Summary
Dynein recruitment to the nuclear envelope is required for pre-mitotic nucleus-centrosome interactions in nonneuronal cells, and for apical nuclear migration in neural stem cells. In each case, dynein is recruited to the nuclear envelope (NE) specifically during G2, via two nuclear pore-mediated mechanisms involving RanBP2-BicD2 and Nup133-CENP-F. The mechanisms responsible for cell cycle control of this behavior are unknown. We now find that Cdk1 serves as a direct master controller for NE dynein recruitment in neural stem cells and HeLa cells. Cdk1 phosphorylates conserved sites within RanBP2 and activates BicD2 binding and early dynein recruitment. Late recruitment is triggered by a Cdk1-induced export of CENP-F from the nucleus. Forced NE targeting of BicD2 overrides Cdk1 inhibition, fully rescuing dynein recruitment and nuclear migration in neural stem cells. These results reveal how NE dynein recruitment is cell cycle regulated, and identify the trigger mechanism for apical nuclear migration in the brain.
Introduction
Cell cycle-mediated recruitment of motor proteins to the nuclear envelope (NE) has emerged as a general and important phenomenon in mitotic progression and brain development. G2-dependent NE dynein recruitment, in particular, contributes to pre-mitotic centrosome separation and proper spindle assembly in nonneuronal cells (Bolhy et al., 2011; Raaijmakers et al., 2012). This mechanism plays an additional, essential role in driving cell cycle-dependent nuclear oscillations and in controlling proliferation of radial glial progenitor cells (RGP cells), the neural stem cells of the neocortex (Hu et al., 2013).
Development of the neocortex is a highly complex process, initiated within a zone of rapidly proliferating RGP cells, followed by long-range migration of newborn neurons to establish the highly ordered cortical neuronal layers. Elaborate cellular mechanisms have evolved to ensure the fidelity of these processes. The RGP cells are important in giving rise to all neurogenic lineages in the mammalian cortex, including adult stem cells (Kriegstein and Alvarez-Buylla, 2009; Noctor et al., 2001; Paridaen and Huttner, 2014). They are highly elongated, spanning the distance from the ventricular (apical) to the pial (basal) surface of the brain. Following mitosis at the ventricular surface, they undergo interkinetic nuclear migration (INM) (Kosodo, 2012; Lee and Norden, 2013; Spear and Erickson, 2012). This involves G1-specific basal nuclear migration, S phase, and G2-specific apical nuclear migration for the subsequent mitotic division.
The mechanisms responsible for this long-mysterious behavior, its biological control, and its developmental purpose have only recently begun to be understood. Microtubule motors and acto-myosin have been implicated in INM in a number of systems (Messier, 1978; Meyer et al., 2011; Norden et al., 2009; Pacary et al., 2013; Rujano et al., 2013; Schenk et al., 2009; Tsai et al., 2005; 2010). In mammalian RGP cells, where microtubules play a key role, the centrosome is localized apically and organizes a polarized microtubule network (Tsai et al., 2010). Our own work in rat brain has identified reciprocal roles for the plus-end-directed kinesin KIF1A in G1 basal nuclear migration, and the minus-end-directed motor cytoplasmic dynein in G2 apical migration (Hu et al., 2013; Tsai et al., 2010) (Figure 1A).
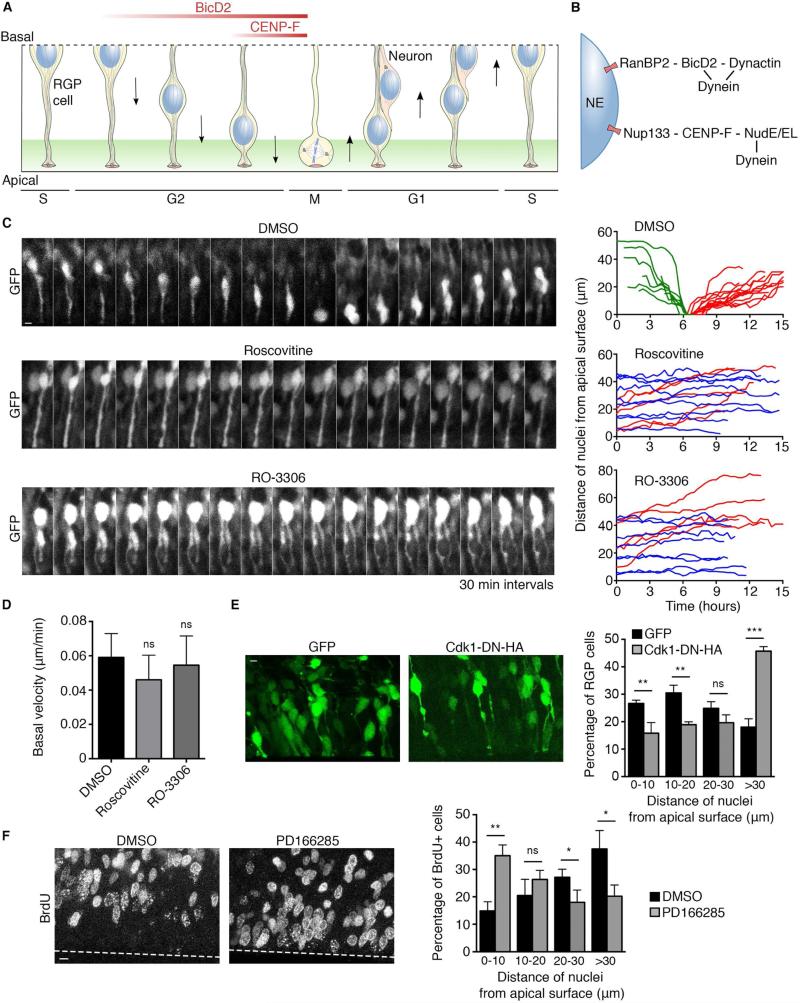
Figure 1. Requirement for Cdk1 in apical nuclear migration in RGP cells.
(A) Schematic representation of interkinetic nuclear migration (INM) in RGP cells (from S-phase to S-phase to correspond to time-lapse imaging). Following S-phase, the G2 nucleus moves to the apical (ventricular) surface, driven by NE-associated cytoplasmic dynein. (B) Dynein is recruited to the NE through an early G2 pathway anchored by the nucleoporin RanBP2, responsible for long-range apical nuclear migration; and a late G2 pathway anchored by the nucleoporin Nup133, responsible for pre-mitotic nuclear transport to the ventricular surface of the brain. All factors are expressed throughout the cell cycle except CENP-F, which is absent in G1 and rises during S and G2 phases. (C) Live imaging of GFP-expressing RGP cells in embryonic rat brain slices 3 days after in utero electroporation at E16. The slices were treated with vehicle (DMSO), 55 μM Roscovitine, or 100 μM RO-3306 and imaged for 16 hours (see methods). DMSO-treated cells showed typical INM behavior. Roscovitine and RO-3306 each blocked apical nuclear migration. Right: Representative tracks of individual nuclei for each condition are presented. Green tracks indicate apically migrating nuclei, red tracks basally migrating nuclei, and blue tracks non-migrating nuclei. (D) Basal nuclear migration velocity showed a small, but insignificant change in response to Roscovitine and RO-3306 treatment. (E) Effect of Cdk1 dominant negative on distribution of RGP cells nuclei. Brains were electroporated at E16 with GFP or with Cdk1-DN-HA and fixed and imaged at E18. Measurement of the distance between the bottom of the nucleus and the apical surface shows strong accumulation of nuclei away from the apical surface in cells expressing Cdk1-DN-HA (>30 μm from the ventricle). (F) Effect of Wee1/Myt1 inhibitor PD166285 on distribution of BrdU+ cells, two hours after BrdU pulse. Live brain slices were incubated with BrdU for 15 minutes, washed, and subsequently treated for 2 hours with 1 μM PD166285 or DMSO. The percentage of BrdU+ nuclei that reached the apical region (0-10 μm from the ventricle) strongly increased in PD166285-treated brain slices, compared to DMSO control. For each experiment, at least three independent brains were imaged. Error bars indicate SD; *p<0.05; **p<0.01; ***p<0.001; ns = not significant, based on a Student's t-test. Scale bar, 5 μm. See also Figure S1 and Movies S1, S2, S3, S4 and S5.
Although INM is essential for normal brain development (Hu et al., 2013), the mechanisms for its cell cycle control remain largely unknown (Liang et al., 2014). The microtubule-associated protein Tpx2 was reported to be enriched in the apical process during G2, and its knockdown reduced the rate of nuclear migration at this stage (Kosodo et al., 2011). The G2-M kinase Cdk1 was also implicated in myosin-dependent nuclear migration in the zebrafish neuroepithelium (Leung et al., 2011; Strzyz et al., 2015). In the rodent neocortex, apical nuclear migration involves G2-specific dynein recruitment to the nuclear envelope (NE) (Hu et al., 2013), potentially a target of cell cycle regulation.
Dynein associates with the G2 NE through two sequential mechanisms, which are active in cultured nonneuronal cells as well as in RGP cells (Figure 1B) (Beaudouin et al., 2002; Bolhy et al., 2011; Hu et al., 2013; Salina et al., 2002; Splinter et al., 2010). In early G2, the nucleoporin RanBP2 binds the dynein regulator BicD2, which in turn recruits dynein, dynactin, and LIS1 to the NE (Splinter et al., 2010; 2012). Later in G2, the nucleoporin Nup133 binds CENP-F, which recruits dynein via NudE and NudEL (Bolhy et al., 2011). In nonneuronal cells, the early and late pathways both contribute to dynein NE recruitment, which controls centrosome separation and early spindle assembly (Bolhy et al., 2011; Raaijmakers et al., 2012). We found that, in the brain, the early RanBP2 pathway was responsible for the initial stage of nuclear migration, whereas the later Nup133 pathway was required for the nucleus to reach the ventricular surface for mitotic entry (Hu et al., 2013). How these early and late mechanisms are activated in a cell cycle stage-specific manner in RGP cells as well as nonneuronal cells remains a mystery.
