Abstract
Interferon-alpha (IFNα) is a cytokine that orchestrates innate and adaptive immune responses, and potently inhibits proliferation of normal and tumor cells. These properties have warranted the use of IFNα in clinical practice for the treatment of several viral infections and malignancies. However, over-expression of IFNα leads to immunopathology observed in the context of chronic viral infections and autoimmune conditions. Thus, it is desirable to develop therapeutic approaches that aim at suppressing excessive IFNα production. To that end, artificial evolution of peptides from phage display libraries represents a strategy that seeks to disrupt the interaction between IFNα and its cell surface receptor, and thus inhibit the ensuing biological effects. Mirror-image phage display that screens peptide libraries against the D-enantiomer is particularly attractive because it allows for identification of proteolysis-resistant D-peptide inhibitors. This approach, however, relies on the availability of chemically synthesized D-IFNα composed entirely of D-amino acids. Here we describe the synthesis and biological properties of IFNα2b of 165 amino acid residues produced by native chemical ligation, which represents an important first step toward the discovery of D-peptide antagonists with potential therapeutic applications.
Introduction
Interferon alpha (IFNα) is a cytokine that plays key roles in innate and adaptive immune responses [1]. It is rapidly produced in response to viral infections and orchestrates innate immune responses by triggering the expression of genes that interfere with virus replication at various stages, and by activating natural killer (NK) cells [2]. Further, IFNα helps shape the adaptive immune response through activation of immature dendritic cells (upregulation of MHC molecules, chemokine receptors, and co-stimulatory molecules such as CD80 and CD86) [3, 4], modulation of B cell function, and promotion of Th1 or Tr1 effector immune responses [5, 6]. Moreover, IFNα exerts potent anti-proliferative effects on T cells by suppressing expression of IL-2 and the high affinity chain of the IL-2 receptor α-chain (IL-2Rα or CD25) [7–9]. Thus, IFNα is used in the treatment of several malignancies (hairy cell leukemia, renal carcinoma, malignant melanoma, follicular lymphoma, etc.), and viral infections (HBV and HCV).
Over-expression of IFNα is a contributing factor in the etiology of several autoimmune disorders, notably systemic lupus erythematosus (SLE) and insulin-dependent diabetes mellitus (IDDM; type 1 diabetes). In SLE patients, formation of immune-complexes containing DNA or RNA released from apoptotic or necrotic cells triggers exacerbated IFNα production [10–12], which in turn induces unabated activation and maturation of monocytes into dendritic cells, and persistent activation of CD4+ and CD8+ T cells, including auto-reactive T cells that may have escaped the mechanisms of central and peripheral tolerance [13]. In addition, IFNα cooperates with IL-6 to promote the activation and maturation of antibody-secreting B cells [13]. Recent reports showed that chronic IFNα production (often in correlation with Coxsackie B virus infection) causes IDDM in humans and in animal models of the disease [14–16], and is involved in other autoimmune conditions such as multiple sclerosis, rheumatoid arthritis, myasthenia gravis and autoimmune hemolytic anemia. IFNα is also a key mediator of immunosuppression in the context of chronic viral infections: its expression is markedly upregulated during human immunodeficiency virus type-1 (HIV-1) infection, and contributes to disease progression [17]. Indeed, over-expression of IFNα distinguishes pathogenic SIV infection of rhesus macaques from non-pathogenic infection of the natural host, sooty mangabey [18]. Infection with human cytomegalovirus (HCMV) affects monocyte differentiation and maturation into dendritic cells, and inhibits proliferation of T cells through chronic IFNα production [19]. Altogether, these studies show that defects in the spatial and temporal localization of IFNα expression cause or contribute to human disease. Thus, the ability to suppress the detrimental effects of chronic exacerbated IFNα expression may have therapeutic applications.
Isolation of peptide ligands from combinatorial libraries is a useful method for the identification of potent antagonists capable of disrupting protein-protein interactions. Particularly powerful is a modified phage display technique termed mirror-image phage display pioneered by Kim and colleagues [20, 21], in which a phage-expressed peptide library is screened against the D-enantiomer of a native L-protein of interest. The resultant L-peptide ligand only binds the unnatural D-protein. However, inversion of the L-peptide to its D-enantiomer creates a D-peptide ligand that, for reasons of symmetry, only binds the native target protein. D-peptides are proteolytically stable, thus ideally suited for therapeutic development [22–26]. This mirror-image phage display approach requires properly folded D-proteins to be used as bait, which can be made only chemically.
In the present report we describe the successful synthesis of IFNα2b of 165 amino acid residues using native chemical ligation [27–29]. In addition, we show that the purified protein displays the same biological properties and activities as the commercially available recombinant counterpart. The synthetic IFNα2b we produced is correctly folded and fully functional, thereby demonstrating the feasibility of the synthesis of D-enantiomeric IFNα2b suitable for use in mirror-image phage display.
Materials and Methods
Materials
All Boc-(L)-amino acids were purchased from Peptides International (Louisville, KY); Boc-Leu-OCH2-PAM and Boc-Glu(OcHex)-OCH2-PAM resin were obtained from Applied Biosystems (Foster City, CA); Dichloromethane, N,N-dimethylformamide and HPLC grade acetonitrile were purchased from Fisher Scientific (Pittsburgh, PA), and 2-(1H-benzotriazol-l-yl)-1,1,3,3-tetramethyluroniumhexafluorophosphate (HBTU) was purchased from American Bioanalytical (Natrick, MA). Trifluoroacetic acid (TFA) was acquired from Halocarbon (River Edge, NJ) and hydrogen fluoride (HF) from Matheson Tri-gas (Montegomeryville, PA). N,N-Diisopropylethylamine (DIEA), thiophenol, and p-cresol were from Fluka (Switzerland), and ultrapure guanidinium hydrochloride was from ICN Biochemicals (Irvine, CA).
