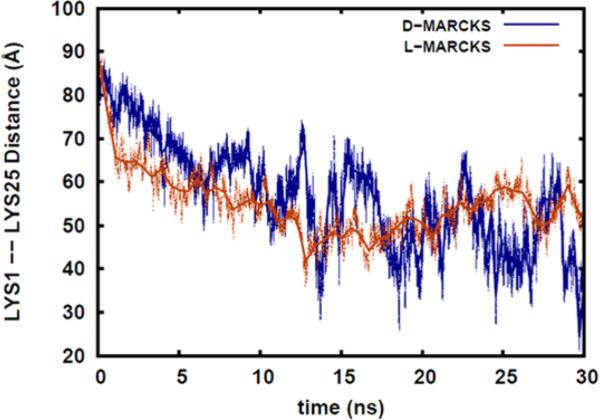
Distance Between the Ends of the MARCKS-ED Strand
During the course of simulation, as the MARCKS-ED peptide starts moving away from its initial, fully stretched conformation and move towards the boat conformation, it is expected that the distances between the two ends of the peptide strand will decrease. Measuring this distance opens the way for experimental validation, via fluorescent resonance energy transfer (FRET) for example, of the conformational changes that occur during the simulation. Fig. 4 shows the distance between Lys1 and Lys25, the residues found at the end of the MARCKS-ED strand. This end-Lys distance is calculated from the distances between the centers-of-mass of the side chains of the end Lys residues. As expected, during progress of the simulation time, the distance between the two Lys residues decreases. Though there are a certain level of fluctuations, it can be seen from the Fig. 4 that around the 10-ns time point, the end-Lys distance start to level off around 50 Å for both the L-MARCKS-ED and DMARCKS-ED peptides. Similar to the previous section, this observation suggests hat the D- and L-forms of the peptide behave similarly when these interact with the lipid bilayer. For L-MARCKS-ED, it can be observed that the fluctuation in the end-Lys distances is less than that of D-MARCKS-ED. This is due to the fact that the red curve represents an average of the end-Lys distances of the peptides located in the top and bottom leaflets of the bilayer. This demonstrates that an improvement in sampling can be obtained if one MARCKS-ED peptide is placed in each bilayer leaflet.
Fig 4.
Distance between the centers-of-mass of the side chains of D/L-Lys1 and D/L-Lys 25, the residues found at the ends of the MARCKS-ED strand.
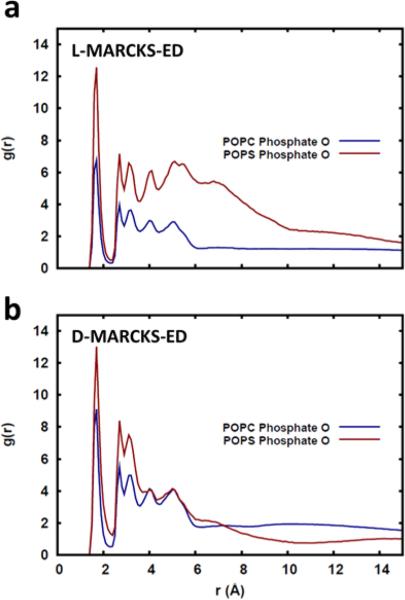
Distribution of Lipid Head Group Components around Lysine Residues
It is known that one of the forces driving the interaction between MARCKS-ED and membranes is the electrostatic interaction between the positively charged residues and the PS lipid head group [41,43,44]. One expected consequence of such an interaction is that the cationic residues would be found close to a PS lipid more frequently. This can be seen from Fig. 5 where it is shown that the cationic Lys residues are, on average, more likely to be surrounded by the phosphate oxygen of POPS lipids compared to POPC lipid. This observation is true for both the L- and DMARCKS-ED peptides (Figs. 5a and b, respectively), once more suggesting that the two peptides interact similarly with the lipid bilayer.
Fig 5.
Radial distribution of phosphate oxygen around the Lysine side chains of the a) L-MARCKS-ED and b) D-MARCKS-ED.