We have now tested the role of cell cycle regulators in the control of dynein recruitment to the HeLa and RGP cell NE and in the regulation of INM in the brain. We find that dynein recruitment and apical nuclear migration are each triggered by the G2/M regulator Cdk1. The protein kinase phosphorylates a unique cluster of sites within RanBP2, and activates BicD2 binding and recruitment of its downstream interactors dynein, dynactin, and LIS1. We find that late G2 dynein recruitment to the NE and late apical nuclear migration are initiated by a Cdk1-induced redistribution of CENP-F from inside the nucleus to the NE. Finally, we show that forced targeting of BicD2 to the NE is sufficient to rescue dynein recruitment in Cdk1-inhibited HeLa cells and to restore apical nuclear migration in Cdk1-inhibited brain tissue.
Results
Requirement for Cdk1 in apical nuclear migration in rat brain
As a first step in understanding cell cycle control of NE dynein recruitment, we tested the role of the G2-M phase-specific protein kinase Cdk1 (Deibler and Kirschner, 2010; Lindqvist et al., 2009) in apical nuclear migration in rat brain. GFP was expressed in E16 RGP cells using in utero electroporation. Live brain slices were prepared at E19, exposed to small molecule inhibitors, and RGP cells behavior then monitored by live imaging. Cells exposed to the DMSO vehicle alone exhibited typical INM behavior, with apical nuclear migration occurring at normal rates (0.147 ± 0.042 μm/min), followed by mitosis at the apical surface and basal nuclear migration (0.059 ± 0.014 μm/min) (Figure 1C and Movie S1). Incubation of the rat brain slices with the commonly used Cdk inhibitor Roscovitine (Meijer et al., 1997), which has a potent effect on Cdk1, strongly impaired apical nuclear migration (Figure 1C and Movie S2). No significant effect on basal nuclear migration was detected (0.046 ± 0.014 μm/min) (Figures 1D and S1A and Movie S3). As expected, the cells showed no sign of mitotic entry. Because Roscovitine also affects Cdk2 and Cdk5, we tested the selective Cdk1 inhibitor RO-3306 (Lapenna and Giordano, 2009; Vassilev et al., 2006). This compound also arrested apical nuclear migration, with no effect on basal nuclear migration (0.054 ± 0.017 μm/min) (Figures 1C, 1D and S1A and Movies S4 and S5). As controls for inhibitor specificity, we tested their effect on the migration of post-mitotic neurons, which are generated from asymmetric RGP cell divisions. These cells have exited the cell cycle and no longer depend on Cdk1 activity, though Cdk5 is critical in their behavior (Smith et al., 2001). Consistent with these considerations, Roscovitine potently reduced the rate of neuronal migration, whereas RO-3306 had no significant effect (Figures S1B and S1C).
To confirm a role for Cdk1 in apical nuclear migration, we expressed an HA-tagged Cdk1 kinase-dead dominant-negative construct (van den Heuvel and Harlow, 1993) in RGP cells and measured its effect on nuclear positioning. In agreement with our small molecule inhibition data, this construct induced a strong accumulation of nuclei away from the apical surface of the brain (>30 μm from the ventricle) (Fig 1E), a phenotype reminiscent of that for dynein and BicD2 knockdown (Hu et al., 2013; Tsai et al., 2010). Finally, we tested the effect of Cdk1 stimulation on apical nuclear migration using PD166285, a compound that inhibits the Cdk1 negative regulators Wee1 and Myt1 (Potapova et al., 2009). To quantitatively measure a potential stimulatory effect of this inhibitor on apical migration, we traced the fraction of RGP cells in S phase using a 5-bromo-2’ deoxyuridine (BrdU) labeling experiment and analyzed the localization of their nuclei two hours after the BrdU pulse. In DMSO-treated brain slices, only 14.8 ± 3.4% of BrdU+ nuclei had reached the apical region (0-10 μm from the ventricle) while this percentage was increased to 35 ± 3.9% in PD166285-treated slices, indicating that Wee1/Myt1 inhibition stimulated apical nuclear migration (Fig 1F). Together, our results show that Cdk1 activity controls apical nuclear migration in RGP cells.
Requirement for Cdk1 in nuclear envelope dynein recruitment in HeLa cells
In view of the role of dynein in apical INM, we investigated the effect of the cell cycle inhibitors on its recruitment to the NE. We first focused on HeLa cells, which use the same adaptor proteins as RGP cells to recruit dynein to the NE during G2 (Hu et al., 2013). Incubation of HeLa cells with Roscovitine caused dramatic displacement of dynein from the NE of G2 cells (identified by the expression of cyclin B1), an effect readily reversed by drug washout (Figure 2A and 2E). Very similar results were observed using RO-3306 or the Cdk1/Cdk2 inhibitor III (Gavet and Pines, 2010) (Figure 2A and 2E). The Aurora A inhibitor VX680 (Harrington et al., 2004) had no significant effect, but a mild effect was observed with the Plk1 inhibitor BTO-1 (Bader et al., 2011) (Figure 2E). This could reflect a role for Plk1 in Cdk1 activation, though a more direct role in dynein recruitment is also possible (Li et al., 2010).
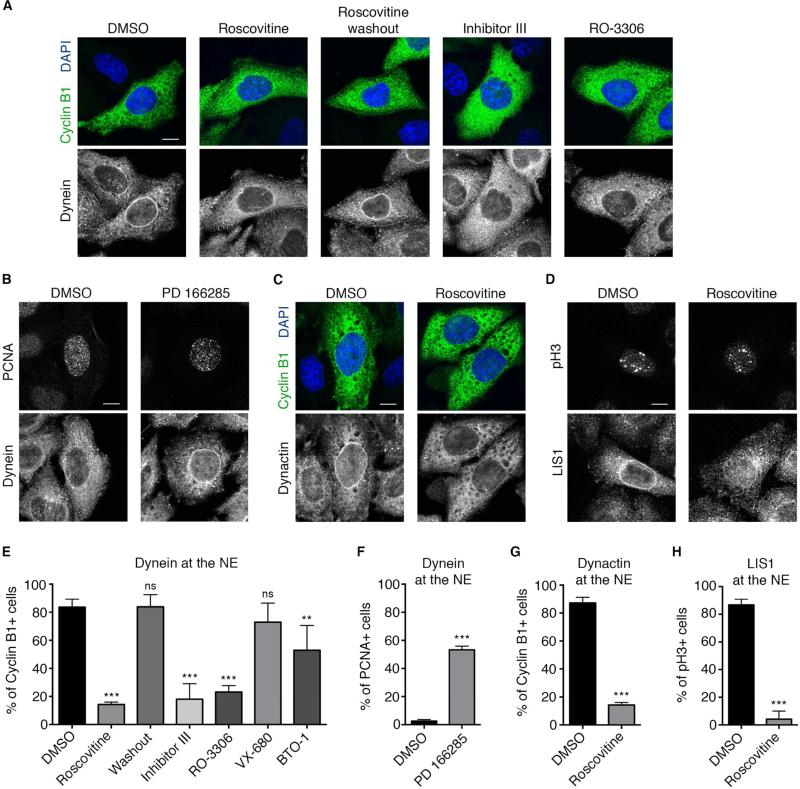
Figure 2. Requirement for Cdk1 in HeLa cell nuclear envelope (NE) dynein recruitment.
(A) Effect of Cdk1 inhibitors on NE dynein (anti-IC) in G2 (cyclin B1-positive) HeLa cells. Roscovitine (55 μM, 30 min) markedly reduced cytoplasmic dynein staining at the NE, an effect reversed by 30 minutes of Roscovitine washout. Incubation with other Cdk1 inhibitors: inhibitor III (0.9 μM) or RO-3306 (28 μM), for 30 minutes, also blocked dynein accumulation at the NE. (B) Effect of Wee1/Myt1 inhibitor PD166285 on dynein recruitment to the NE. HeLa cells were immunostained for dynein (IC) and the S phase marker PCNA (which forms intranuclear foci at this stage). Exposure to PD166285 (0.5 μM, 30 min) strongly increased the fraction of S phase cells exhibiting NE dynein. (C) Roscovitine (55 μM, 30 min) strongly inhibited NE dynactin (anti-p150) staining in Cyclin B1-positive HeLa cells and (D) NE LIS1 staining in phospho-Histone 3 (pH3)-positive HeLa cells (For antibody compatibility, phospho-histone H3 (pH3) was used as a marker for G2/prophase cells, see methods). (E) Quantification of the effects of Cdk1 inhibitors Roscovitine, Inhibitor III, and RO-3306; Aurora A inhibitor VX-680 (0.3 μM); and Plk1 inhibitor BTO-1 (22 μM) on dynein recruitment to the NE of cyclin B1+ cells. (F) Quantification of the effect of Wee1/Myt1 inhibitor PD166285 on dynein recruitment to the NE of S-phase (PCNA+) cells. (G) Quantification of the effect of Cdk1 inhibitor Roscovitine on dynactin recruitment to the NE of cyclin B1+ cells. (H) Quantification of the effect of Cdk1 inhibitor Roscovitine on LIS1 recruitment to the NE of pH3+ cells. Each experiment was reproduced three independent times (over 50 cells per condition and per experiment were counted). Error bars indicate SD; **p<0.01; ***p<0.001; ns = not significant, based on a Student's t-test. Scale bar, 10 μm.
