Abstract
Physical inactivity reduces mechanical load on the skeleton, which leads to losses of bone mass and strength in non-hibernating mammalian species. Although bears are largely inactive during hibernation, they show no loss in bone mass and strength. To obtain insight into molecular mechanisms preventing disuse bone loss, we conducted a large-scale screen of transcriptional changes in trabecular bone comparing winter hibernating and summer non-hibernating black bears using a custom 12,800 probe cDNA microarray. A total of 241 genes were differentially expressed (P<0.01 and fold change >1.4) in the ilium bone of bears between winter and summer. The Gene Ontology and Gene Set Enrichment Analysis showed an elevated proportion in hibernating bears of overexpressed genes in six functional sets of genes involved in anabolic processes of tissue morphogenesis and development including skeletal development, cartilage development, and bone biosynthesis. Apoptosis genes demonstrated a tendency for downregulation during hibernation. No coordinated directional changes were detected for genes involved in bone resorption, although some genes responsible for osteoclast formation and differentiation (Ostf1, Rab9a, and c-Fos) were significantly underexpressed in bone of hibernating bears. Elevated expression of multiple anabolic genes without induction of bone resorption genes, and the down regulation of apoptosis-related genes, likely contribute to the adaptive mechanism that preserves bone mass and structure through prolonged periods of immobility during hibernation.
Keywords: Hibernation, Bone biosynthesis, Gene expression, Apoptosis
Introduction
Mammalian hibernation is an adaptation involving metabolic suppression to conserve energy during periods of low food availability in highly seasonal or unpredictable environments. Black bears hibernate for up to 6 months each year, and during hibernation, bears remain largely immobile and do not eat, drink, urinate, defecate, and they reduce metabolic rate by 20–50%, yet maintain core body temperatures above 30°C (Nelson 1980; Tøien et al. 2011).
Physical inactivity decreases mechanical load on skeleton, which when prolonged leads to losses of muscle and bone mass and strength in non-hibernating mammalian species (Kaneps et al. 1997; Harlow et al. 2001). Disuse-induced bone loss occurs by increases in osteoclastic bone resorption and/or decreases in osteoblastic bone formation (Zerwekh et al. 1998). This unbalance in resorption/formation leads to increased serum and urinary calcium concentrations (Watanabe et al. 2004). Although bears are largely inactive during hibernation, they show no loss in bone mass and less loss in muscle mass and strength than would be anticipated over such a prolonged period of physical inactivity (Harlow et al. 2001; Floyd et al. 1990; McGee et al. 2008; McGee-Lawrence et al. 2008, 2009; Nelson et al. 1975). An important adaptive consequence is that bears maintain skeleton function and preserve mobility during and after winter hibernation. This suggests that bears have unique mechanisms to prevent disuse-induced bone loss and reduce muscle atrophy during the inactivity of hibernation. Hibernating bears prevent bone loss by maintaining balanced bone resorption/formation, and they maintain normal serum calcium concentration despite anuria during this long period of disuse (Floyd et al. 1990; McGee et al. 2008; McGee-Lawrence et al. 2008, 2009). The molecular mechanisms that sustain balanced bone turnover during hibernation are unknown, but there is some evidence forendocrine or paracrine mechanisms (Donahue et al. 2006). For example, serum from hibernating bears decreases apoptotic signaling in osteoblastic (bone forming) cells compared to serum from non-hibernating bears (Bradford et al. 2009). Furthermore, bear stem cells may have the unusual ability to spontaneously differentiate down an osteoblastic lineage and form bone-like nodules (Fink et al. 2011). However, there has not before been a genome-wide screening of transcriptional changes in bone of hibernating bears to identify functional groups of co-regulated genes and provide insight into the molecular mechanisms that prevent disuse-induced bone loss.