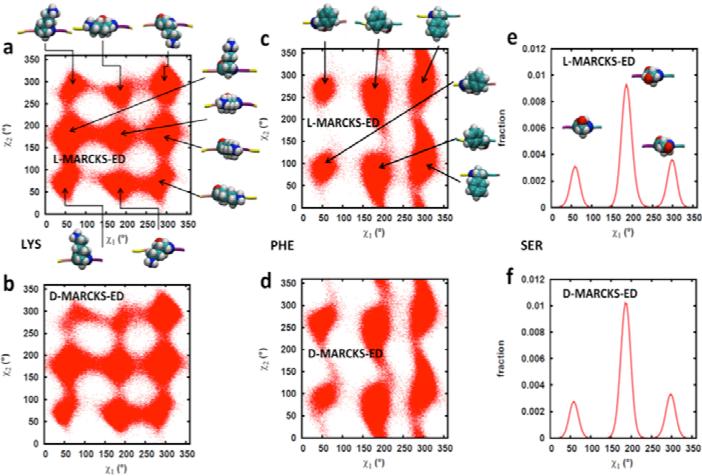
Dihedral Angles of the Amino Acid Side Chains
The rotameric states visited by the side chains of the Lys, Phe and Ser residues during the course of the simulation are shown in Fig. 6. This Figure shows a map of the side chain dihedrals χ1 and χ2. χ1 is defined by the dihedral angle formed by the atoms N-Cα-Cβ-Cγ (-O for Ser), where N is the nitrogen atom along the backbone. On the other hand, χ2 is defined by the atoms Cα-Cβ-Cγ-Cδ. In general, the residues of MARCKS-ED are able to access the stable rotameric states predicted by Dunbrack and Karplus [73]. From Fig. 6a and 6b, it can be seen that Lys is able to attain different side chain orientations. This is expected as Lys residues are able to go into the solvent, and thus are relatively free to move without being constrained by the lipid bilayer. The Phe residues comprising MARCKS-ED are likewise able to access the predicted rotameric states (Fig. 6c and 6d) for Phe. In addition, it can also be observed from the general orientations shown in the Figure that a number of these rotamers show the plane of the aromatic ring to be parallel to the membrane normal. Such conformations are also observed for Trp that also has an aromatic moiety [74]. These orientations are not unexpected since this allows the bulky aromatic ring to fit better between the lipid tails compared to a state where the aromatic ring is perpendicular to the membrane normal. Ser residues are generally not considered to have a χ2 dihedral angle and thus, a distribution of the χ1 dihedral angles is shown (Fig. 6e and 6f). As with Lys and Phe, the rotameric states accessed by Ser during the simulation also conforms to the predicted side chain conformations. In comparing the top with the bottom panels, it is also evident that the residues comprising L- and D-MARCKS-ED are behaving similarly.
Fig 6.
Side chain dihedral angle values (χ1 and χ2) accessed by Lys (a and b), Phe (c and d) and Ser (e and f) residues during the course of the simulation. Starting from the backbone, χ1 is defined by the atoms N-Cα-Cβ-Cγ (-O for Ser) while χ2 is defined by Cα-Cβ-Cγ-Cδ. The pictures show the general orientation of the side chains at the indicated rotamer center.
Fluorescence Enhancement Validating the Binding between MARCKS-ED and Liposomes
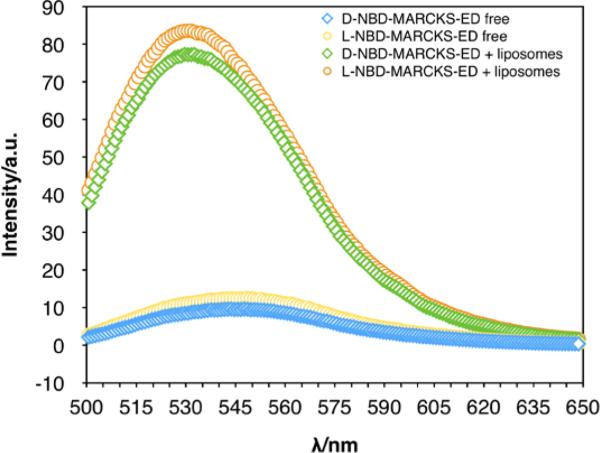
The L- and D-MARCKS-ED peptides were prepared using previously established methods [43]. We conduct a fluorescence enhancement assay to validate the binging behavior of MARCKS-ED to liposomes. Upon binding to liposomes, the fluorescence intensity of the NBD- labeled MARCKS-ED peptide increases and the maximum emission wavelength blue shifts due to the elevated hydrophobicity of the surrounding environment of the fluorophore (Fig. 7). Besides proving the binding, the fluorescence spectra also reveal the similar affinity of D- and L-NBD-MARCKS-ED for liposomes.