To test the effect of Cdk1 stimulation on the recruitment of dynein to the NE, we again used the Wee1/Myt1 inhibitor PD166285. This reagent markedly increased the number of total cells positive for perinuclear dynein. Strikingly, NE dynein could now be observed not only in G2 cells, but, remarkably, in more than half of S-phase cells as well, a condition not previously reported (Figure 2B and 2F). Finally, Roscovitine also displaced the major dynein regulators dynactin, LIS1, and NudE/EL from the NE (Figure 2C, 2D, 2G, 2H and S3) (Raaijmakers et al., 2013).
Cdk1 independently regulates each of the two dynein recruitment pathways in HeLa cells
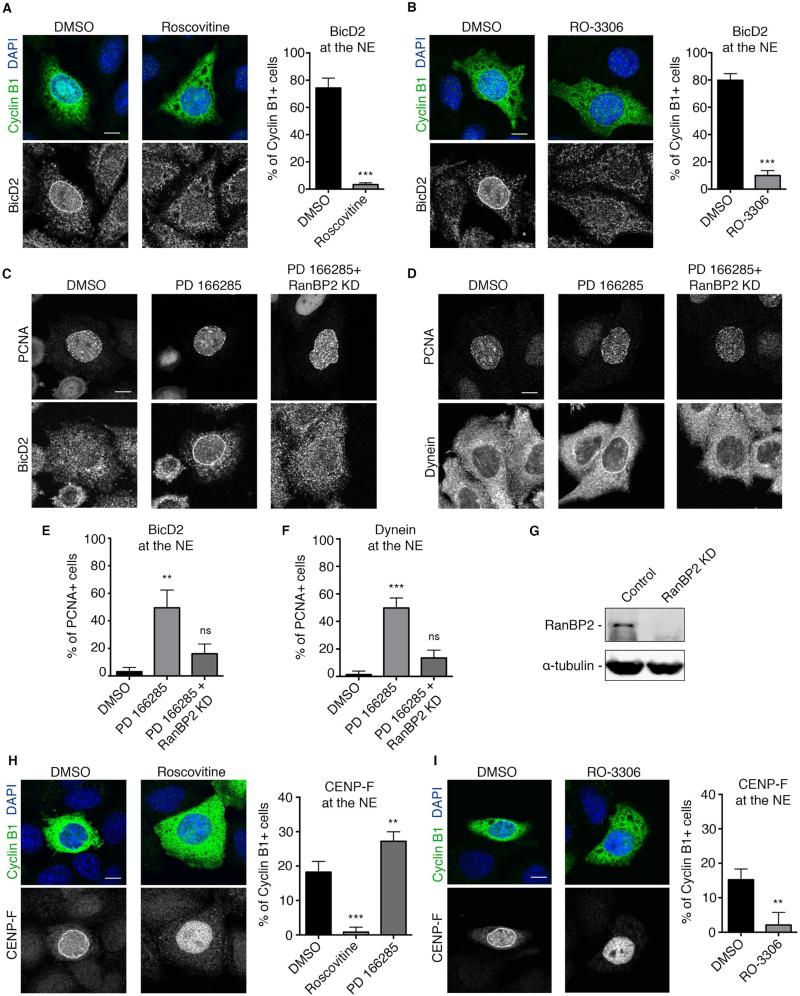
Our previous results suggested that the RanBP2-BicD2 dynein recruitment pathway is activated earlier during G2 than the Nup133-CENP-F pathway (Hu et al., 2013). To determine whether Cdk1 acts at a unique control point or independently activates each mechanism, we stained inhibitor-treated HeLa cells for BicD2 and CENP-F as upstream markers for each pathway. Roscovitine as well as RO-3306 potently and dramatically inhibited recruitment of BicD2 to the NE of cyclin B1-positive cells (Figures 3A and 3B). Treating cells with the Wee1/Myt1 inhibitor PD166285 resulted, as for dynein, in the appearance of BicD2 at the NE of a substantial subset of S phase cells (49.5 ± 12.9%) (Figure 3C and 3E). siRNA-mediated knockdown of RanBP2 abolished this premature S phase recruitment of BicD2 ad dynein (Figures 3C-3G), suggesting that PD166285 acts by activating the RanBP2-BicD2 pathway.
Figure 3. Requirement for Cdk1 in both early and late dynein NE recruitment pathways.
(A) Roscovitine (55 μM, 30 min) strongly inhibited BicD2 recruitment to the NE of Cyclin B1+ cells. (B) RO-3306 (28 μM, 30 min) also strongly impaired BicD2 recruitment to the NE of Cyclin B1+ cells. (C) The Wee1/Myt1 inhibitor PD166285 (0.5 μM, 30 min) induced premature BicD2 accumulation at the S-phase NE marked by PCNA. siRNA-mediated knockdown of RanBP2 inhibited this premature BicD2 recruitment in PCNA+ cells. (D) siRNA-mediated knockdown of RanBP2 also inhibited the premature recruitment of dynein to the NE of S phase cells (marked by PCNA). (E and F) Quantification of the effect of Wee1/Myt1 inhibitor PD166285 and RanBP2 knockdown on (E) BicD2 and (F) dynein recruitment to the nuclear envelope of S phase cells (marked by PCNA). (G) Western Blot analysis of RanBP2 levels in HeLa cells reveals strong knockdown 3 days post-transfection with RanBP2 siRNA. (H) Effect of Roscovitine and PD166285 on CENP-F recruitment to the NE of cyclin B1+ cells. NE CENP-F decreased from 16.75 ± 1.7 % in DMSO-treated cells to 0.82 ± 1.4 % in Roscovitine-treated cells but increased to 27.2 ± 1.6% in PD166285 treated cells. Roscovitine treatment caused CENP-F to remain inside the nucleus. (I) RO-3306 (28 μM, 30 min) also strongly impaired CENP-F recruitment to the NE of Cyclin B1+ cells, which remained inside the nucleus. Each experiment was reproduced three independent times (over 50 cells per condition and per experiment were counted). Error bars indicate SD; **p<0.01, ***p<0.001, ns = not significant, based on a Student's t-test. Scale bar, 10 μm.
In contrast to the other NE proteins, CENP-F is concentrated within the nucleus during G2 but exits to accumulate at the NE prior to prophase nuclear envelope breakdown, before relocalizing to the kinetochores during mitosis (Hussein and Taylor, 2002). Strikingly, CENP-F remained inside the nucleus in virtually all Roscovitine- and RO-3306-treated G2 cells (Figures 3H and 3I), suggesting a role for Cdk1 in controlling the redistribution CENP-F to the cytoplasm, though whether this is a direct or indirect function is uncertain. PD166285 increased the fraction of NE CENP-F-positive G2 cells (Figure 3H), though, in this case, S-phase cells were unaffected (data not shown). These results reveal that Cdk1 plays a critical role in each of the two G2-specific dynein recruitment pathways and acts primarily at the level of BicD2 and CENP-F recruitment to the nucleoporins.
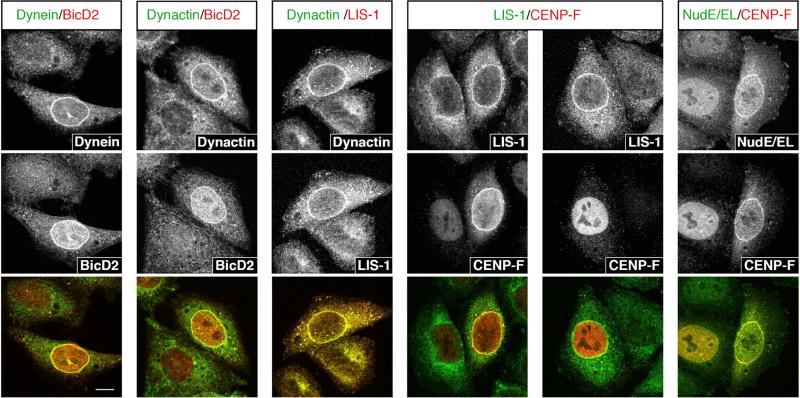
To better define the involvement of dynein regulatory factors in the two pathways, we performed a series of pairwise colocalization tests (Figure 4). BicD2, dynein, dynactin, and LIS1 always colocalized when any one was detected at the NE, in support of a shared role in the early RanP2-BicD2 pathway. Conversely, most cells with NE LIS1 exhibited intranuclear CENP-F (76.4% ±2.4), suggesting that LIS1 can be recruited independently of the Nup133-CENP-F pathway. Finally, CENP-F and NudE/EL always coincided at the NE, supporting NudE/EL recruitment through the late pathway only. These results suggest that BicD2 recruits dynein, dynactin, and LIS1 to the NE, while NudE/EL is recruited specifically though CENP-F.
Figure 4. Colocalization analysis between dynein and dynein regulatory factors at the NE.
Stainings for dynein and known dynein partners at the NE. Factors were stained two by two and colocalization was assessed. Dynein, dynactin, BicD2 and LIS1 always colocalize to the nuclear envelope, suggesting that they are all part of the early dynein recruitment pathway. LIS1 and CENP-F only colocalize in 23.6% (± 2.4%) of the case, while in 76.4% (± 2.4%) of LIS1-positive cells, CENP-F is still intranuclear. CENP-F and NudE/EL always colocalize at the NE, suggesting that they are both specific to the late dynein recruitment pathway.