Solid-Phase Synthesis of Thz-Cys(2–28)IFNα-COSR, Thz-Cys(30–97)IFNα-COSR and (98–165)IFNα
The three peptide fragments Thz-Cys(2–28)IFNα-COSR, Thz-Cys(30–97)IFNα-COSR and (98–165)IFNα (Thz = 1,3-thiazolidine-4-carboxo; R = CH2CO-Leu-OH or CH2CH2CO-Leu-OH) were synthesized separately on appropriate resins [30] on an automated ABI 433A peptide synthesizer using an optimized HBTU activation/DIEA in situ neutralization protocol developed by Kent and coworkers for Boc chemistry [31]. Crude peptides were purified to homogeneity by reversed-phase (RP) HPLC and their molecular masses ascertained by electrospray ionization mass spectrometry (ESI-MS). To avoid intra-molecular ligation (head-to-tail cyclization) of thioester peptides containing an N-terminal Cys residue, Thz-Cys was incorporated in Thz-Cys(2–28)IFNα-COSR and Thz-Cys(30–97)IFNα-COSR as the N-terminal residue in place of Cys as described [32].
Native Chemical Ligation and Disulfide Bond Formation
Native chemical ligation between Thz-Cys(30–97)IFNα-COSR and (98–165)IFNα was performed at a total peptide concentration of 20 mg/ml in 0.2 M phosphate buffer containing 6 M guanidine hydrochloride and 2% thiophenol, pH 7.4. The reaction proceeded to completion overnight as monitored by analytical HPLC. Addition of 0.2 M methoxyamine to the crude ligation mixture resulted, after 2 h at room temperature, in conversion of Thz-Cys(30–165)IFNα to the desired intermediate (29–165)IFNα. This product was purified by preparative RP-HPLC and its molecular mass verified by ESI-MS. The ligation reaction between Thz-Cys(2–28)IFNα-COSR and (29–165)IFNα was carried out under similar conditions. After deprotection of the N-terminal Thz-Cys by methoxyamine, the final product (1–165)IFNα was purified by preparative HPLC and its molecular mass confirmed by ESI-MS.
Oxidative folding of IFNα2b was performed through thiol-disulfide shuffling aided by reduced and oxidized thiol pairs in the presence of GuHCl. In brief, the reduced polypeptide was dissolved in 6 M GuHCl at a concentration of ~1 mg/ml, followed by a 6-fold dilution into 50 mM Tris/HCl, 3 mM reduced glutathione, 0.3 mM oxidized glutathione, pH 8.3. After stirring overnight at room temperature, the folded protein was purified by preparative RP-HPLC. ESI-MS analysis showed a loss of four mass units, indicative of the formation of two disulfide bonds in the folded synthetic IFNα2b. Protein quantification was carried out spectroscopically by UV absorbance measurements at 280 nm using a molar extinction coefficient calculated from a published algorithm [33].
Circular Dichroism (CD) Spectroscopy
The CD spectrum of IFNα2b at 10 μM in 10 mM phosphate buffer, pH 7.4, was collected at room temperature on a Jasco J-810 spectropolarimeter using a 0.1-cm path length.
Biological assays
Peripheral blood mononuclear cells (PBMC) were isolated by Ficoll-Paque (GE Healthcare) centrifugation from the peripheral blood of healthy volunteers after approval by the Institutional Review Board of the University of Maryland, Baltimore, and with signed informed consent forms. Cells were plated in triplicate samples at 105/well in 200 μl of RPMI 1640 medium and 10% human serum AB containing 100 ng/ml Staphylococcal enterotoxin B (Sigma Aldrich) and 20 U/ml IL-2 (Roche Biochemicals). Cells were cultured in the absence or presence of recombinant (PBL InterferonSource, Piscataway, NJ) or synthetic IFNα2b (0.01–10 ng/ml) for 3 days at 37°C. [3H]-Thymidine was added at 0.5 μCi/well for the last 16 hours of incubation, and then incorporation of the radioactive nucleotide precursor was measured using a liquid scintillation counter. Cellular proliferation was also measured by cell counts with trypan blue exclusion. The antiviral assay was based on a visual read of the protection of cells from cytopathic effect (CPE) due to viral challenge [34]. Samples were run in duplicate in a viral challenge assay using EMCV on A549 cells. Recombinant and synthetic IFNα2b were titrated in 96-well plates, and protection was determined in comparison to untreated A549 cells infected with ECMV (no IFNα) and uninfected, untreated cells (no virus, no IFNα) controls. After maturation of the viral CPE the live cells were fixed and stained with a Crystal Violet solution. The dye was then solubilized and absorbance was read. The wells were also examined under the microscope to determine the IFNα2b dilution that protects 50% of the cells from CPE. This dilution was calibrated to a standard interferon solution to obtain Units/ml of the sample.