We previously developed genomic resources for the black bear (Zhao et al. 2010) and detected transcriptional changes at the genomic scale in liver, heart, and skeletal muscle during hibernation (Fedorov et al. 2009; Fedorov et al. 2011). In the context of preserving skeleton functionality, an important finding was that protein biosynthesis genes demonstrated elevated expression in hibernating bears. This implies induction of translation and suggests activation of energy expensive anabolic mechanisms that contribute to the adaptive ability to reduce muscle atrophy over long periods of fasting and immobility during hibernation (Fedorov et al. 2009). In the present study, we use a custom 12,800 cDNA microarray to reveal transcriptional changes in trabecular bone of hibernating bears compared to animals sampled during summer. We conducted pathway analyses to identify functional groups of co-regulated genes and assessed the biological significance of the transcriptional changes. We also assessed differences in the expression of individual genes involved in bone formation, resorption, and apoptosis. Transcriptional changes are considered in light of previous findings such as balanced trabecular bone turnover (McGee-Lawrence et al. 2009) and reduced apoptosis (Bradford et al. 2009) that prevent disuse osteoporosis during hibernation.
Material and methods
Animals
We sampled the ilium bone from black bears including animals reported on in a previous study (Fedorov et al. 2009). Bears (51–143 kg) were captured May–July (two bears sampled in hibernation were captured in October) from the field in Alaska and transferred to Fairbanks. In order to decrease intragroup variation in gene expression, only males >2 years old are compared in the experiments. Summer active bears that were still feeding and active were euthanized and sampled for tissues between late May and early July (n=5) and one bear was sampled on October 2 (Fig. 1). Food was withdrawn 24 h before these animals were sacrificed. Bears in the hibernating condition were euthanized for tissue sampling between March 1 and 11 (n=5), about 1 month before expected emergence from hibernation. These animals were without food since October 27. Animal protocols were approved by the University of Alaska Fairbanks Institutional Animal Care and Use Committee (protocol nos. 02–39, 02–44, 05–55, and 05–56) and USAMRMC Animal Care and Use Review Office (proposal number 05178001).
Fig. 1.
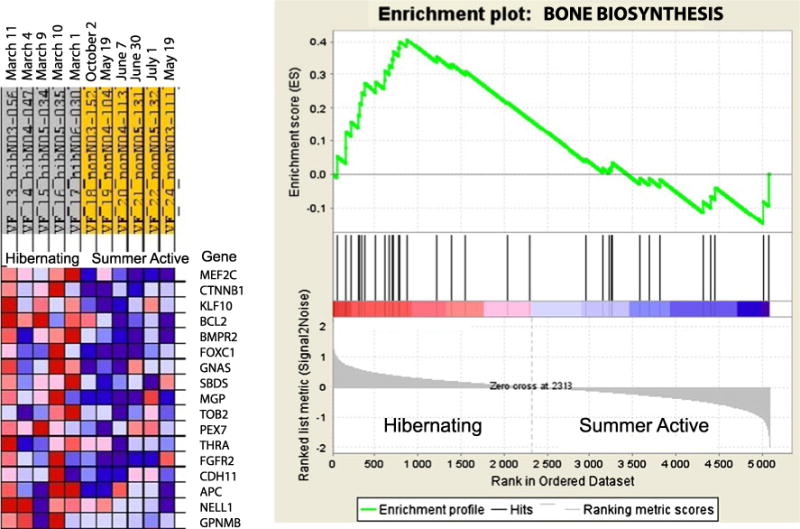
Gene set enrichment analysis results for the bone biosynthesis (ossification) category. The ossification category is enriched by upregulated genes in the ilium bone of hibernating black bears. An expression data set sorted by correlation with hibernating phenotype and the corresponding heat map with red for upregulated and blue for downregulated genes during hibernation are shown on the left. Dates on the top indicate time of tissue sampling from each bear. Plot of the running sum for enrichment score (ES) in the data set (top) and location of genes (hits) from the GO category in the list ranked according to expression differences (middle) and the ranked list metric (bottom) are shown on the right
Physiological monitoring and tissue harvesting
For monitoring of physiological conditions, bears were instrumented as previously described (Fedorov et al. 2009). Briefly, core body temperature, ECG, and EMG were monitored with radio telemetry. Beginning in late November, bears were housed in individual undisturbed outdoor enclosures that had dens with straw material for nests. Oxygen consumption and respiratory quotient were monitored with open flow respirometry by drawing air from the closed dens. On the day of tissue harvesting, bears were immobilized using Telazol (8–10 mg/kg) and transported to a necropsy suite in a nearby building. Oxygen consumption in the immobilized state was checked on a subsample of animals, with an open flow respirometry system during blood sampling, just prior to euthanasia via a tracheal tube. Between the first disturbance of bears and the beginning of tissue sampling, 41–65 min elapsed. Bears were euthanized by an intravenous injection of pentobarbital with death assessed by termination of heart beats as assessed with a stethoscope. Tissues collection followed immediately with samples frozen in liquid nitrogen within 12 min of death. One trabecular bone sample 80×20 mm was cut from the ilium tuber coxae of each bear.