Forced recruitment of BicD2 to the NE rescues dynein localization in Cdk1-inhibited cells
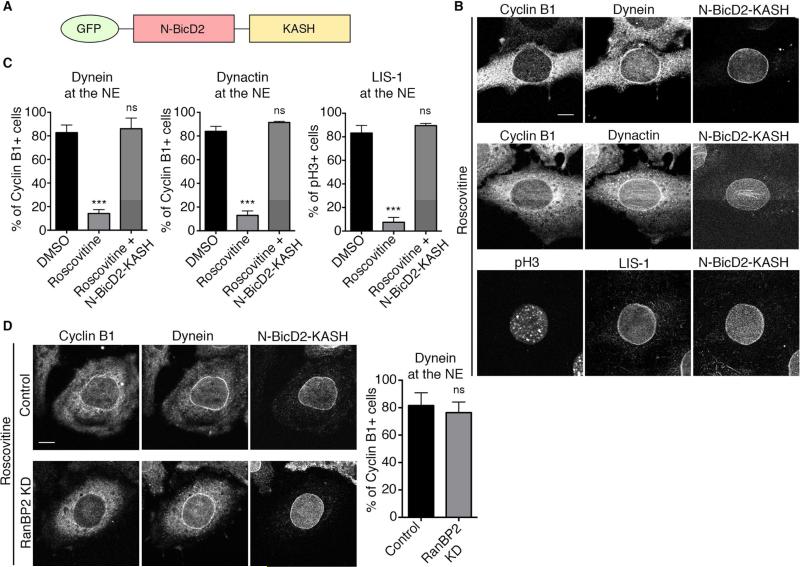
We focused our further analysis on the early pathway (RanBP2-BicD2) for its role in the initiation and early stages of apical nuclear migration in RPG cells. Our data revealed a role for Cdk1 in NE BicD2 recruitment, but whether downstream factors were also under direct Cdk1 control remained unclear. To resolve this issue, we expressed the N-BicD2-KASH fusion construct (Splinter et al., 2010), which targets the N-terminal dynein/dynactin-binding domain of BicD2 to the nuclear envelope via the Nesprin-3 KASH domain (Figure 5A). Expression of this construct in Roscovitine-treated cells restored dynein, dynactin, and LIS1 to the NE of G2/prophase cells (Figure 5B and 5C), indicating that NE BicD2 is sufficient to recruit these factors, independently of Cdk1 activity. NE NudE/EL recruitment was not restored, supporting a distinct role in late NE dynein recruitment (Figure S2). As expected, knockdown of RanBP2 did not affect dynein recruitment to the NE by N-BicD2-KASH (Figure 5D). Expression of this construct also induced the recruitment of the dynein/dynactin complex to the NE of Cyclin B1 negative cells (Figure S3A and S3B), implying that G1/S cells and Cdk1-inhibited G2 cells are equally competent for dynein recruitment by BicD2. Together, these experiments indicate that Cdk1 is the main regulator of the early dynein recruitment pathway and suggest that it mainly acts at the level of BicD2 binding to RanBP2.
Figure 5. Forced recruitment of BicD2 to the NE rescues dynein localization in Cdk1-inhibited cells.
(A) Schematic representation of N-BicD2-KASH fusion protein (Splinter et al., 2010). The C-terminal RanBP2-binding domain has been replaced by the KASH nuclear envelope targeting motif from Nesprin 3 (yellow). (B) Effect of NE-targeted BicD2 on NE envelope recruitment of dynein and its cofactors in Cdk1-inhibited HeLa cells. Cells were transfected with N-BicD2-KASH, treated with Roscovitine (55 μM, 30 min), and stained for dynein, dynactin, and LIS1. N-BicD2-KASH decorates the NE and restores dynein, dynactin, and LIS1 localization to the NE of cyclin B1+ cells in the first two cases, and of pH3+ cells in the case of LIS1. (C) Quantification of the effect of N-BicD2-KASH expression on dynein, dynactin, and LIS1 localization to the NE in Roscovitine treated cells. (D) RanBP2 knockdown did not affect N-BicD2-KASH-mediated recruitment of dynein/dynactin to the NE of Cyclin B1+ cells. Each experiment was reproduced three independent times (over 50 cells per condition and per experiment were counted). Error bars indicate SD; ***p<0.001; ns = not significant, based on a Student's t-test. Scale bar, 10 μm. See also Figures S2 and S3.
Regulation of RanBP2-BicD2 Interaction by Cdk1 phosphorylation
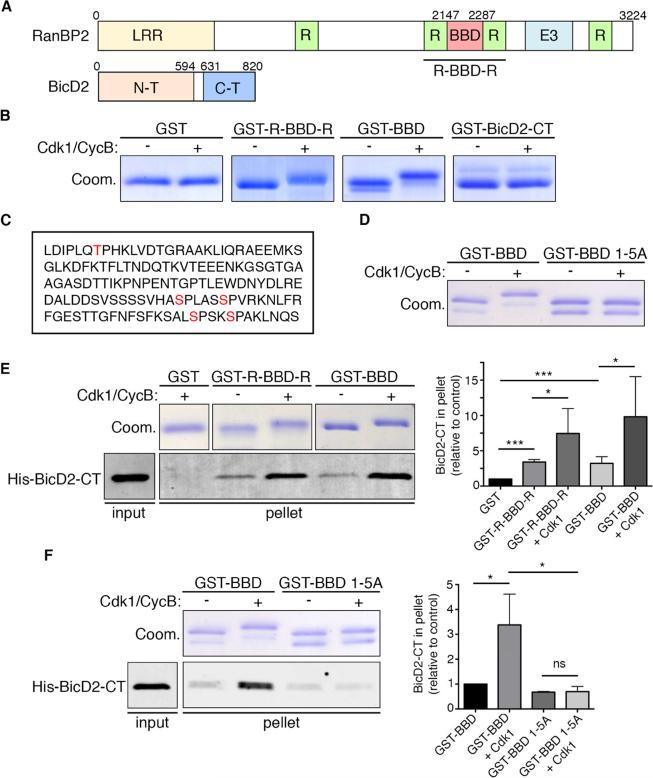
RanBP2 is a large, multi-functional nucleoporin (Pichler et al., 2002; Walther et al., 2002). It interacts directly through an internal fragment (hereafter BBD) with the BicD2 C-terminus (Splinter et al., 2010) (Figure 6A). Sequence analysis of the RanBP2 BBD domain revealed four consensus Cdk1 phosphorylation sites (S/T-P-X-K/R) and one lacking the K/R residue in position +2. In contrast, BicD2-CT contains no predicted Cdk1 sites. To test whether the RanBP2 BBD domain is, indeed, a Cdk1 substrate, we purified recombinant GST-BBD and a larger domain containing flanking Ran binding domains (R-BBD-R), and incubated each polypeptide in vitro with recombinant Cdk1 in the presence of Cyclin B and ATP. A clear electrophoretic gel migration shift was observed for both RanBP2 fragments, but not for GST-BicD2-CT or GST alone (Figure 6B). We analyzed the shifted GST-BBD fragment by mass spectrometry, which revealed phosphorylation at T2153, S2246, S2251, S2276, and S2280 (Figure 6C). Mutating all five sites to alanines abolished the Cdk1/Cyclin B-induced gel shift (Figure 6D). As a further test of this conclusion, we performed the same experiment using phosphate binding tag (Phos-Tag)-containing gels, which further delay the migration of phosphorylated proteins (Kinoshita, 2005). Under these conditions, even the much stronger migration shift induced by Cdk1/Cyclin B treatment was completely abolished by mutation of residues 1-5A (Fig S4A).
Figure 6. Effect of Cdk1 phosphorylation on RanBP2-BicD2 interaction.
(A) Schematic representation of RanBP2 and BicD2. RanBP2 is a large, multi-functional nucleoporin containing a leucine-rich region (LRR), four Ran-binding domains (R), an E3 Sumo ligase domain (E3), and a BicD2-bindng domain (BBD). BicD2 contains substantial regions of α-helical coiled-coil and binds the dynein and dynactin complexes through its N-terminal region and other factors, including RanBP2, through its C-terminal cargo-binding domain (Liu et al., 2013). (B) In vitro assay for Cdk1 phosphorylation of GST-tagged RanBP2 BBD, RanBP2 R-BBD-R, and BicD2-CT. Both of the BicD2-binding RanBP2 fragments displayed a shift in migration on Coomassie-stained electrophoretic gels (Coom.) upon exposure to in vitro purified recombinant Cdk1 and Cyclin B in the presence of ATP. GST-BicD2-CT and GST alone showed no such effect. (C) RanBP2 BBD sequence (amino acids 2147 to 2287) showing the five Cdk1 phosphorylation sites (red) identified on the basis of the known Cdk1 consensus motif and directly demonstrated to be phosphorylated by Cdk1 in vitro using mass spectrometry. (D) In vitro Cdk1 phosphorylation of GST-BBD WT and mutated to alanine in all five identified Cdk1 phosphorylation sites (GST-BBD 1-5A). The electrophoretic gel shift observed for the WT fragment was abolished in the BBD 1-5A fragment. (E) GST-pull down assay with indicated RanBP2 fusions and purified His-BicD2-CT in the presence or absence of purified Cdk1/cyclin B. GST fragments were visualized by Coomassie Blue staining, and His-BicD2-CT was detected by Western blotting with anti-His tag antibody. 10% of the input and 50% of the bound fractions were loaded on the gel. Right: Quantification of BicD2-CT bound fraction relative to amount bound to GST (n=5 independent experiments). Phosphorylation of the RanBP2 fusions by Cdk1 dramatically increased binding to BicD2-CT. (F) GST-pull down assay with indicated RanBP2 fusions and purified His-BicD2-CT in the presence or absence of purified Cdk1 + cyclin B. 10% of the input and 50% of the bound fractions were loaded on gel. Right: Quantification BicD2-CT bound fraction relative to amount bound to GST-BBD (n=4 independent experiments). Pull down with GST-BBD 1-5A shows loss of Cdk1-dependant affinity increase for BicD2-CT. Error bars indicate SD; *p<0.05; ***p<0.001; ns = not significant, based on a Student's t-test. See also Figure S4.