Results
Synthesis of IFNα2b fragments
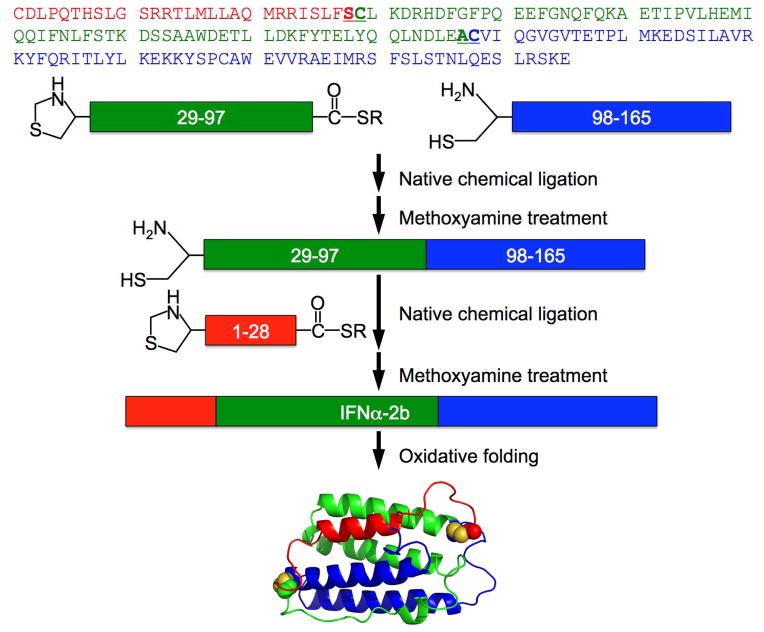
The human IFNα2b protein consists of 165 amino acid residues with two disulfide bonds. Shown in Fig 1 is a three-segment ligation strategy for the synthesis of IFNα2b. To establish chemical access to the ligation sites, the N-terminal and middle segments, i.e., Thz-Cys(2–28)IFNα-COSR and Thz-Cys(30–97)IFNα-COSR, were functionalized with a thioester moiety at the C-terminus. In order to prevent cyclization caused by an intramolecular ligation with the thioester moiety, the N-terminal Cys residues of these two segments were protected with a 1,3-thiazolidine-4-carboxo group (Thz), which can be converted to Cys through methoxyamine treatment. The C-terminal segment (98–165)IFNα was separately synthesized with a free N-terminal Cys residue.
Fig 1.
Strategy of native chemical ligation for the total chemical synthesis of human interferon alpha-2b protein, IFNα2b. The N-terminal 28-residue (red) and middle 69-residue peptide-thioester (green) fragments and C-terminal 68-residue fragment (blue) were synthesized by stepwise SPPS techniques using Boc-chemistry protocols. Folded synthetic IFNα2b is depicted by the NMR structure of human interferon alpha-2a (PDB code 1ITF) [35].
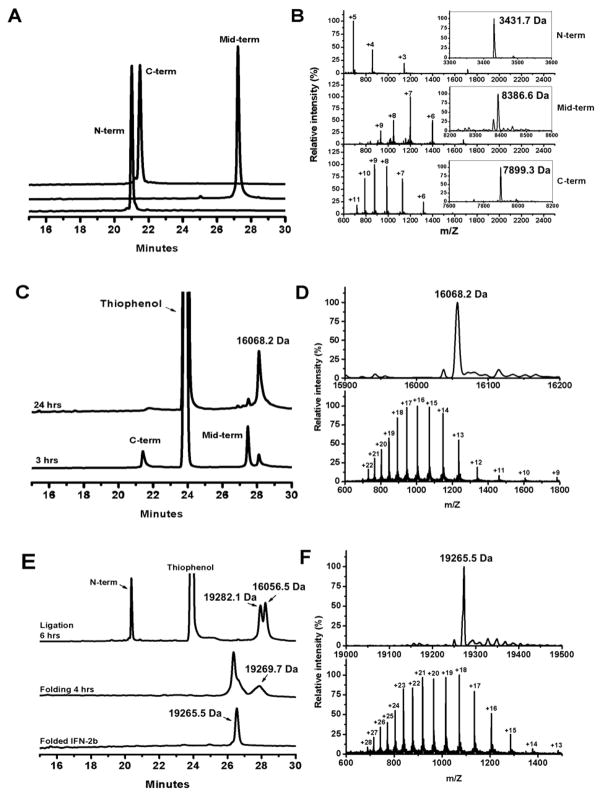
All three peptide segments were synthesized on a 0.25-mmol scale using an optimized in situ neutralization protocol for Boc chemistry. Crude peptides were purified to homogeneity by preparative reversed-phase HPLC, typically yielding 50–70 mg of purified product for each segment. Shown in Fig 2A are these peptide fragments characterized by analytical RP-HPLC. Their molecular masses were determined by electrospray ionization mass spectroscopy (ESI-MS), giving rise to 3431.7, 8386.6 and 7899.3 Da (Fig 2B), in agreement, within experimental error, with the theoretical values calculated for Thz-Cys(2–28)IFNα-COSR (3431.1 Da, R=CH2CO-Leu-OH), Thz-Cys(30–97)IFNα-COSR (8387.4 Da, R=CH2CH2CO-Leu-OH) and (98–165)IFNα (7899.2 Da), respectively.
Fig 2.