Fig 7.
Emission spectra from fluorescence enhancement binding assays for MARCKS-ED. The blue diamond plot represents free D-NBD-MARCKS-ED. The green diamond plot represents D-BND-MARCKS-ED in the presence of liposomes. The yellow circle plot represents free L-NBD-MARCKS-ED. The orange circle plot represents L-MARCKS-ED in the presence of liposomes. A blue shift was observed for both peptide-vesicle samples. [Total Lipid] = 500 μM, [NBD-MARCKS-ED] = 1 μM, λex = 480 nm.
Change of Conformation Demonstrated in CD Spectroscopy
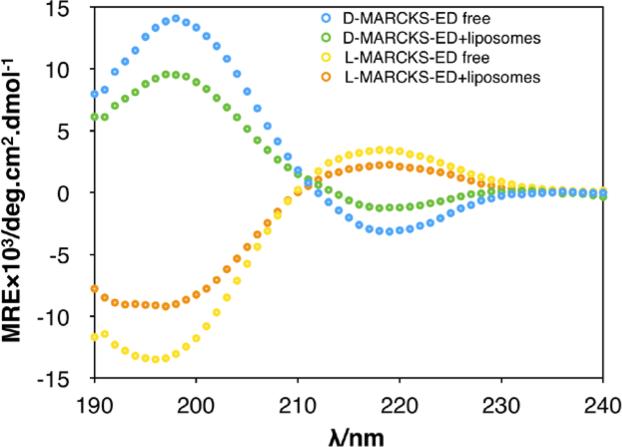
Different secondary structures of peptides can be detected using CD spectroscopy [78]. As shown in Fig. 8, CD spectrum of free D-MARCKS-ED (blue) indicated a random coil secondary structure in aqueous solution, with two main peaks observed in 198 nm (positive) and 220 nm (negative). Upon addition of the liposomes into the aqueous peptide solution, CD intensity decreases significantly in both 198 nm and 220 nm (green) perhaps due to elevated rigidity of the peptide. This transformation suggests that the conformation of D-MARCKS-ED changes in the presence of liposomes and indicates that binding occurred between D-MARCKS-ED peptide and liposomes. Similar result was observed in the untreated and liposomes-treated LMARCKS-ED peptide (Fig. 8).
Fig 8.
CD spectrum of D-/L- MARCKS-ED in the presence/absence of liposomes. [Total Lipid] = 500 μM. [MARCKS-ED] = 10 μM.
Discussion
In order to study the behaviors of the unnatural D-MARCKS-ED peptides in membrane binding, we conducted both experimental and computational studies. Computational simulations provide a useful method to explain how MARCKS-ED peptides bind to the bilayer liposomes. The initial placement of the D-MARCKS-ED peptide placed the side chains approximately parallel to the surface of membrane. After 20 nanoseconds, the Lys side chains at both ends of D-MARCKS-ED stretch from the membrane surface towards the solvent. In contrast, central Phe residues of D-MARCKS-ED are already beginning to move deeper and bury into the bilayer. Finally, D-MARCKS-ED adopts a boat-like shape upon membrane association (Fig. 1). In a good agreement, changes of CD spectra of D-MARCKS-ED are observed upon addition of liposomes, which could probably cause by the movement of the residues and adoption of a more rigid conformation. More importantly, we observed that the CD spectrum of D- and L-MARCKS-ED is almost symmetrical via X-axis (Fig. 8) in the presence or absence of liposomes. This can be rationalized by the exactly symmetric configuration of amino acid residues. Furthermore, these results also confirmed similar behavior of D- and L-MARCKS-ED when binding to curved membrane. Finally, fluorescence enhancement assay conducted under the similar conditions also indicated similar affinities of D- and L- MARCKS-ED toward the membranes. Taken together, our results demonstrate that D- and L-MARCKS-ED work through very similar mechanisms of binding to the liposomes, which is highly consistent with the computational simulation results.
ACKNOWLEDGMENTS
We thank the National Institutes of Health (R01GM103843) for the financial supports.
Footnotes
‡ Special issue of contributions presented at the 13th Chinese International Peptide Symposium, Peptides: Treasure of Chemistry and Biology, June 30 - July 4, 2014 in Datong, Shanxi, China.