We then used a pull-down assay to determine whether Cdk1 phosphorylation of the RanBP2 BBD domain affects its affinity for BicD2-CT. Untreated BBD-containing fragments showed a weak interaction with BicD2-CT (Splinter et al., 2010), whereas exposure of GST-BBD or GST-R-BBD-R to Cdk1/Cyclin B clearly increased BicD2-CT pull-down, indicating that Cdk1 enhances the interaction (Figure 6E). To confirm that this effect was dependent on Cdk1 phosphorylation, we tested the consequence of Cdk1 mutations on BicD2-CT binding. Mutation of all five sites (1-5A) abolished enhancement of BicD2-CT binding by Cdk1 (Figure 6F). Mutation of sites 2 to 5 (2-5A) had a similar effect to that of 1-5A whereas mutation of site 1 alone (1A) had no detectable effect (Figure S4B). Mutation of sites 2 and 3 (2-3A) and 4 and 5 (4-5A) each resulted in partial reduction in BicD2-CT binding (not shown). These results indicate that Cdk1 phosphorylation of a cluster of serine residues within RanBP2 BBD increases its affinity for BicD2, providing a potential molecular basis for Cdk1-dependent recruitment of BicD2 and dynein to nuclear pores. We note that, in this system as in some others, phospho-mimetic RanBP2 fragments (1-5E and 1-5D) proved unable to mimic the functional effects of enzymatic phosphorylation (not shown) (Paleologou et al., 2008; Whyte et al., 2008).
Cdk1 targets BicD2 to the NE of RGP cells to induce apical nuclear migration
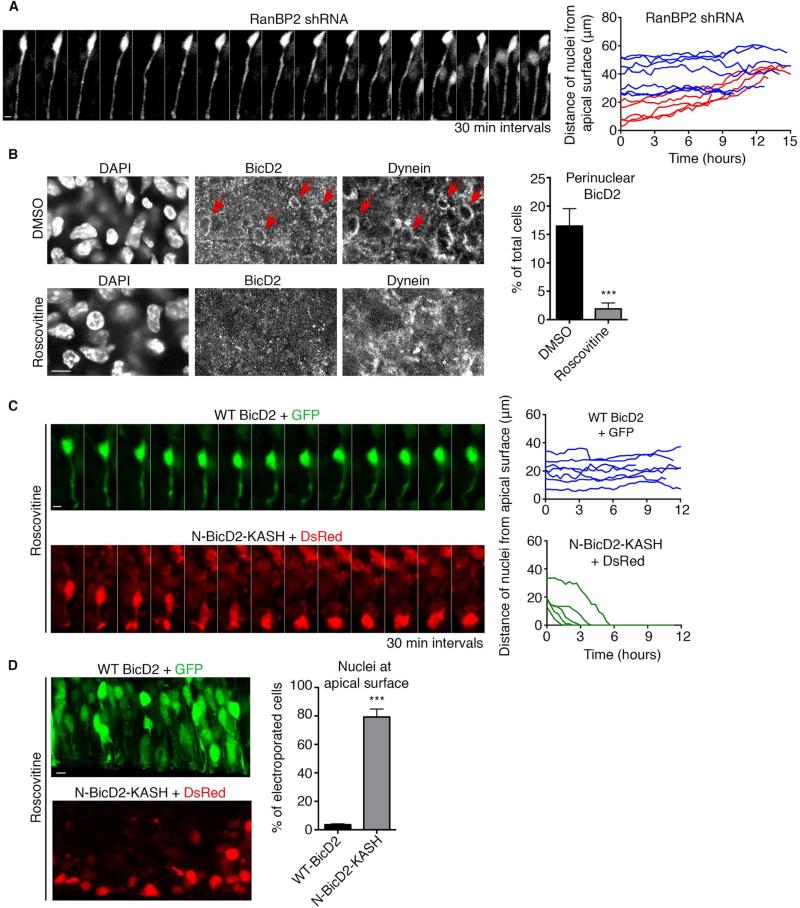
Our results raised the possibility that Cdk1 might initiate apical nuclear migration in RGP cells by activating BicD2 recruitment to RanBP2. As an initial test of this hypothesis, we asked whether depletion of RanBP2 itself affects nuclear migration. This was accomplished by in utero electroporation with shRNA-expressing plasmid, shown to interfere with RanBP2 expression by immunoblotting (Figure S5A). As previously reported for BicD2, nuclei of RanBP2-depleted cells arrested at a considerable distance (25-50 μm) from the apical brain surface, consistent with a role in the early dynein NE recruitment pathway. Live imaging revealed severe inhibition of apical nuclear migration (Figure 7A and Movie S6), whereas basal nuclear migration was largely unaffected (0.046 ± 0.003 μm/min) (Figure S5B and Movie S7). Mitotic index was strongly reduced, consistent with failure of RGP cell nuclei to reach the apical surface of the brain, where mitosis is initiated (Figure S5C). We also tested whether Cdk1 affects BicD2 and dynein recruitment to the RGP cell NE. Both could be detected at the NE in a subset of control cells (Figure 7B) (Hu et al., 2013). In contrast, NE BicD2 and dynein staining were undetectable after exposure of the brain slices to Roscovitine prior to fixation (Figure 7B).
Figure 7. Role of RanBP2-regulated recruitment of BicD2 to the NE of RGP cells.
(A) Live imaging of RanBP2 shRNA-expressing RGP cells in embryonic rat brain slices 4 days after in utero electroporation at E16. The nuclei of electroporated cells were unable to undergo apical nuclear migration. Right: Representative tracings of nuclei. Red tracks indicate basally migrating nuclei and blue tracks non-migrating nuclei. Scale bar, 5 μm. (B) NE BicD2 and dynein labeling within the ventricular zone (VZ) of E19 rat brain sections. Top: NE BicD2 and dynein staining was observed in a subset of cells in control DMSO-treated brain slices (red arrows). Bottom: NE BicD2 and dynein staining was absent from the NE of RGP cells in brain slices treated with Roscovitine (55 μM, 60 min prior to fixation). Right: Quantification of the percentage of total cells with NE BicD2 staining. Only nuclei located within 40 μm from the ventricular surface were evaluated. (C) Live imaging of RGP cell expressing wild type BicD2 + GFP or N-BicD2-KASH + DsRed and treated with Roscovitine (55 μM). Brains were subjected to in utero electroporation at E18 and sliced and put into culture at E19. Top: Wild type BicD2 expression does not restore apical nuclear migration in the presence of Roscovitine. Bottom: N-BicD2-KASH restores apical nuclear migration in the presence of Roscovitine. Green tracks indicate apically migrating nuclei and blue tracks non-migrating nuclei. (D) Brains were subjected to electroporation at E19 and directly sliced and put into culture in the presence of Roscovitine (55 μM) for 24 hours, before fixation. Top: Nuclei of RGP cells expressing wild type BicD2 + GFP do not reach the apical surface in the presence of Roscovitine. Bottom: A high proportion of nuclei are at the apical surface in RGP cells expressing N-BicD2-KASH + DsRed. Right: Quantification of the percentage of nuclei reaching the apical surface. For each experiment, at least three independent brains were imaged. Error bars indicate SD; ***p<0.001, based on a Student's t-test. Scale bar, 5 μm. See also Figure S5 and Movies S6, S7, S8 and S9.
To test whether restoration of NE BicD2 can rescue the effects of Cdk1 inhibition, we expressed either wild-type BicD2 or the N-BicD2-KASH fusion construct in RGP cells by in utero electroporation. We previously found that BicD2 expression could restore apical INM in RGP cells depleted in components of either the early or late NE recruitment pathways (Hu et al., 2013), but this treatment failed to restore apical nuclear migration in brain slices exposed to Roscovitine (Figure 7C and Movie S8). This result is consistent with the sensitivity of BicD2 localization to Cdk1 activity. In contrast, N-BicD2-KASH expression induced nuclei to migrate all the way to the ventricular surface of the brain (Figure 7C and Movie S9). We note that cell division failed to occur, consistent with a role for Cdk1 in mitotic entry.
We also fixed electroporated brain slices 24 hours after Cdk1 inhibition and determined the fraction of RGP nuclei that reached the apical surface (Figure 7D). Consistent with the live imaging data, a very low percentage of nuclei were located at the apical surface in cells expressing wild-type BicD2. In contrast, over 80% of nuclei were found at the apical surface in N-BicD2-KASH-expressing cells (Figure 7D). Therefore, targeting of BicD2 to the NE is entirely sufficient to drive nuclei to the apical surface in the absence of Cdk1 activity.
Discussion
NE recruitment of cytoplasmic dynein is proving to be a widespread and important phenomenon in cells, though much remains to be understood about its regulation (Gundersen and Worman, 2013). How and through what signals NE dynein recruitment is activated to induce nuclear migration in neural stem cell has remained largely unaddressed. We find that apical migration depends on Cdk1 activity, which our data show controls both early and late mechanisms for dynein recruitment to the NE. Cdk1 phosphorylates conserved sites within the nucleoporin RanBP2 and activates BicD2 binding, which, in turn, recruits dynein, dynactin, and LIS1 to the NE, inducing apical nuclear movement. Cdk1 independently induces CENP-F redistribution from inside the nucleus to the NE to allow migration all the way to the apical surface for mitosis (Hu et al., 2013).
The specific effect of Cdk1 inhibitors on apical but not basal nuclear migration is consistent with the G2-M role for this protein kinase. In our system, therefore, basal movement cannot simply reflect displacement of nuclei by apically migrating ones (Kosodo et al., 2011; Leung et al., 2011; Okamoto et al., 2013). We find that Cdk1 inhibition arrests apically-migrating nuclei at a range of distances from the ventricular surface, which may reflect the dual role we have discovered for Cdk1 in both early and late NE dynein recruitment pathways.
