Abstract
Water molecules that mediate protein–ligand interactions or are released from the binding site on ligand binding can contribute both enthalpically and entropically to the free energy of ligand binding. To elucidate the thermodynamic profile of individual water molecules and their potential contribution to ligand binding, a hydration site analysis program WATsite was developed together with an easy-to-use graphical user interface based on PyMOL. WATsite identifies hydration sites from a molecular dynamics simulation trajectory with explicit water molecules. The free energy profile of each hydration site is estimated by computing the enthalpy and entropy of the water molecule occupying a hydration site throughout the simulation. The results of the hydration site analysis can be displayed in PyMOL. A key feature of WATsite is that it is able to estimate the protein desolvation free energy for any user specified ligand. The WATsite program and its PyMOL plugin are available free of charge from http://people.pnhs.purdue.edu/~mlill/software.
Keywords: hydration site, graphical user interface, PyMOL, WATsite
Introduction
Computer-aided drug design (CADD) has become an integral part of modern drug development in recent decades.[1,2] One most fundamental goal of CADD is to predict new chemical entities binding to a given therapeutic target and estimate their free energy of binding. However, due to the computational complexity of searching the protein–ligand conformational space and locating the optimal binding solution, various simplifications and assumptions have to be introduced in the energetic evaluation of protein–ligand complex structures to make the problem computationally tractable.[2] In particular, protein desolvation and water molecules engaged in the protein–ligand binding interface are often neglected or only implicitly represented. However, water molecules can contribute both enthalpically and entropically to the free energy of ligand binding. They may be directly involved in mediating interactions between ligand and protein or be displaced by the ligand on binding. Both of these mechanisms have been shown to be of importance in drug discovery.[3]
To explicitly include water molecules in CADD models, the position of potential water sites and the free energy profile associated with each water molecule need to be known. When a sufficient number of experimental protein–ligand complex structures are available, the positions of conserved water molecules can be used to define the water sites. However, there is currently no experimental method to probe the thermodynamic profile of individual water molecules in the protein binding site. As an alternative, various computational methods[4–10] have been developed to predict the locations of the hydration sites and estimate the desolvation energies associated with the hydration sites. One of the earliest empirical methods, GRID, uses the water molecule as a chemical probe to locate the energetically favorable hydration site positions in the protein binding site.[4] The knowledge-based method AQUARIUS[5,6] predicts the most probable positions for hydration sites in the first hydration shell of the protein based on the solvent distributions surrounding the 20 different amino acids derived from the analysis of protein structures.[11] Molecular dynamics (MD) simulations are also used to predict the location of hydration sites with explicit water molecules.[9] The method WaterMap[12,13] combines the explicit water simulations with the inhomogeneous solvation theory[14] to predict the entropic and enthalpic contribution of individual hydration sites on ligand binding.
In our effort of including water molecules in protein–ligand docking studies, we have developed a hydration site prediction program, named WATsite. WATsite solvates the protein with explicit water molecules and performs MD simulations to sample the fluctuations of the water molecules within the protein binding site. Quality threshold (QT) clustering algorithm is used to identify the locations with high water molecule occupancy, defining the “hydration sites.” The free energy profile of each hydration site is estimated by analyzing the enthalpy and entropy of the water molecule occupying a hydration site throughout the MD simulation.[15] In a previous study, we have combined the hydration site information provided by WATsite with our in-house pharmacophore modeling tool to define the optimal protein-based pharmacophore models for virtual screening.[15] We found that the hydration site information can be useful in constructing pharmacophore models with higher computational efficiency and enrichment quality compared to the models that did not use the hydration site information. To make WATsite more readily useful for researchers interested in analyzing key water molecules in the protein binding pocket, we developed a graphical user interface (GUI) for WATsite based on PyMOL. The GUI allows users to prepare and submit the WATsite underlying MD simulations, analyze the results to predict hydration sites and estimate the desolvation free energy of the protein for a given ligand replacing water molecules on binding to the protein. To the best of our knowledge, this is the first freely available MD-based hydration site prediction program combined with an easy-to-use GUI. The individual steps of the WATsite procedure, the underlying methodological details and their implementation in the context of the PyMOL plugin are described in the following Methods section.