Native chemical ligation reactions monitored by analytical RP-HPLC and ESI-MS. All HPLC chromatograms were obtained at 40 °C on a Waters Symmetry 300 C18 column (4.6×150 mm, 5 μm) running a linear gradient of 5% - 65 % of acetonitrile containing 0.1% TFA at a flow rate of 1 ml/min over 30 min. (A) HPLC traces of the three synthetic peptide fragments, Thz-Cys(2–28)IFNα-COSR, Thz-Cys(30–97)IFNα-COSR and (98–165)IFNα, and their ESI-MS data (B). (C) The first ligation reaction monitored by HPLC and verified by ESI-MS (D). (E) The second ligation reaction and protein oxidative folding monitored by HPLC and verified by ESI-MS (F). As expected, protein folding shortens retention time of IFNα2b as its hydrophobic residues are buried in the folded structure. The determined molecular mass of folded synthetic IFNα2b of 19265.5 Da is within experimental error of the theoretical value of 19265.1 Da calculated on the basis of the average isotopic compositions of IFNα2b.
Synthesis of IFNα2b protein by native chemical ligation
Native chemical ligation of the C-terminal and middle segments of the IFNα2b protein was performed with 10 mg of (98–165)IFNα and 12 mg of Thz-Cys(30–97)IFNα-COSR in 2 ml ligation buffer with 2.5% thiophenol. After stirring for 3 hours at room temperature, the ligation product could be readily detected by analytical RP-HPLC and ESI-MS (Fig 2C–2D). The ligation reaction was completed after 24 hours (Fig 2C–2D), followed by a 5-hour treatment with 0.2 M methoxyamine-HCl to deprotect the N-terminal Cys residue. After purification by preparative RP-HPLC, 8.5 mg of the target peptide (29–165)IFNα was recovered with a confirmed molecular mass of 16056.5 Da (vs. a theoretical value of 16055.3 Da).
The second ligation reaction to generate the whole IFNα2b molecule was less efficient than the first one. In this case, about 28.5 mg of (29–165)IFNα and 30.5 mg of Thz-Cys(2–28)IFNα-COSR (in large excess), at a total concentration of 10 mg/ml, were dissolved in ligation buffer. After 6 hours, approximately 50% of (29–165)IFNα was converted to a full-length product (Fig 2E). After overnight ligation, methoxyamine treatment for 5 hours and RP-HPLC purification, we obtained 9 mg of IFNα2b in its reduced form with a measured molecular mass of 19269.7 Da (19269.1 Da, calculated).
Folding of IFNα2b protein
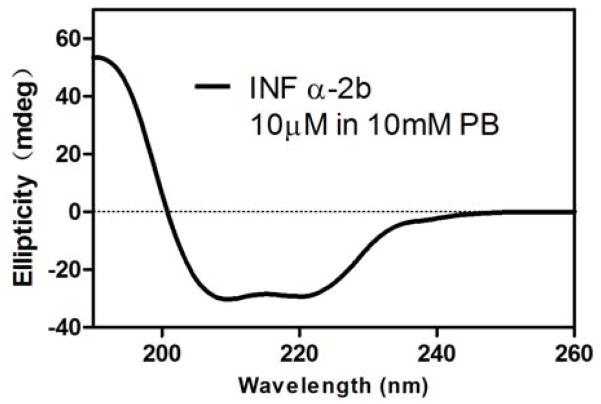
The 165-residue polypeptide chain of IFNα2b was oxidatively folded in a mildly denaturing condition favoring thiol-disulfide exchanges. The folding reaction proceeded to completion within 24 hours, accompanied by a significant shortening of retention time on analytical RP-HPLC (Fig 2E). ESI-MS showed a decrease in mass of the folded protein from 19269.7 to 19265.5 Da (Fig 2F). The loss of 4 mass units is indicative of the formation of two native disulfide bonds in folded IFNα2b protein. To verify the correct folding of IFNα2b, we characterized the synthetic protein using CD spectroscopy. As shown in Fig 3, folded IFNα2b protein displayed a CD spectrum with double negative peaks at 208 and 222 nm and a single positive peak at 195 nm, characteristic of alpha-helical secondary structure and consistent with the known structural features of IFNα [35].
Fig 3.
The CD spectrum of IFNα2b obtained at room temperature. The double negative peaks at 208 and 222 nm and single positive peak at 195 nm are indicative of alpha-helical secondary structure, consistent with the known structural features of IFNα.
Biological properties of synthetic IFNα2b
Finally, we performed a series of biological tests to compare the properties of synthetic IFNα2b with its recombinant counterpart. For these studies we used a commercially available source of IFNα2b that is considered to be the gold standard. Among the various effects that IFNα has been shown to possess, we chose to compare the two molecules in two well-established, reliable and relatively simple assays that cover the major biological properties of IFNα: 1) antiviral activity against infection of A549 cells with encephalomyocarditis virus (ECMV), and 2) anti-proliferative activity in growth assays of human primary T cells.
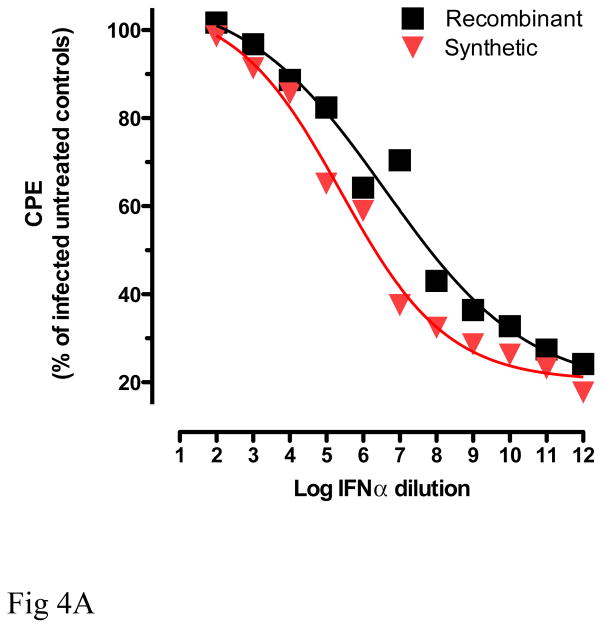
The antiviral assay measures the ability of IFNα to protect A549 cells from cytopathic effect (CPE) after challenge with EMCV [34]. As shown in Fig 4A, treatment of cells with recombinant and synthetic IFNα2b reduced the cytopathic effects of virus in a dose dependent manner. No statistically significant differences on the protection against CPE were observed between recombinant and synthetic IFNα2b.