Our data indicate that Cdk1 plays a similar role in nonneuronal G2-specific NE dynein recruitment. We suspect, therefore, that this is a very general function, which is exaggerated in RGP cells because of their great length and epithelial cytoskeletal organization. Quite possibly, other epithelial cells also use Cdk1-induced NE dynein recruitment to reorganize their contents for mitosis.
To identify primary targets for Cdk1, we employed both cellular and biochemical approaches. We found that Cdk1 inhibition displaces BicD2 from the NE of G2 cells. However, constitutive targeting of the BicD2 N-terminal domain to the NE induced co-recruitment of dynein, dynactin, and LIS1, though not NudE/EL. These results were obtained in cells inhibited for Cdk1, thus allowing us to deduce that BicD2 localization alone is under control of the protein kinase. We tested this possibility by Cdk1 phosphorylation of the BicD2-binding domain of RanBP2, which resulted in a clear increase in its affinity for BicD2 in vitro. We identified 5 consensus Cdk1 sites in the RanBP2 fragment by sequence analysis, all of which were confirmed as targets of Cdk1 by in vitro phosphorylation followed by mass spectrometry. Mutational analysis revealed that four are necessary to stimulate the BicD2 interaction. However, replacement of the target serines with acidic residues to produce phosphomimetic mutations had no effect on BicD2 binding. These results, and the large size of full-length RanBP2 (358 kDa), precluded attempts to rescue RanBP2 knockdown in brain with phosphorylated vs. dephosphorylated forms of the protein. Nonetheless, we were able to bypass the effects of Cdk1 inhibition by constitutive targeting of the BicD2 N-terminus to the RGP NE in embryonic rat brain (Figure 7). This further supports a primary role for Cdk1 phosphorylation in regulating recruitment of BicD2 to the NE and apical nuclear migration.
Two other dynein binding partners, NudE/EL and the light intermediate chain 1 (LIC1), are known Cdk1 targets (Dell et al., 2000; Yan et al., 2003). NudEL phosphorylation, in particular, was reported to stimulate prophase NE invagination (PNEI), thought to be an indicator of increased dynein forces at the NE (Hebbar et al., 2008). Our data indicate that phosphorylation of these substrates is not involved in regulating early dynein NE recruitment, but a role in the late mechanism remains a likely possibility.
An intriguing question is whether BicD2 exclusively serves as a recruiting factor for dynein at the NE or if it also participates in dynein activation. BicD2 is emerging as a key regulator of dynein, with important roles in the nervous system. In humans, dominant mutations in BicD2 cause spinal muscular atrophy and hereditary spastic paraplegia (Lipka et al., 2013), while BicD2-deficient mice have disrupted laminar organization in the cortex and cerebellum (Jaarsma et al., 2014). At the molecular level, BicD2 was recently shown to form a complex with dynein and dynactin, dramatically stimulating processive dynein movement on microtubules in vitro (Mckenney et al., 2014; Schlager et al., 2014; Splinter et al., 2012). These observations suggest that the Cdk1-dependence of the BicD2-RanBP2 interaction found here may not only regulate dynein targeting to the NE, but also stimulate motor activity to facilitate nuclear migration in RGP cells.
Activation of the late NE dynein recruitment pathway involves a mechanism very different from that for the early pathway. In this case, CENP-F is at first intranuclear, and physically segregated from its nucleoporin binding partner Nup133. CENP-F is known to exit the nucleus during late G2 to decorate the NE and early assembling kinetochores (Hussein and Taylor, 2002; Stehman et al., 2007; Vergnolle and Taylor, 2007). Quite strikingly, Cdk1 inhibition blocked CENP-F nuclear exit (Figure 3C), identifying a role for the kinase in CENP-F subcellular localization. How phosphorylation regulates CENP-F redistribution remains to be investigated. We note that farnesylation of CENP-F is also required for NE localization (Hussein and Taylor, 2002), pointing to another potential level of regulation of the late dynein recruitment pathway.
The evolutionary basis for the differences between the early and late NE dynein recruitment mechanisms is unclear. We speculate that the intranuclear sequestration of CENP-F, detected during most of G2, may prevent gradual activation of the late dynein recruitment pathway by the continuously increasing cytoplasmic Cdk1 kinase activity. We note that CENP-F exits the nucleus at about the time of cyclin B entry. This may ensure that the CENP-F-dependent increase in NE dynein levels is coordinated with early mitotic events, such as centrosome separation, NE breakdown, and kinetochore assembly (Raaijmakers et al., 2012; Turgay et al., 2014).
Our data indicate that Cdk1 serves as a master regulator for apical nuclear migration through a direct effect on dynein recruitment to the NE. It therefore appears that apical nuclear migration and mitotic entry are under the control of the same protein kinase, which may ensure that these two processes always occur in proper sequence. Nuclei would, thus, migrate towards the ventricular surface only as cells become ready to divide, thus preventing nuclear overcrowding at the apical surface (Okamoto et al., 2013; Strzyz et al., 2015).
Experimental Procedures
Expression constructs and shRNA
We used the following previously described expression constructs: Full length BicD2 in pIRES2 DsRed-Express2 (Hu et al., 2013); Cdk1-DN-HA (obtained from addgene) (van den Heuvel and Harlow, 1993); GFP-N-BicD2-KASH, GST-BicD2-CT and GST-BBD (GST-RanBP2 fragment 3) (gifts from Dr. Anna Akhmanova) (Hu et al., 2013; Splinter et al., 2010); GST-R-BBD-R (GST-RBD2-JX2-RBD3, a gift from Dr. Paulo Ferreira) (Cai et al., 2001). His-BicD2-CT was generated by gene synthesis (Genescript) and inserted into pGex-6p-1 (BamH1-EcoRI). Mutagenesis of GST-BBD was performed using QuickChange II site-directed mutagenesis kit (Agilent technologies). For RanBP2 knockdown in the brain, shRNA construct inserted in the pGFP-V-RS vector was used (Origene). We used the following shRNA sequence: 5’-CTTGTGATGCCTCCAAGCCAACTCATAAG-3’. Empty vectors of pEGFP-C1 and pDsRed-Express2-C1 (Clontech) were used as controls. For RanBP2 knockdown in HeLa cells, we used Thermo Scientific siGenome SMARTpool: 5’-CGAAACAGCUGUCAAGAAA-3’, 5’-GAAAGAAGGUCACUGGGAU-3’, 5’-GAAAGGACAUGUAUCACUG-3’ and 5’-GAAUAACUAUCACAGAAUG-3’. As a control, we used ON-Target plus non-targeting siRNA #1 (Thermo Scientific): 5’-UGGUUUACAUGUCGACUAA-3’.
Antibodies
Antibodies used in this study were: Mouse monoclonal against dynein intermediate chain clone 74.1 (a gift from Dr. Kevin Pfister), CENP-F (BD Biosciences), p150 (BD Biosciences), cyclin B1 (BD Biosciences), phospho Histone H3 (Abcam), His tag (Genescript), PCNA (Proteintech); Rabbit polyclonal against LIS-1 (Santa Cruz), Cyclin B1 (Santa Cruz), PCNA (Abcam), BicD2 (Abcam), BicD2 (a gift from Dr. Anna Akhmanova), RanBP2 (Abcam), NudE/EL (Stehman et al., 2007); Chicken polyclonal against GFP (Millipore); Rat monoclonal against BrdU (Abcam).
Cell culture, transfection, immunostaining, and drug treatment
HeLa cells were cultured in DMEM supplemented with 10% FBS. Effectene reagent (QIAGEN) was used for plasmid transfection and HiPerFect (QIAGEN) for siRNA transfection. For immunostaining, cells were washed in PBS, fixed in -20°C methanol, 10 mM EGTA (10 min), permeabilized in PBS, 0.1% Triton X-100 (5 min), and stained in PBS, 0.05% Tween supplemented with donkey serum. For LIS-1 and NudE/EL stainings, cells were pre-extracted with 0.5% triton (in PHEM buffer) for 1 minute and fixed in 3% PFA (in PHEM buffer) for 20 minutes. Cells were then washed in PBS, permeabilized in 0.1% triton (25 minutes), and incubated in NH4Cl 50 mM for 10 minutes. Cells were then stained in PBS, 0.05% Tween supplemented with donkey serum. Because pre-extraction is not compatible with cyclin B1 staining, we used phospho-Histone 3 as a late G2/prophase marker. For better visualization of NE dynein and dynein partners, cells were incubated for 1 hr in Nocodazol (10 μM) prior to fixation (Bolhy et al., 2011; Hu et al., 2013; Raaijmakers et al., 2013; Splinter et al., 2012). The following drugs were used for protein kinase inhibition: 55 μM Roscovitine (Meijer et al., 1997) (Selleck Chemicals); 0.9 μM Cdk1/2 inhibitor III (Gavet and Pines, 2010) (Calbiochem); 28 μM RO-3306 (Deibler and Kirschner, 2010; Vassilev et al., 2006) (Tocris Bioscience); 0.5 μM PD166285 (Potapova et al., 2009) (Tocris Bioscience); 0.3 μM VX-680 (Harrington et al., 2004) (Selleck Chemicals); 22 μM BTO-1 (Bader et al., 2011) (EMD Millipore).