Methods
MD simulation with hydrated binding sites
The WATsite process starts with a MD simulation of the protein within an octahedron of explicit water molecules. The WATsite program can be executed through the command line on a Linux OS or using a PyMOL GUI. We will focus our description of WATsite through the use of the PyMOL plugin in this article. The readers are referred to our online user guide for the details of using WATsite on the command line. In the PyMOL plugin, the user specifies a protein structure from a file or a structure already displayed in the current PyMOL session (Fig. 1). By default, all water molecules in the user-provided protein structure are kept during the preparation of the MD simulation. The user can remove the water molecules in the original crystal structures within PyMOL if those water positions should not be directly included in the MD simulation. To define the protein binding site, the user needs to provide a ligand molecule positioned within the binding site and a margin (in Å) for defining the binding pocket enclosing the ligand. The binding pocket is then defined as a box surrounding the ligand with the minimum distance between any ligand heavy atom to the edge of the box equal to the margin value. The binding site box specifies the volume to which the hydration site analysis is restricted. It is worth mentioning that the ligand provided in this step is only intended for defining the binding site. It is not included in the MD simulation and the subsequent hydration site identification process. Therefore, the user can also construct a “pseudo-ligand” using the binding site residues to define the binding site.
Figure 1.

Screenshot of dialog utilized to select input protein and possible ligand structure, and the option for defining the binding site. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
WATsite allows the identification of hydration sites for both ligand-free (apo) and ligand-bound (holo) protein structures. If the holo-simulation option is selected, the file location of the bound ligand is specified within the PyMOL plugin. The specified ligand will be included in the MD simulation and the following hydration site identification process. In the current version, no docking service for the user-specified ligand is provided. Therefore, the provided ligand conformation needs to be a meaningful binding pose for the protein.
Once the location of all necessary files has been specified, the MD simulation is performed in the background following the WATsite protocol: for each protein structure, the side-chain conformations of ASN, GLN, and HIS, and tautomers and protonation states of HIS were adjusted using the Reduce[16] program. Hydrogen atoms were added to the structures using the pdb2gmx module of GROMACS.[17] The protein is then solvated in an octahedron water box using the SPC water model[18] with a water layer of a minimum of 10 Å between any protein atom to the edge of the box. Chlorine and sodium ions are added to neutralize the system. MD simulation is performed using GROMACS[17] with the AMBER03 force field.[19–21] The system is energy minimized for 5000 steps using the steepest descent algorithm. The water molecules of the system are then equilibrated for 250 ps with periodic boundary conditions and with all protein heavy atoms and potentially present ligand heavy atoms harmonically restrained (spring constants of 1000 kJ mol−1 nm−2). The Nose–Hoover[22,23] thermostat is used for temperature coupling at 300 K, and the Parrinello–Rahman[24] approach is used for pressure coupling at 1 bar. The electrostatic interactions are calculated exactly for atom pairs within a spatial separation less than 10 Å and by using the Particle Mesh Ewald[25,26] method for pairs beyond this cutoff. The Lennard-Jones interactions are truncated at 14 Å. Under the same settings, 2 ns of MD simulations are performed and the coordinates are saved every 1 ps for the last 1 ns of the simulation to generate 1000 snapshots for subsequent analysis.
Hydration site identification and free energy estimation
Using the 1000 snapshots generated throughout the MD simulation, the hydration sites are identified. The detailed method has been described elsewhere.[15] Briefly, a three-dimensional (3D) grid is placed over the user-defined binding site using a grid spacing of 0.25 Å. In each snapshot, the positions of all the waters' oxygen atoms in the binding site are determined. A Gaussian distribution function centered on the oxygen atom centroid is used to distribute the occupancy of the water molecule onto the 3D grid. The occupancy distribution is then averaged over the MD trajectory and a QT clustering algorithm is used to identify the pronounced peaks that define the hydration site locations.