REFERENCES
- 1.Baumgart T, Capraro BR, Zhu C, Das SL. Thermodynamics and Mechanics of Membrane Curvature Generation and Sensing by Proteins and Lipids. Annu. Rev. Phys. Chem. 2011;62:83–506. doi: 10.1146/annurev.physchem.012809.103450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Callan-Jones A, Bassereau P. Curvature-driven membrane lipid and protein distribution. Curr. Opin. Solid. St. M. 2013;17:143–150. [Google Scholar]
- 3.Aimon S, Callan-Jones A, Berthaud A, Pinot M, Toombes GES, Bassereau P. Membrane Shape Modulates Transmembrane Protein Distribution. Dev. Cell. 2014;28:212–218. doi: 10.1016/j.devcel.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koller D, Lohner K. The role of spontaneous lipid curvature in the interaction of interfacially active peptides with membranes. Bba-Biomembranes. 2014;1838:2250–2259. doi: 10.1016/j.bbamem.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Bigay J, Antonny B. Curvature, Lipid Packing, and Electrostatics of Membrane Organelles: Defining Cellular Territories in Determining Specificity. Dev. Cell. 2012;23:886–895. doi: 10.1016/j.devcel.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Janmey PA, Kinnunen PKJ. Biophysical properties of lipids and dynamic membranes. Trends. Cell. Biol. 2006;16:538–546. doi: 10.1016/j.tcb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 7.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 8.Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell. Bio. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 9.Haney EF, Nathoo S, Vogel HJ, Prenner EJ. Induction of non-lamellar lipid phases by antimicrobial peptides: a potential link to mode of action. Chem. Phys. Lipids. 2010;163:82–93. doi: 10.1016/j.chemphyslip.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Antonny B. Mechanisms of Membrane Curvature Sensing. Annu. Rev. Biochem. 2011;80:101–123. doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- 11.Hatzakis NS, Bhatia VK, Larsen J, Madsen KL, Bolinger PY, Kunding AH, Castillo J, Gether U, Hedegard P, Stamou D. How curved membranes recruit amphipathic helices and protein anchoring motifs. Nat. Chem. Biol. 2009;5:835–841. doi: 10.1038/nchembio.213. [DOI] [PubMed] [Google Scholar]
- 12.Graham TR, Kozlov MM. Interplay of proteins and lipids in generating membrane curvature. Curr. Opin. Cell. Biol. 2010;22:430–436. doi: 10.1016/j.ceb.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jao CC, Hegde BG, Gallop JL, Hegde PB, McMahon HT, Haworth IS, Langen R. Roles of Amphipathic Helices and the Bin/Amphiphysin/Rvs (BAR) Domain of Endophilin in Membrane Curvature Generation. J. Biol. Chem. 2010;285:20164–20170. doi: 10.1074/jbc.M110.127811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farsad K, Ringstad N, Takei K, Floyd SR, Rose K, De Camilli P. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J. Cell. Biol. 2001;155:193–200. doi: 10.1083/jcb.200107075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJG, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 16.Hui E, Johnson CP, Yao J, Dunning FM, Chapman ER. Synaptotagmin-mediated bending of the target membrane is a critical step in Ca(2+)-regulated fusion. Cell. 2009;138:709–721. doi: 10.1016/j.cell.2009.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ford MGJ, Mills IG, Peter BJ, Vallis Y, Praefcke GJK, Evans PR, McMahon HT. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- 18.Roux A, Koster G, Lenz M, Sorre B, Manneville JB, Nassoy P, Bassereau P. Membrane curvature controls dynamin polymerization. P. Natl. Acad. Sci. USA. 2010;107:4141–4146. doi: 10.1073/pnas.0913734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA. Mechanisms Determining the Morphology of the Peripheral ER. Cell. 2010;143:774–788. doi: 10.1016/j.cell.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat. Struct. Mol. Bio. 2007;14:138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 21.Zimmerberg J, McLaughlin S. Membrane curvature: How BAR domains bend bilayers. Curr. Biol. 2004;14:250–252. doi: 10.1016/j.cub.2004.02.060. [DOI] [PubMed] [Google Scholar]