Fig 4.
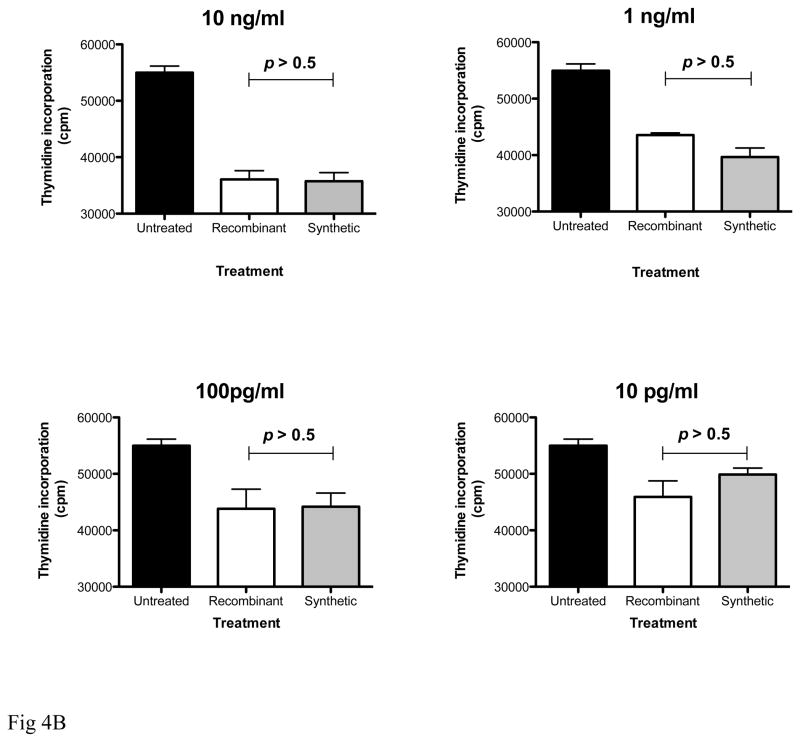
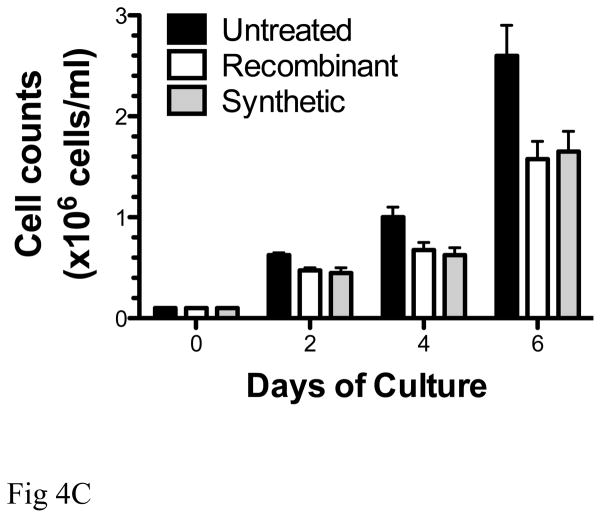
Comparative analyses of the biological properties of recombinant and synthetic IFNα2b. (A): Antiviral activity of the two molecules was determined based on a visual read of the protection of A549 cells from cytopathic effect (CPE) due to viral challenge with encephalomyocarditis virus (ECMV). Recombinant and synthetic IFNα2b were titrated in duplicate samples, and protection from CPE due to ECMV infection was determined in comparison to untreated A549 cells (no IFNα), and to uninfected, untreated cells (no virus, no IFNα) controls. (B, C): Inhibition of cell proliferation. Peripheral blood mononuclear cells (PBMC) of healthy volunteers were plated in triplicate samples, and proliferation was stimulated with Staphylococcal enterotoxin B and IL-2 (Roche Biochemicals). Cells were cultured in the presence of recombinant or synthetic IFNα2b (0.01–10 ng/ml) for 3 days at 37°C. Proliferation was determined by incorporation of [3H]-Thymidine compared to untreated samples (B). In samples treated with 10 ng/ml IFNα2b, cellular proliferation was also measured by cell counts with trypan blue exclusion (C).
Next, we compared the effect of recombinant and synthetic IFNα2b on the proliferation of human T cells following stimulation with SEB and IL-2. We used two alternative readouts to measure the effect of IFNα2b molecules on T cell growth: incorporation of radioactive thymidine into genomic DNA, and cell counts after staining with the viral dye trypan blue. Both assays demonstrate that recombinant and synthetic IFNα2b suppress proliferation of T cells in a dose-dependent fashion, and with similar potencies (Fig 4B, C).