The desolvation free energy of each hydration site is determined by analyzing the enthalpy and entropy of the water molecules inside a hydration site
| (1) |
where ΔHhs and ΔShs are the enthalpic and entropic change of transferring a water molecule from the bulk solvent into the hydration site of the protein cavity. The change of the pressure–volume work associated with a volume change can be neglected.[27] Thus, the enthalpic change can be estimated by the change of the average interaction energies:
| (2) |
where Ehs is the interaction energy of a water molecule in the hydration site with the surrounding protein and water atoms. It is determined based on the average sum of van der Waals and electrostatic interactions between each water molecule inside a given hydration site with the protein and all the other water molecules. Ebulk is the interaction energy of a water molecule with its surrounding environment in the bulk solvent. The detailed calculation of Ebulk can be found in our previous work.[15]
Assuming no change in the momenta part of the partition function on transferring a water molecule from the bulk solvent into the protein cavity, ΔShs can be estimated by[28]
| (3) |
where C° is the concentration of pure water (1 molecule/29.9 Å3), R is the gas constant, and pext(q) is the external mode probability density function of the water molecules’ translational and rotational motions during the MD simulation. Please refer to our previous publication[15] for the detailed calculation of pext(q).
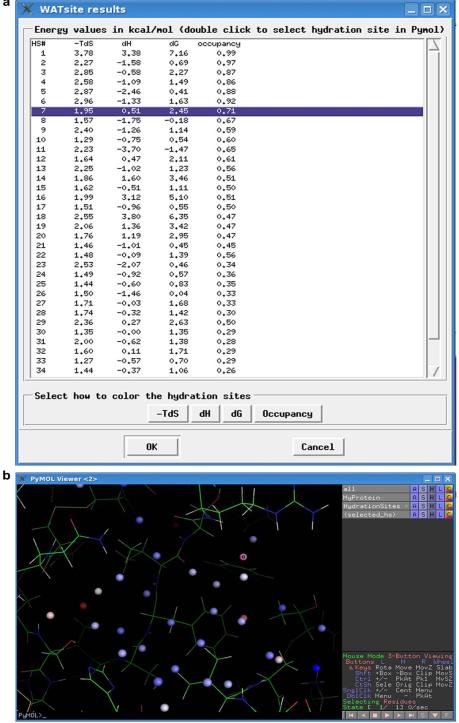
After completion of the WATsite simulation, the directory to the location of the prediction results is stored in the “WATsite.out” file. User can import the results through the “Import Results” command under the WATsite menu. Figure 2 displays an example of the imported results. The “WATsite results” window (Fig. 2a) displays the ΔG, ΔH, and −TΔS values for each hydration site estimated as described above. A hydration site with a positive ΔG value indicates an unfavorable environment of the water molecule in the binding site. Therefore, a gain in free energy of binding can be expected if the water in that hydration site is replaced by a ligand. The “occupancy” values indicate the probability a water molecule is observed in the given hydration site during the MD simulation.
Figure 2.
Example of WATsite results as displayed in the PyMOL plugin. a) “WATsite results” window listing the estimated desolvation free energy, entropy, enthalpy, and occupancy for each hydration site. By double clicking a line in the list (e.g., hydration site 7), an individual hydration site is selected in the PyMOL viewer (selected_hs selection in B). The user can also choose according to which descriptor the hydration sites are colored: free energy, entropy, enthalpy, or occupancy. b) The PyMOL viewer window showing the predicted hydration sites in the protein binding site. The hydration sites are shown as small spheres and colored in this example based on their ΔG values.
The location of the hydration sites in the protein binding site is displayed in the PyMOL viewer (Fig. 2b). When first loaded, the hydration sites are shown as nonbonded spheres and are color-coded based on their ΔG values, in a blue-to-red spectrum where blue indicates relatively low ΔG values and red indicates relatively high ΔG values. In addition, the user has the options to color the hydration sites based on ΔH, −TΔS or occupancy values in the WATsite results window (Fig. 2a). The user can also focus on individual hydration sites by double-clicking on a hydration site in the WATsite results window (Fig. 2a) to select the specific site in the PyMOL viewer.