In vitro kinase assay, GST pull-down, and Western Blotting
Cdk1 kinase assay was performed as follow: 10 μg of GST-tagged protein were incubated for 45 minutes at 30°C, in the presence of 0.2 μg active Cdk1/CyclinB (EMD Millipore) and 400 μM ATP (Sigma Aldrich) in PK buffer (NEB), to a final volume of 50 μL. For GST pull downs, the entire kinase reaction was incubated with 40 μL Glutathione Magnetic beads (Pierce) in binding buffer (50 mM Tris pH 7.4; 5 mM EDTA; 0.1% NP40; 125 mM NaCl) for 1 hour at 4°C. Beads were washed and incubated with 1 μg His-BicD2-CT in binding buffer for 1 hour at 4°C. Beads were then washed, transferred to a new tube, and boiled in 1X Laemli buffer. The proteins were analyzed by SDS-PAGE using standard procedures followed by Western Blotting or Coomassie brilliant blue gel staining. For phosphate binding tag (Phos-Tag)-containing gels, 10% acrylamide gels were prepared in the presence of 40 μM Phos-Tag™ (Wako chemicals). Immunoblots were incubated with fluorescently tagged secondary antibodies (Rockland Immunochemicals) and developed at subsaturating conditions using a scanning device (Odyssey; LI-COR Biosciences) and Odyssey software version 3.0.
Mass spectrometry for identification of Cdk1 phosphorylation sites
GST-tagged RanBP2 BBD fragment was incubated with Cdk1/CyclinB and ATP at 30°C for 45 minutes as described above. Proteins were then loaded on an SDS-PAGE gel and analyzed by mass spectrometry to identify Cdk1 phosphorylation sites (see supplemental experimental procedures).
In utero electroporation, live imaging, and drug treatment of brain slices
Plasmids were transfected by intraventricular injection in embryonic rats in utero, followed by electroporation as described previously (Tsai et al., 2010). In summary, 1-2 μL of cDNA (1-3 μg/μL) was injected into the ventricle of embryonic E16 brains. A series of five electric pulses at 50 V for 50 ms at 1 s intervals was applied by a platinum-plated electrode (Harvard Apparatus) through the uterine walls. Coronal slices from embryos were sectioned at 300 μm on a Vibratome (Leica microsystems) 3-4 days post electroporation, and then imaged live for 10-16 hours at 10 minutes intervals. Throughout the live imaging duration, coronal slices were placed on Millicell-CM inserts (Millipore) in culture medium containing 25% Hanks balanced salt solution, 47% basal MEM, 25% normal horse serum, 1× penicillin/streptomycin/glutamine (GIBCO BRL), and 0.66% glucose. Slices remained incubated in 5% CO2 at 37°C. For drug treatments, brain slices were incubated in culture medium containing 55 μM Roscovitine or 100 μM RO-3306 for 45 minutes prior to imaging and throughout the duration of the movie. For analysis of PD166285 effect on apical nuclear migration, live brain slices were incubated with BrdU (Sigma-Aldrich) at 20 μg/ml for 15 minutes, washed, and subsequently treated for 2 hours with 1 μM PD166285 before fixation and immunostaining. All animals were maintained according to protocols approved by the Institutional Animal Care and Use Committee at Columbia University.
Analysis and quantification of live imaging movies
Time-lapse images were captured by IX80 scanning confocal microscope using Fluoview 100 software (Olympus), with each image taken every 10 minutes. Movies were created and processed at 12 fps using ImageJ software. Tracings of RGP cell nuclear movements were obtained by measuring the distance between the RGP cell endfoot (bottom of the apical process) to the bottom of the nucleus. This controlled any overall movement of the entire brain slice. Nuclear distances for tracings were measured every 20 minutes, which equated 2 intervals during live imaging. RGP cells were distinguished from neurons by the presence of the apical process, which neurons lacked, and by focusing within the ventricular zone. The velocity of nuclear movement was calculated by measuring the total distance the nucleus traveled from when the nucleus first moved to when it last moved and dividing that by the time the nucleus took to travel that distance.
Immunostaining of brain slices
Rat embryos were perfused transcardially with chilled saline and 4% paraformaldehyde (PFA) (EMS, wt/vol), and then incubated in 4% PFA overnight. Brain were then coronally sectioned at a thickness of 100 μm, washed with PBS, and incubated in blocking solution containing PBS, 0.3% Triton X-100 supplemented with donkey serum for 1 hourr. Primary antibodies were incubated overnight in blocking solution at 4°C and secondary antibodies in blocking solution were incubated for 2 hours at room temperature. For BrdU immunostaining, brain slices were first incubated in 2N HCl for 25 minutes at 37°C. Slices were then washed in 0.1 M sodium borate for 10 minutes and then PBS, prior to antibody incubation. For dynein and BicD2 immunostaining, rat brains were coronally sectioned prior to fixation, and incubated in Nocodazol (10 μM) for 2 hours at 37°C for better visualization (Hu et al., 2013). Brain slices were then fixed in methanol at -20°C for 20 minutes. Immunostaining proceeded as described above.
Microscopy and image analysis
All images were collected with an IX80 laser scanning confocal microscope (Olympus FV100 Spectral Confocal System). HeLa cells were imaged using a 100x 1.40 N.A. oil objective. Brain sections were imaged using a 60x 1.42 N.A. oil objective or a 10x 0.40 N.A. air objective. Images were analyzed using ImageJ and prepared for publication using Adobe Photoshop.
Supplementary Material
Acknowledgement
We thank Drs. A. Akhmanova and P. Ferreira for advice and reagents and all the members of the Vallee lab for technical expertise and feedback. The mass spectrometry analysis was performed at the Proteomics Shared Resource at the Herbert Irving Comprehensive Cancer Center at Columbia University. This project was supported by NIH HD40182 and GM102347 to RBV and AHA postdoctoral fellowship to ADB.
Abbreviations
- RGP
Radial Glial Progenitor
- INM
Interkinetic Nuclear Migration
- NE
Nuclear Envelope
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors Contributions
ADB and RBV conceived the project and wrote the manuscript; ADB and DJH performed experiments and analyzed data.
References
- Bader JR, Kasuboski JM, Winding M, Vaughan PS, Hinchcliffe EH, Vaughan KT. Polo-like kinase1 is required for recruitment of dynein to kinetochores during mitosis. Journal of Biological Chemistry. 2011;286:20769–20777. doi: 10.1074/jbc.M111.226605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudouin J, Gerlich D, Daigle N, Eils R, Ellenberg J. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell. 2002;108:83–96. doi: 10.1016/s0092-8674(01)00627-4. [DOI] [PubMed] [Google Scholar]
- Bolhy S, Bouhlel I, Dultz E, Nayak T, Zuccolo M, Gatti X, Vallee R, Ellenberg J, Doye V. A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase. J Cell Biol. 2011;192:855–871. doi: 10.1083/jcb.201007118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Singh BB, Aslanukov A, Zhao H, Ferreira PA. The docking of kinesins, KIF5B and KIF5C, to Ran-binding protein 2 (RanBP2) is mediated via a novel RanBP2 domain. J Biol Chem. 2001;276:41594–41602. doi: 10.1074/jbc.M104514200. [DOI] [PubMed] [Google Scholar]
- Deibler RW, Kirschner MW. Quantitative reconstitution of mitotic CDK1 activation in somatic cell extracts. Mol Cell. 2010;37:753–767. doi: 10.1016/j.molcel.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell KR, Turck CW, Vale RD. Mitotic phosphorylation of the dynein light intermediate chain is mediated by cdc2 kinase. Traffic. 2000;1:38–44. doi: 10.1034/j.1600-0854.2000.010107.x. [DOI] [PubMed] [Google Scholar]
- Gavet O, Pines J. Progressive Activation of CyclinB1-Cdk1 Coordinates Entry to Mitosis. Dev Cell. 2010;18:533–543. doi: 10.1016/j.devcel.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen GG, Worman HJ. Nuclear positioning. Cell. 2013;152:1376–1389. doi: 10.1016/j.cell.2013.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington EA, Bebbington D, Moore J, Rasmussen RK, Ajose-Adeogun AO, Nakayama T, Graham JA, Demur C, Hercend T, Diu-Hercend A, et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med. 2004;10:262–267. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- Hebbar S, Mesngon MT, Guillotte AM, Desai B, Ayala R, Smith DS. Lis1 and Ndel1 influence the timing of nuclear envelope breakdown in neural stem cells. J Cell Biol. 2008;182:1063–1071. doi: 10.1083/jcb.200803071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu DJ-K, Baffet AD, Nayak T, Akhmanova A, Doye V, Vallee RB. Dynein recruitment to nuclear pores activates apical nuclear migration and mitotic entry in brain progenitor cells. Cell. 2013;154:1300–1313. doi: 10.1016/j.cell.2013.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein D, Taylor SS. Farnesylation of Cenp-F is required for G2/M progression and degradation after mitosis. J Cell Sci. 2002;115:3403–3414. doi: 10.1242/jcs.115.17.3403. [DOI] [PubMed] [Google Scholar]
- Jaarsma D, van den Berg R, Wulf PS, van Erp S, Keijzer N, Schlager MA, de Graaff E, de Zeeuw CI, Pasterkamp RJ, Akhmanova A, et al. A role for Bicaudal-D2 in radial cerebellar granule cell migration. Nature Communications. 2014;5:3411. doi: 10.1038/ncomms4411. [DOI] [PubMed] [Google Scholar]
- Kinoshita E. Phosphate-binding Tag, a New Tool to Visualize Phosphorylated Proteins. Molecular & Cellular Proteomics. 2005;5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- Kosodo Y. Interkinetic nuclear migration: beyond a hallmark of neurogenesis. Cell Mol Life Sci. 2012;69:2727–2738. doi: 10.1007/s00018-012-0952-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosodo Y, Suetsugu T, Suda M, Mimori-Kiyosue Y, Toida K, Baba SA, Kimura A, Matsuzaki F. Regulation of interkinetic nuclear migration by cell cycle-coupled active and passive mechanisms in the developing brain. Embo J. 2011;30:1690–1704. doi: 10.1038/emboj.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapenna S, Giordano A. Cell cycle kinases as therapeutic targets for cancer. Nat Rev Drug Discov. 2009;8:547–566. doi: 10.1038/nrd2907. [DOI] [PubMed] [Google Scholar]