- 22.Fleming A, Sampey G, Chung MC, Bailey C, van Hoek ML, Kashanchi F, Hakami RM. The carrying pigeons of the cell: exosomes and their role in infectious diseases caused by human pathogens. Pathog. Dis. 2014;71:107–118. doi: 10.1111/2049-632X.12135. [DOI] [PubMed] [Google Scholar]
- 23.Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer. Metast. Rev. 2013;32:623–642. doi: 10.1007/s10555-013-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belting M, Wittrup A. Nanotubes, exosomes, and nucleic acid-binding peptides provide novel mechanisms of intercellular communication in eukaryotic cells: implications in health and disease. J. Cell. Biol. 2008;183:1187–1191. doi: 10.1083/jcb.200810038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: A Common Pathway for a Specialized Function. J. Biochem. 2006;140:13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- 26.Luga V, Zhang L, Viloria-Petit AM, Ogunjimi AA, Inanlou MR, Chiu E, Buchanan M, Hosein AN, Basik M, Wrana JL. Exosomes Mediate Stromal Mobilization of Autocrine Wnt-PCP Signaling in Breast Cancer Cell Migration. Cell. 2012;151:1542–1556. doi: 10.1016/j.cell.2012.11.024. [DOI] [PubMed] [Google Scholar]
- 27.Ono M, Kosaka N, Tominaga N, Yoshioka Y, Takeshita F, Takahashi RU, Yoshida M, Tsuda H, Tamura K, Ochiya T. Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Sci. Signal. 2014;7:1–10. doi: 10.1126/scisignal.2005231. [DOI] [PubMed] [Google Scholar]
- 28.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 29.Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal MicroRNA: A Diagnostic Marker for Lung Cancer. Clin. Lung. Cancer. 2009;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 30.Taylor DD, Gercel-Taylor C. Exosomes/microvesicles: mediators of cancer-associated immunosuppressive microenvironments. Semin. Immunopathol. 2011;33:441–454. doi: 10.1007/s00281-010-0234-8. [DOI] [PubMed] [Google Scholar]
- 31.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, Garcia-Santos G, Ghajar CM, Nitadori-Hoshino A, Hoffman C, Badal K, Garcia BA, Callahan MK, Yuan JD, Martins VR, Skog J, Kaplan RN, Brady MS, Wolchok JD, Chapman PB, Kang YB, Bromberg J, Lyden D. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miranda KC, Bond DT, McKee M, Skog J, Paunescu TG, Da Silva N, Brown D, Russo LM. Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney. Int. 2010;78:191–199. doi: 10.1038/ki.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Izquierdo-Useros N, Naranjo-Gomez M, Erkizia I, Puertas MC, Borras FE, Blanco J, Martinez-Picado J. HIV and Mature Dendritic Cells: Trojan Exosomes Riding the Trojan Horse? Plos. Pathog. 2010;6:e1000740. doi: 10.1371/journal.ppat.1000740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lenassi M, Cagney G, Liao MF, Vaupotic T, Bartholomeeusen K, Cheng YF, Krogan NJ, Plemenitas A, Peterlin BM. HIV Nef is Secreted in Exosomes and Triggers Apoptosis in Bystander CD4 (+) T Cells. Traffic. 2010;11:110–122. doi: 10.1111/j.1600-0854.2009.01006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kadiu I, Narayanasamy P, Dash PK, Zhang W, Gendelman HE. Biochemical and Biologic Characterization of Exosomes and Microvesicles as Facilitators of HIV-1 Infection in Macrophages. J. Immunol. 2012;189:744–754. doi: 10.4049/jimmunol.1102244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Record M, Poirot M, Silvente-Poirot S. Emerging concepts on the role of exosomes in lipid metabolic diseases. Biochimie. 2014;96:67–74. doi: 10.1016/j.biochi.2013.06.016. [DOI] [PubMed] [Google Scholar]
- 37.Vella LJ, Sharples RA, Nisbet RM, Cappai R, Hill AF. The role of exosomes in the processing of proteins associated with neurodegenerative diseases. Eur. Biophys. J. Biophy. 2008;37:323–332. doi: 10.1007/s00249-007-0246-z. [DOI] [PubMed] [Google Scholar]
- 38.Vella LJ, Sharples RA, Lawson VA, Masters CL, Cappai R, Hill AF. Packaging of prions into exosomes is associated with a novel pathway of PrP processing. J. pathol. 2007;211:582–590. doi: 10.1002/path.2145. [DOI] [PubMed] [Google Scholar]