Altogether, these assays prove that synthetic IFNα2b presents the same biological properties and similar potency as the recombinant, commercially available counterpart. Thus, the synthetic molecule is suitable for the screening of phage display libraries with the purpose of isolating peptides capable of blocking the interaction between IFNα2b and its receptor, and to suppress the detrimental effects due to chronic stimulation of the IFNα system.
Discussion
In the present study we have successfully produced IFNα2b using solid-phase synthesis and native chemical ligation. The final 165-residue polypeptide was able to fold into its native-like conformation containing two disulfide bonds (Cys1-Cys98, Cys29-Cys138). The molecular mass of folded IFNα2b protein as measured by electrospray ionization mass spectrometry was in agreement with the theoretical average isotropic mass. Moreover, the synthetic IFNα2b protein displayed the same spectrum of biological properties and activities as its recombinant counterpart.
Of note, ligation between Ser and Cys residues has been reported to cause minor racemization at Ser [36, 37]. We previously found in the total chemical synthesis of HIV-1 protease that racemization occurred at the C-terminal ligation site Ser89-Cys90, but did not occur at the N-terminal ligation site Ser46-Cys47, suggesting that this event is sequence-dependent [38]. In this work, we did not observe detectable amounts of racemization at the Ser28-Cys29 ligation site, certainly insufficient to inform a structural or functional detriment.
IFNα is a class 2 α-helical cytokine comprising a five-helix bundle, and a functionally important long loop that connects helices A and B [39]. The human genome contains at least 13 different IFNα genes clustered on the short arm of chromosome 9 [40]. Since IFNα1 and IFNα13 give rise to identical polypeptides, there are only 12 IFNα subtypes [41]. All IFNα subtypes are very similar in amino acid sequence and chemical structure, and exert their effects through the same IFNα/β receptor (IFNAR) receptor, constituted by two subunits – IFNAR1 and IFNAR2 – that comprise extracellular, transmembrane and intracellular domains [2]. IFNα binds its receptor by contacting first IFNAR2 with an affinity in the nanomolar range, and subsequently IFNAR1 with an affinity in the micromolar range [42]. The residues involved in the formation of the ternary complex have been determined for all three molecules [42]. IFNAR1 and IFNAR2 contact IFNα on opposite sides of the molecule [43, 44], forming a 1:1:1 ternary complex with an architecture that is very similar for all IFNα subtypes [42]. The initial interaction is driven by continuous hydrophobic patches on binding sites, whereas the association is maintained by electrostatic complementarity [45]. The highly variable range of binding affinities involved in the interaction between each IFNα molecule and the receptor account for the differences in signal activation triggered by various IFNα subtypes [42]. Indeed, the binding affinities of various IFNα subtypes correlate well with their potencies in biological assays [46].
IFNα is a key player in innate immunity against viral pathogens, and contributes to shape Th1 adaptive immune responses. However, chronic production of IFNα plays a role in the immunopathogenesis of certain viral infections and autoimmune conditions. Therefore, the ability to suppress the effects of unabated IFNα expression might have important clinical implications. This concept has been explored through active vaccination of HIV-1 infected individuals with an immunogenic, but biologically inactive form of IFNα2b [47]. Alternative strategies for the identification of specific binders against a target protein have been developed, including rational design and isolation of compounds from a collection of small-molecule chemicals. An additional approach is the isolation of antagonistic peptides from phage display libraries. The ability to produce large amounts of pure, biologically active IFNα represents the first, critical step toward the accomplishment of this goal.
Acknowledgments
We thank Dr. Bryan Ericksen for useful comments on this manuscript and Ms Weirong Yuan for technical assistance. This work was partially supported by National Institutes of Health Grants AI072732 and AI061482 (to WL).
References
- 1.Tough DF. Type I interferon as a link between innate and adaptive immunity through dendritic cell stimulation. Leukemia & lymphoma. 2004;45:257–64. doi: 10.1080/1042819031000149368. [DOI] [PubMed] [Google Scholar]
- 2.Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annual review of immunology. 2005;23:307–36. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- 3.Luft T, Pang KC, Thomas E, Hertzog P, Hart DN, Trapani J, et al. Type I IFNs enhance the terminal differentiation of dendritic cells. Journal of immunology. 1998;161:1947–53. [PubMed] [Google Scholar]
- 4.Luft T, Luetjens P, Hochrein H, Toy T, Masterman KA, Rizkalla M, et al. IFN-alpha enhances CD40 ligand-mediated activation of immature monocyte-derived dendritic cells. International immunology. 2002;14:367–80. doi: 10.1093/intimm/14.4.367. [DOI] [PubMed] [Google Scholar]