- Lee HO, Norden C. Mechanisms controlling arrangements and movements of nuclei in pseudostratified epithelia. Trends Cell Biol. 2013;23:141–150. doi: 10.1016/j.tcb.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Leung L, Klopper AV, Grill SW, Harris WA, Norden C. Apical migration of nuclei during G2 is a prerequisite for all nuclear motion in zebrafish neuroepithelia. Development. 2011;138:5003–5013. doi: 10.1242/dev.071522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Liu XS, Yang X, Song B, Wang Y, Liu X. Polo-like kinase 1 phosphorylation of p150Glued facilitates nuclear envelope breakdown during prophase. Proc Natl Acad Sci USA. 2010;107:14633–14638. doi: 10.1073/pnas.1006615107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Haug JS, Seidel CW, Gibson MC. Functional genomic analysis of the periodic transcriptome in the developing Drosophila wing. Dev Cell. 2014;29:112–127. doi: 10.1016/j.devcel.2014.02.018. [DOI] [PubMed] [Google Scholar]
- Lindqvist A, Rodríguez-Bravo V, Medema RH. The decision to enter mitosis: feedback and redundancy in the mitotic entry network. J Cell Biol. 2009;185:193–202. doi: 10.1083/jcb.200812045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka J, Kuijpers M, Jaworski J, Hoogenraad CC. Mutations in cytoplasmic dynein and its regulators cause malformations of cortical development and neurodegenerative diseases. Biochem Soc Trans. 2013;41:1605–1612. doi: 10.1042/BST20130188. [DOI] [PubMed] [Google Scholar]
- Liu Y, Salter HK, Holding AN, Johnson CM, Stephens E, Lukavsky PJ, Walshaw J, Bullock SL. Bicaudal-D uses a parallel, homodimeric coiled coil with heterotypic registry to coordinate recruitment of cargos to dynein. Genes & Development. 2013;27:1233–1246. doi: 10.1101/gad.212381.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mckenney RJ, Huynh W, Tanenbaum ME, Bhabha G, Vale RD. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science. 2014;345:337–341. doi: 10.1126/science.1254198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer L, Borgne A, Mulner O, Chong JP, Blow JJ, Inagaki N, Inagaki M, Delcros JG, Moulinoux JP. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5. Eur. J. Biochem. 1997;243:527–536. doi: 10.1111/j.1432-1033.1997.t01-2-00527.x. [DOI] [PubMed] [Google Scholar]
- Messier PE. Microtubules, interkinetic nuclear migration and neurulation. Experientia. 1978;34:289–296. doi: 10.1007/BF01922992. [DOI] [PubMed] [Google Scholar]
- Meyer EJ, Ikmi A, Gibson MC. Interkinetic nuclear migration is a broadly conserved feature of cell division in pseudostratified epithelia. Curr Biol. 2011;21:485–491. doi: 10.1016/j.cub.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Norden C, Young S, Link BA, Harris WA. Actomyosin is the main driver of interkinetic nuclear migration in the retina. Cell. 2009;138:1195–1208. doi: 10.1016/j.cell.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Namba T, Shinoda T, Kondo T, Watanabe T, Inoue Y, Takeuchi K, Enomoto Y, Ota K, Oda K, et al. TAG-1-assisted progenitor elongation streamlines nuclear migration to optimize subapical crowding. Nat Neurosci. 2013 doi: 10.1038/nn.3525. [DOI] [PubMed] [Google Scholar]
- Pacary E, Azzarelli R, Guillemot F. Rnd3 coordinates early steps of cortical neurogenesis through actin-dependent and -independent mechanisms. Nature Communications. 2013;4:1635. doi: 10.1038/ncomms2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paleologou KE, Schmid AW, Rospigliosi CC, Kim H-Y, Lamberto GR, Fredenburg RA, Lansbury PT, Fernandez CO, Eliezer D, Zweckstetter M, et al. Phosphorylation at Ser-129 but not the phosphomimics S129E/D inhibits the fibrillation of alpha-synuclein. J Biol Chem. 2008;283:16895–16905. doi: 10.1074/jbc.M800747200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paridaen JT, Huttner WB. Neurogenesis during development of the vertebrate central nervous system. EMBO Rep. 2014;15:351–364. doi: 10.1002/embr.201438447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- Potapova TA, Daum JR, Byrd KS, Gorbsky GJ. Fine tuning the cell cycle: activation of the Cdk1 inhibitory phosphorylation pathway during mitotic exit. Mol Biol Cell. 2009;20:1737–1748. doi: 10.1091/mbc.E08-07-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JA, Tanenbaum ME, Medema RH. Systematic dissection of dynein regulators in mitosis. J Cell Biol. 2013;201:201–215. doi: 10.1083/jcb.201208098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JA, van Heesbeen RGHP, Meaders JL, Geers EF, Fernandez-Garcia B, Medema RH, Tanenbaum ME. Nuclear envelope-associated dynein drives prophase centrosome separation and enables Eg5-independent bipolar spindle formation. Embo J. 2012;31:4179–4190. doi: 10.1038/emboj.2012.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujano MA, Sanchez-Pulido L, Pennetier C, le Dez G, Basto R. The microcephaly protein Asp regulates neuroepithelium morphogenesis by controlling the spatial distribution of myosin II. Nat Cell Biol. 2013;15:1294–1306. doi: 10.1038/ncb2858. [DOI] [PubMed] [Google Scholar]
- Salina D, Bodoor K, Eckley DM, Schroer TA, Rattner JB, Burke B. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 2002;108:97–107. doi: 10.1016/s0092-8674(01)00628-6. [DOI] [PubMed] [Google Scholar]
- Schenk J, Wilsch-Bräuninger M, Calegari F, Huttner WB. Myosin II is required for interkinetic nuclear migration of neural progenitors. Proc Natl Acad Sci USA. 2009;106:16487–16492. doi: 10.1073/pnas.0908928106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlager MA, Hoang HT, Urnavicius L, Bullock SL, Carter AP. In vitro reconstitution of a highly processive recombinant human dynein complex. Embo J. 2014;33:1855–1868. doi: 10.15252/embj.201488792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DS, Greer PL, Tsai LH. Cdk5 on the brain. Cell Growth Differ. 2001;12:277–283. [PubMed] [Google Scholar]
- Spear PC, Erickson CA. Interkinetic nuclear migration: a mysterious process in search of a function. Develop. Growth Differ. 2012;54:306–316. doi: 10.1111/j.1440-169X.2012.01342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter D, Razafsky DS, Schlager MA, Serra-Marques A, Grigoriev I, Demmers J, Keijzer N, Jiang K, Poser I, Hyman AA, et al. BICD2, dynactin, and LIS1 cooperate in regulating dynein recruitment to cellular structures. Mol Biol Cell. 2012;23:4226–4241. doi: 10.1091/mbc.E12-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter D, Tanenbaum ME, Lindqvist A, Jaarsma D, Flotho A, Yu KL, Grigoriev I, Engelsma D, Haasdijk ED, Keijzer N, et al. Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol. 2010;8:e1000350. doi: 10.1371/journal.pbio.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehman SA, Chen Y, Mckenney RJ, Vallee RB. NudE and NudEL are required for mitotic progression and are involved in dynein recruitment to kinetochores. J Cell Biol. 2007;178:583–594. doi: 10.1083/jcb.200610112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzyz PJ, Lee HO, Sidhaye J, Weber IP, Leung LC, Norden C. Interkinetic nuclear migration is centrosome independent and ensures apical cell division to maintain tissue integrity. Dev Cell. 2015;32:203–219. doi: 10.1016/j.devcel.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Tsai J-W, Chen Y, Kriegstein AR, Vallee RB. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J Cell Biol. 2005;170:935–945. doi: 10.1083/jcb.200505166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J-W, Lian W-N, Kemal S, Kriegstein AR, Vallee RB. Kinesin 3 and cytoplasmic dynein mediate interkinetic nuclear migration in neural stem cells. Nat Neurosci. 2010;13:1463–1471. doi: 10.1038/nn.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgay Y, Champion L, Balazs C, Held M, Toso A, Gerlich DW, Meraldi P, Kutay U. SUN proteins facilitate the removal of membranes from chromatin during nuclear envelope breakdown. J Cell Biol. 2014;204:1099–1109. doi: 10.1083/jcb.201310116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel S, Harlow E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science. 1993;262:2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, Heimbrook DC, Chen L. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci USA. 2006;103:10660–10665. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnolle MAS, Taylor SS. Cenp-F links kinetochores to Ndel1/Nde1/Lis1/dynein microtubule motor complexes. Curr Biol. 2007;17:1173–1179. doi: 10.1016/j.cub.2007.05.077. [DOI] [PubMed] [Google Scholar]
- Walther TC, Pickersgill HS, Cordes VC, Goldberg MW, Allen TD, Mattaj IW, Fornerod M. The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J Cell Biol. 2002;158:63–77. doi: 10.1083/jcb.200202088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte J, Bader JR, Tauhata SBF, Raycroft M, Hornick J, Pfister KK, Lane WS, Chan GK, Hinchcliffe EH, Vaughan PS, et al. Phosphorylation regulates targeting of cytoplasmic dynein to kinetochores during mitosis. J Cell Biol. 2008;183:819–834. doi: 10.1083/jcb.200804114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Li F, Liang Y, Shen Y, Zhao X, Huang Q, Zhu X. Human Nudel and NudE as regulators of cytoplasmic dynein in poleward protein transport along the mitotic spindle. Mol Cell Biol. 2003;23:1239–1250. doi: 10.1128/MCB.23.4.1239-1250.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.