- 39.Bhatia VK, Madsen KL, Bolinger PY, Kunding A, Hedegard P, Gether U, Stamou D. Amphipathic motifs in BAR domains are essential for membrane curvature sensing. Embo. J. 2009;28:3303–3314. doi: 10.1038/emboj.2009.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mesmin B, Drin G, Levi S, Rawet M, Cassel D, Bigay J, Antonny B. Two lipid-packing sensor motifs contribute to the sensitivity of ArfGAP1 to membrane curvature. Biochemistry. 2007;46:1779–1790. doi: 10.1021/bi062288w. [DOI] [PubMed] [Google Scholar]
- 41.Morton LA, Yang HW, Saludes JP, Fiorini Z, Beninson L, Chapman ER, Fleshner M, Xue D, Yin H. MARCKS-ED Peptide as a Curvature and Lipid Sensor. ACS. Chem. Biol. 2013;8:218–225. doi: 10.1021/cb300429e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saludes JP, Morton LA, Ghosh N, Beninson LA, Chapman ER, Fleshner M, Yin H. Detection of Highly Curved Membrane Surfaces Using a Cyclic Peptide Derived from Synaptotagmin-I. ACS. Chem. Biol. 2012;7:1629–1635. doi: 10.1021/cb3002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morton LA, Tamura R, de Jesus AJ, Espinoza A, Yin H. Biophysical investigations with MARCKS-ED: dissecting the molecular mechanism of its curvature sensing behaviors. Bba-Biomembrane. 2014;1838:3137–3144. doi: 10.1016/j.bbamem.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang JY, Gambhir A, Hangyas-Mihalyne G, Murray D, Golebiewska U, McLaughlin S. Lateral sequestration of phosphatidylinositol 4,5-bisphosphate by the basic effector domain of myristoylated alanine-rich C kinase substrate is due to nonspecific electrostatic interactions. J. Biol. Chem. 2002;277:34401–34412. doi: 10.1074/jbc.M203954200. [DOI] [PubMed] [Google Scholar]
- 45.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 46.Vanni S, Vamparys L, Gautier R, Drin G, Etchebest C, Fuchs PFJ, Antonny B. Amphipathic lipid packing sensor motifs: probing bilayer defects with hydrophobic residues. Biophys. J. 2013;104:575–584. doi: 10.1016/j.bpj.2012.11.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang WY, Crocker E, McLaughlin S, Smith SO. Binding of peptides with basic and aromatic residues to bilayer membranes - Phenylalanine in the myristoylated alanine-rich C kinase substrate effector domain penetrates into the hydrophobic core of the bilayer. J. Biol. Chem. 2003;278:21459–21466. doi: 10.1074/jbc.M301652200. [DOI] [PubMed] [Google Scholar]
- 48.Bigay J, Gounon P, Robineau S, Antonny B. Lipid packing sensed by ArfGAP1 couples COPI coat disassembly to membrane bilayer curvature. Nature. 2003;426:563–566. doi: 10.1038/nature02108. [DOI] [PubMed] [Google Scholar]
- 49.Bigay J, Casella JF, Drin G, Mesmin B, Antonny B. ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. Embo. J. 2005;24:2244–2253. doi: 10.1038/sj.emboj.7600714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McGregor DP. Discovering and improving novel peptide therapeutics. Curr. Opin. Pharmacol. 2008;8:616–619. doi: 10.1016/j.coph.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 51.Croft NP, Purcell AW. Peptidomimetics: modifying peptides in the pursuit of better vaccines. Expert. Rev. Vaccines. 2011;10:211–226. doi: 10.1586/erv.10.161. [DOI] [PubMed] [Google Scholar]
- 52.Lien S, Lowman HB. Therapeutic peptides. Trends. Biotechnol. 2003;21:556–562. doi: 10.1016/j.tibtech.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 53.Pujals S, Sabido E, Tarrago T, Giralt E. all-D proline-rich cell-penetrating peptides: a preliminary in vivo internalization study. Biochem. Soc. T. 2007;35:794–796. doi: 10.1042/BST0350794. [DOI] [PubMed] [Google Scholar]
- 54.Pappenheimer JR, Karnovsky ML, Maggio JE. Absorption and excretion of undegradable peptides: Role of lipid solubility and net charge. J. Pharmacol. Exp. Ther. 1997;280:292–300. [PubMed] [Google Scholar]