- 5.Montoya M, Schiavoni G, Mattei F, Gresser I, Belardelli F, Borrow P, et al. Type I interferons produced by dendritic cells promote their phenotypic and functional activation. Blood. 2002;99:3263–71. doi: 10.1182/blood.v99.9.3263. [DOI] [PubMed] [Google Scholar]
- 6.Levings MK, Sangregorio R, Galbiati F, Squadrone S, de Waal Malefyt R, Roncarolo MG. IFN-alpha and IL-10 induce the differentiation of human type 1 T regulatory cells. Journal of immunology. 2001;166:5530–9. doi: 10.4049/jimmunol.166.9.5530. [DOI] [PubMed] [Google Scholar]
- 7.Zella D, Romerio F, Curreli S, Secchiero P, Cicala C, Zagury D, et al. IFN-alpha 2b reduces IL-2 production and IL-2 receptor function in primary CD4+ T cells. Journal of immunology. 2000;164:2296–302. doi: 10.4049/jimmunol.164.5.2296. [DOI] [PubMed] [Google Scholar]
- 8.Romerio F, Riva A, Zella D. Interferon-alpha2b reduces phosphorylation and activity of MEK and ERK through a Ras/Raf-independent mechanism. British journal of cancer. 2000;83:532–8. doi: 10.1054/bjoc.2000.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romerio F, Zella D. MEK and ERK inhibitors enhance the anti-proliferative effect of interferon-alpha2b. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2002;16:1680–2. doi: 10.1096/fj.02-0120fje. [DOI] [PubMed] [Google Scholar]
- 10.Blomberg S, Eloranta ML, Cederblad B, Nordlin K, Alm GV, Ronnblom L. Presence of cutaneous interferon-alpha producing cells in patients with systemic lupus erythematosus. Lupus. 2001;10:484–90. doi: 10.1191/096120301678416042. [DOI] [PubMed] [Google Scholar]
- 11.Lovgren T, Eloranta ML, Bave U, Alm GV, Ronnblom L. Induction of interferon-alpha production in plasmacytoid dendritic cells by immune complexes containing nucleic acid released by necrotic or late apoptotic cells and lupus IgG. Arthritis and rheumatism. 2004;50:1861–72. doi: 10.1002/art.20254. [DOI] [PubMed] [Google Scholar]
- 12.Lovgren T, Eloranta ML, Kastner B, Wahren-Herlenius M, Alm GV, Ronnblom L. Induction of interferon-alpha by immune complexes or liposomes containing systemic lupus erythematosus autoantigen- and Sjogren’s syndrome autoantigen-associated RNA. Arthritis and rheumatism. 2006;54:1917–27. doi: 10.1002/art.21893. [DOI] [PubMed] [Google Scholar]
- 13.Ronnblom L, Pascual V. The innate immune system in SLE: type I interferons and dendritic cells. Lupus. 2008;17:394–9. doi: 10.1177/0961203308090020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang X, Yuang J, Goddard A, Foulis A, James RF, Lernmark A, et al. Interferon expression in the pancreases of patients with type I diabetes. Diabetes. 1995;44:658–64. doi: 10.2337/diab.44.6.658. [DOI] [PubMed] [Google Scholar]
- 15.Chehadeh W, Weill J, Vantyghem MC, Alm G, Lefebvre J, Wattre P, et al. Increased level of interferon-alpha in blood of patients with insulin-dependent diabetes mellitus: relationship with coxsackievirus B infection. The Journal of infectious diseases. 2000;181:1929–39. doi: 10.1086/315516. [DOI] [PubMed] [Google Scholar]
- 16.Stewart TA, Hultgren B, Huang X, Pitts-Meek S, Hully J, MacLachlan NJ. Induction of type I diabetes by interferon-alpha in transgenic mice. Science. 1993;260:1942–6. doi: 10.1126/science.8100367. [DOI] [PubMed] [Google Scholar]
- 17.Calabrese LH, Proffitt MR, Gupta MK, Easley KA, Walker JR, Rehm SJ, et al. Serum beta 2-microglobulin and interferon in homosexual males: relationship to clinical findings and serologic status to the human T lymphotropic virus (HTLV-III) AIDS research. 1984;1:423–38. doi: 10.1089/aid.1.1983.1.423. [DOI] [PubMed] [Google Scholar]
- 18.Mandl JN, Barry AP, Vanderford TH, Kozyr N, Chavan R, Klucking S, et al. Divergent TLR7 and TLR9 signaling and type I interferon production distinguish pathogenic and nonpathogenic AIDS virus infections. Nature medicine. 2008;14:1077–87. doi: 10.1038/nm.1871. [DOI] [PubMed] [Google Scholar]
- 19.Noraz N, Lathey JL, Spector SA. Human cytomegalovirus-associated immunosuppression is mediated through interferon-alpha. Blood. 1997;89:2443–52. [PubMed] [Google Scholar]
- 20.Eckert DM, Malashkevich VN, Hong LH, Carr PA, Kim PS. Inhibiting HIV-1 entry: discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket. Cell. 1999;99:103–15. doi: 10.1016/s0092-8674(00)80066-5. [DOI] [PubMed] [Google Scholar]
- 21.Schumacher TN, Mayr LM, Minor DL, Jr, Milhollen MA, Burgess MW, Kim PS. Identification of D-peptide ligands through mirror-image phage display. Science. 1996;271:1854–7. doi: 10.1126/science.271.5257.1854. [DOI] [PubMed] [Google Scholar]
- 22.Liu M, Li C, Pazgier M, Li C, Mao Y, Lv Y, et al. D-peptide inhibitors of the p53-MDM2 interaction for targeted molecular therapy of malignant neoplasms. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14321–6. doi: 10.1073/pnas.1008930107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu M, Pazgier M, Li C, Yuan W, Li C, Lu W. A left-handed solution to peptide inhibition of the p53-MDM2 interaction. Angewandte Chemie. 2010;49:3649–52. doi: 10.1002/anie.201000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhan C, Zhao L, Wei X, Wu X, Chen X, Yuan W, et al. An ultrahigh affinity d-peptide antagonist Of MDM2. Journal of medicinal chemistry. 2012;55:6237–41. doi: 10.1021/jm3005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welch BD, VanDemark AP, Heroux A, Hill CP, Kay MS. Potent D-peptide inhibitors of HIV-1 entry. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16828–33. doi: 10.1073/pnas.0708109104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao L, Lu W. Mirror image proteins. Current opinion in chemical biology. 2014;22C:56–61. doi: 10.1016/j.cbpa.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dawson PE, Kent SB. Synthesis of native proteins by chemical ligation. Annu Rev Biochem. 2000;69:923–60. doi: 10.1146/annurev.biochem.69.1.923. [DOI] [PubMed] [Google Scholar]