Prediction of protein desolvation free energies on ligand binding using WATsite
A major application of WATsite is to use the predicted hydration sites to estimate the desolvation free energies involved in replacing water molecules in the protein binding site on ligand binding. For this purpose, a ligand library can be imported into PyMOL using the plugin and the desolvation free energy associated with replacing the binding site water molecules with each ligand is computed (Fig. 3). Within the plugin, the user can specify the directory containing all the ligands of interest and the radius used to select the hydration sites overlapping with the ligand heavy atoms (Fig. 3a). Once the ligands are successfully imported into PyMOL, the WATsite plugin will identify the hydration sites that are within the user-specified distance (default value is 1 Å) to any of the ligand's heavy atoms and add up the free energies associated with the selected hydration sites. The estimated desolvation free energies will be displayed for all ligands in a separate window (Fig. 3b). The hydration sites that were replaced by each individual ligand will also be shown as named selections in the PyMOL viewer (Fig. 3c; cmp1_replacedHS). It is worth noting that the estimated desolvation free energy only includes the energy of releasing the water molecules from the protein binding site into the bulk solvent. It cannot be used for a direct comparison of the ligands' free energies of binding. The ligand's free energy of binding includes other important contributions such as the direct protein–ligand interaction energy or desolvation energy of the ligand. The provided information, however, can guide further optimization of a lead compound rationally stabilizing or replacing additional water molecules in the binding site.
Figure 3.
Snapshot of example for estimating the protein's desolvation free energy associated with replacing the water molecules in the protein binding site on ligand binding. a) Dialog for importing ligand library and define hydration site selection radius. b) Results for desolvation free energy estimation. c) Snapshot of PyMOL viewer window associated with the desolvation free energy estimation. One ligand (cmp1) is shown in stick representation. The hydration sites that are within 1 Å to any of cmp1's heavy atoms were selected as “cmp1_replacedHS” and shown as spheres.
Conclusions
Water molecules can contribute both enthalpically and entropically to the free energy of ligand binding.[3] This contribution can be as important as the direct protein–ligand interactions in determining the thermodynamics of binding.[29] The availability of various computational methods[10] in hydration site analysis allows to elucidate the thermodynamic profile of individual water molecules to ligand binding.[29,30] In this article, we have developed a free hydration site analysis program WATsite that comes with a user-friendly PyMOL interface. WATsite is able to predict the position and thermodynamic profile of the hydration sites for both ligand-free and ligand-bound protein structure. The PyMOL interface allows users to intuitively view the hydration sites based on their thermodynamics profiles. Finally, a key feature of WATsite is that WATsite is able to estimate the protein desolvation free energies for the user specified ligands. The WATsite program, the PyMOL plugin and a user guide are available free of charge from http://people.pnhs.purdue.edu/~mlill/software.