- 55.Pappenheimer JR, Dahl CE, Karnovsky ML, Maggio JE. Intestinal-Absorption and Excretion of Octapeptides Composed of D-Amino-Acids. P. Natl. Acad. Sci. USA. 1994;91:1942–1945. doi: 10.1073/pnas.91.5.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jo S, Lim JB, Klauda JB, Im W. CHARMM-GUI Membrane Builder for mixed bilayers and its application to yeast membranes. Biophys. J. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ellena JF, Burnitz MC, Cafiso DS. Location of the myristoylated alanine-rich C-kinase substrate (MARCKS) effector domain in negatively charged phospholipid bicelles. Biophys. J. 2003;85:2442–2448. doi: 10.1016/s0006-3495(03)74667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.MacCallum JL, Bennett WFD, Tieleman DP. Distribution of amino acids in a lipid bilayer from computer simulations. Biophys. J. 2008;94:3393–3404. doi: 10.1529/biophysj.107.112805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. Charmm - a Program for Macromolecular Energy, Minimization, and Dynamics Calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 60.Klauda JB, Venable RM, Freites JA, O'Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Jr., Pastor RW. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mackerell AD, Jr., Feig M, Brooks CL., 3rd Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 62.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 63.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 64.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical-Integration of Cartesian Equations of Motion of a System with Constraints - Molecular-Dynamics of NAlkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 65.Darden TA, Pedersen LG. Molecular modeling: an experimental tool. Environ. Health. Perspect. 1993;101:410–412. doi: 10.1289/ehp.93101410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li LB, Vorobyov I, Allen TW. Potential of mean force and pK(a) profile calculation for a lipid membrane-exposed arginine side chain. J. Phys. Chem. B. 2008;112:9574–9587. doi: 10.1021/jp7114912. [DOI] [PubMed] [Google Scholar]
- 67.Dorairaj S, Allen TW. On the thermodynamic stability of a charged arginine side chain in a transmembrane helix. P. Natl. Acad. Sci. USA. 2007;104:4943–4948. doi: 10.1073/pnas.0610470104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Allen TW, Andersen OS, Roux B. Structure of gramicidin A in a lipid bilayer environment determined using molecular dynamics simulations and solid-state NMR data. J. Am. Chem. Soc. 2003;125:9868–9877. doi: 10.1021/ja029317k. [DOI] [PubMed] [Google Scholar]
- 69.de Jesus AJ, Allen TW. The determinants of hydrophobic mismatch response for transmembrane helices. Bba-Biomembrane. 2013;1828:851–863. doi: 10.1016/j.bbamem.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 70.Hoover WG. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 71.Feller SE, Zhang YH, Pastor RW, Brooks BR. Constant-Pressure Molecular-Dynamics Simulation - the Langevin Piston Method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 72.Zhang LL, Yethiraj A, Cui Q. Free Energy Calculations for the Peripheral Binding of Proteins/Peptides to an Anionic Membrane. 1. Implicit Membrane Models. J. Chem. Theory. Comput. 2014;10:2845–2859. doi: 10.1021/ct500218p. [DOI] [PubMed] [Google Scholar]
- 73.Dunbrack RL, Karplus M. Backbone-Dependent Rotamer Library for Proteins - Application to Side-Chain Prediction. J. Mol. Biol. 1993;230:543–574. doi: 10.1006/jmbi.1993.1170. [DOI] [PubMed] [Google Scholar]
- 74.de Jesus AJ, Allen TW. The role of tryptophan side chains in membrane protein anchoring and hydrophobic mismatch. Bba-Biomembranes. 2013;1828:864–876. doi: 10.1016/j.bbamem.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 75.Gentine Philippe, Bourel-Bonnet Line, Frisch Benoît. Modified and derived ethanol injection toward liposomes: development of the process. J. Liposome. Res. 2013;23:11–19. doi: 10.3109/08982104.2012.717298. [DOI] [PubMed] [Google Scholar]
- 76.Morton Leslie A., Saludes Jonel P., Yin Hang. Constant Pressure-controlled Extrusion Method for the Preparation of Nano-sized Lipid Vesicles. J. Vis. Exp. 2012;64:e415. doi: 10.3791/4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kastelowitz Noah, Yin Hang. Exosomes and Microvesicles: Identification and Targeting By Particle Size and Lipid Chemical Probes. ChemBioChem. 2014;15:923–928. doi: 10.1002/cbic.201400043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kelly Sharon M., Jess Thomas J., Price Nicholas C. How to study proteins by circular dichroism. Bba-Proteins. Proteom. 2005;1751:119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]