- 28.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–9. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 29.Kent SB. Total chemical synthesis of proteins. Chem Soc Rev. 2009;38:338–51. doi: 10.1039/b700141j. [DOI] [PubMed] [Google Scholar]
- 30.Hackeng TM, Griffin JH, Dawson PE. Protein synthesis by native chemical ligation: expanded scope by using straightforward methodology. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:10068–73. doi: 10.1073/pnas.96.18.10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schnolzer M, Alewood P, Jones A, Alewood D, Kent SB. In situ neutralization in Boc-chemistry solid phase peptide synthesis. Rapid, high yield assembly of difficult sequences. International journal of peptide and protein research. 1992;40:180–93. doi: 10.1111/j.1399-3011.1992.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 32.Bang D, Chopra N, Kent SB. Total chemical synthesis of crambin. Journal of the American Chemical Society. 2004;126:1377–83. doi: 10.1021/ja0385078. [DOI] [PubMed] [Google Scholar]
- 33.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein science: a publication of the Protein Society. 1995;4:2411–23. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rubinstein S, Familletti PC, Pestka S. Convenient assay for interferons. Journal of virology. 1981;37:755–8. doi: 10.1128/jvi.37.2.755-758.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klaus W, Gsell B, Labhardt AM, Wipf B, Senn H. The three-dimensional high resolution structure of human interferon alpha-2a determined by heteronuclear NMR spectroscopy in solution. Journal of molecular biology. 1997;274:661–75. doi: 10.1006/jmbi.1997.1396. [DOI] [PubMed] [Google Scholar]
- 36.Kawakami T, Aimoto S. The use if a cysteinyl prolyl ester (CPE) autoactivating unit in peptide ligation reactions. Tetrahedron. 2009;65:3871–7. [Google Scholar]
- 37.Sato K, Shigenaga A, Kitakaze K, Sakamoto K, Tsuji D, Itoh K, et al. Chemical synthesis of biologically active monoglycosylated GM2-activator protein analogue using N-sulfanylethylanilide peptide. Angewandte Chemie. 2013;52:7855–9. doi: 10.1002/anie.201303390. [DOI] [PubMed] [Google Scholar]
- 38.Li C, Li X, Lu W. Total chemical synthesis of human T-cell leukemia virus type 1 protease via native chemical ligation. Biopolymers. 2010;94:487–94. doi: 10.1002/bip.21375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pestka S. The interferons: 50 years after their discovery, there is much more to learn. The Journal of biological chemistry. 2007;282:20047–51. doi: 10.1074/jbc.R700004200. [DOI] [PubMed] [Google Scholar]
- 40.Allen G, Diaz MO. Nomenclature of the human interferon proteins. Journal of interferon research. 1994;14:223–6. doi: 10.1089/jir.1994.14.223. [DOI] [PubMed] [Google Scholar]
- 41.Foster GR, Finter NB. Are all type I human interferons equivalent? Journal of viral hepatitis. 1998;5:143–52. doi: 10.1046/j.1365-2893.1998.00103.x. [DOI] [PubMed] [Google Scholar]
- 42.Uze G, Schreiber G, Piehler J, Pellegrini S. The receptor of the type I interferon family. Current topics in microbiology and immunology. 2007;316:71–95. doi: 10.1007/978-3-540-71329-6_5. [DOI] [PubMed] [Google Scholar]
- 43.Roisman LC, Piehler J, Trosset JY, Scheraga HA, Schreiber G. Structure of the interferon-receptor complex determined by distance constraints from double-mutant cycles and flexible docking. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:13231–6. doi: 10.1073/pnas.221290398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roisman LC, Jaitin DA, Baker DP, Schreiber G. Mutational analysis of the IFNAR1 binding site on IFNalpha2 reveals the architecture of a weak ligand-receptor binding-site. Journal of molecular biology. 2005;353:271–81. doi: 10.1016/j.jmb.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 45.Piehler J, Roisman LC, Schreiber G. New structural and functional aspects of the type I interferon-receptor interaction revealed by comprehensive mutational analysis of the binding interface. The Journal of biological chemistry. 2000;275:40425–33. doi: 10.1074/jbc.M006854200. [DOI] [PubMed] [Google Scholar]
- 46.Langer JA. Interferon at 50: new molecules, new potential, new (and old) questions. Science’s STKE: signal transduction knowledge environment. 2007:pe53. doi: 10.1126/stke.4052007pe53. [DOI] [PubMed] [Google Scholar]
- 47.Gringeri A, Musicco M, Hermans P, Bentwich Z, Cusini M, Bergamasco A, et al. Active anti-interferon-alpha immunization: a European-Israeli, randomized, double-blind, placebo-controlled clinical trial in 242 HIV-1--infected patients (the EURIS study) Journal of acquired immune deficiency syndromes and human retrovirology: official publication of the International Retrovirology Association. 1999;20:358–70. doi: 10.1097/00042560-199904010-00006. [DOI] [PubMed] [Google Scholar]