Acknowledgments
Contract grant sponsor: NIH; Contract grant number: GM092855
References
- [1].Shoichet BK, McGovern SL, Wei B, Irwin JJ. Curr. Opin. Chem. Biol. 2002;6:439. doi: 10.1016/s1367-5931(02)00339-3. [DOI] [PubMed] [Google Scholar]
- [2].Sousa SF, Fernandes PA, Ramos MJ. Proteins. 2006;65:15. doi: 10.1002/prot.21082. [DOI] [PubMed] [Google Scholar]
- [3].Ladbury JE. Chem. Biol. 1996;3:973. doi: 10.1016/s1074-5521(96)90164-7. [DOI] [PubMed] [Google Scholar]
- [4].Goodford PJ. J. Med. Chem. 1985;28:849. doi: 10.1021/jm00145a002. [DOI] [PubMed] [Google Scholar]
- [5].Pitt WR, Goodfellow JM. Protein Eng. 1991;4:531. doi: 10.1093/protein/4.5.531. [DOI] [PubMed] [Google Scholar]
- [6].Pitt WR, Murray-Rust J, Goodfellow JM. J. Comput. Chem. 1993;14:1007. [Google Scholar]
- [7].Young T, Abel R, Kim B, Berne BJ, Friesner RA. Proc. Natl. Acad. Sci. USA. 2007;104:808. doi: 10.1073/pnas.0610202104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ehrlich L, Reczko M, Bohr H, Wade R. Protein Eng. 1998;11:11. doi: 10.1093/protein/11.1.11. [DOI] [PubMed] [Google Scholar]
- [9].Henchman RH, McCammon JA. J. Comput. Chem. 2002;23:861. doi: 10.1002/jcc.10074. [DOI] [PubMed] [Google Scholar]
- [10].de Beer S, Vermeulen NPE, Oostenbrink C. Curr. Top. Med. Chem. 2010;10:55. doi: 10.2174/156802610790232288. [DOI] [PubMed] [Google Scholar]
- [11].Thanki N, Thornton J, Goodfellow J. J. Mol. Biol. 1988;202:637. doi: 10.1016/0022-2836(88)90292-6. [DOI] [PubMed] [Google Scholar]
- [12].Young RJ, Campbell M, Borthwick AD, Brown D, Burns-Kurtis CL, Chan C, Convery MA, Crowe MC, Dayal S, Diallo H. Bioorg. Med. Chem. Lett. 2006;16:5953. doi: 10.1016/j.bmcl.2006.09.001. [DOI] [PubMed] [Google Scholar]
- [13].Abel R, Young T, Farid R, Berne BJ, Friesner RA. J. Am. Chem. Soc. 2008;130:2817. doi: 10.1021/ja0771033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lazaridis T. J. Phys. Chem. B. 1998;102:3531. [Google Scholar]
- [15].Hu B, Lill MA. J. Chem. Inf. Model. 2012;52:1046. doi: 10.1021/ci200620h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Word J, Lovell S, Richardson J, Richardson D. J. Mol. Biol. 1999;285:1735. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- [17].Lindahl E, Hess B, van der Spoel D. J. Mol. Model. 2001;7:306. [Google Scholar]
- [18].Berendsen H, Postma J, Van Gunsteren W, Hermans J. In Intermolecular Forces. Dordrecht, The Netherlands; Reidel: 1981. p. 331. [Google Scholar]
- [19].Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T. J. Comput. Chem. 2003;24:1999. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- [20].Sorin EJ, Pande VS. Biophys. J. 2005;88:2472. doi: 10.1529/biophysj.104.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].DePaul AJ, Thompson EJ, Patel SS, Haldeman K, Sorin EJ. Nucleic Acids Res. 2010;38:4856. doi: 10.1093/nar/gkq134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nose S. Mol. Phys. 1984;52:255. [Google Scholar]
- [23].Hoover WG. Phys. Rev. A: At. Mol. Opt. Phys. 1985;31:1695. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- [24].Parrinello M, Rahman A. J. Appl. Phys. 1981;52:7182. [Google Scholar]
- [25].Darden T, York D, Pedersen L. J. Chem. Phys. 1993;98:10089. [Google Scholar]
- [26].Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. J. Chem. Phys. 1995;103:8577. [Google Scholar]
- [27].Ben-Naim A. Statistical Thermodynamics for Chemists and Biochemists. Plenum Press; New York: 1992. [Google Scholar]
- [28].Minh D, Bui J, Chang C, Jain T, Swanson J, McCammon J. Biophys. J. 2005;89:L25. doi: 10.1529/biophysj.105.069336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Breiten B, Lockett MR, Sherman W, Fujita S, Al-Sayah MH, Lange H, Bowers CM, Heroux A, Krilov G, Whitesides GM. J. Am. Chem. Soc. 2013;135:15579. doi: 10.1021/ja4075776. [DOI] [PubMed] [Google Scholar]
- [30].Higgs C, Beuming T, Sherman W. ACS Med. Chem. Lett. 2010;1:160. doi: 10.1021/ml100008s. [DOI] [PMC free article] [PubMed] [Google Scholar]