RNA preparation
Bone tissue was pulverized in a metal cylinder cooled in liquid nitrogen with a piston and then transferred into Trizol reagent with 0.1 volume of chloroform. The mix was centrifuged at 13,000×g for 20 min at 4°C, and a clear aqueous phase was added to 0.5 volume of 2-propanol and left for 10 min at room temperature. Then the mix was centrifuged at 13,000×g for 20 min at 4°C and the pellet was washed with ethanol twice and resuspended in RNase-free water. Additional RNA cleanup was performed with the Qiagen RNeasy kit. All RNA samples were processed by DNase I (Qiagen) treatment. RNA quality was evaluated with an Agilent 2100 Bioanalyzer and concentration was measured by using Nanodrop ND-1000.
Hybridization
RNA samples were hybridized with the two bear arrays (BA01 and BA02) that contain 3,200 and 9,600 cDNA probes representing unique annotated genes in the black bear expressed sequence tags (ESTs) collection (Zhao et al. 2010; Fedorov et al. 2011). Samples of total RNA were linearly amplified with Illumina TotalPrep RNA Amplification Kit (Ambion), and 1.6 μg of the amplified RNA was labeled with 65 μCi of [33P]dCTP as previously described (Kari et al. 2003). All RNA samples were amplified, labeled, and hybridized in the same batch. The hybridization was carried out for 18 h at 42°C in 4 ml of MicroHyb buffer (Invitrogen). Filters were rinsed at room temperature with 2× SSC/1% SDS to remove residual probe and MicroHyb solution and then transferred to preheated wash solutions in a temperature-controlled shaking water bath. Filters were washed twice for 30 min in 1.5 l of 2× SSC/1% SDS at 50°C and then once for 30 min in 1.5 l of 0.5× SSC/1% SDS at 55°C and once for 30 min in 1.5 l of 0.1× SSC/0.5% SDS at 55°C. Filters were then exposed to phosphorimager screens for 4 days and scanned at 50-μm resolution in a Storm Phosphorimager. Image analysis was performed with the ImaGene program (Biodiscovery).
Microarray data analysis
Hybridization signals were corrected for background, normalized and one-way ANOVA test was used to select genes that exhibited significant differences between hibernating and summer active bears (Fedorov et al. 2011). A P value <0.01 and |log2 fold change|>0.5 were set as cutoffs for significant differences in expressed genes, corresponding to the mean false discovery rate (FDR) around 24%. The FDR was calculated using random permutation as described by Storey and Tibshirani (2003). Lists of all significant genes on the array and differentially expressed genes with cutoffs of P value <0.05 and |log2 fold change|>0.5 were uploaded to Gene Ontology (GO) miner (http://discover.nci.nih.gov/gominer/index.jsp). The false discovery rate was assessed by resampling all the significant genes on the array (Zeeberg et al. 2003, 2005). In addition to GO miner analysis, we verified enrichment in significant GO categories of the biological processes by using Gene Set Enrichment Analysis (http://www.broad.mit.edu/gsea/index.jsp). Genes were ranked according to the correlation between their expression values and the phenotype class (hibernating and summer active phenotypes) distinction by using the signal to noise ratio. An enrichment score (ES) that reflects the degree to which genes involved in category are overrepresented at the extremes (upregulated genes at the top and downregulated genes at the bottom) of the entire ranked list of genes was calculated. The ES was normalized to account for the size of the category gene set presented in the experiment, yielding a normalized enrichment score (NES). A cutoff of 25% for false discovery rate of gene set enrichment was used as this value was suggested appropriate for exploratory studies (Subramanian et al. 2005). We also used Gene Set Enrichment Analysis (GSEA) to test enrichment in selected gene sets that were reported to be important for bone metabolism. These gene sets were obtained from Molecular Signatures Database (www.broadinstitute.org/gsea/msigdb/index.jsp). All microarray data series were submitted to NCBI Gene Expression Omnibus with accession number GSE35796.
Quantitative real-time PCR
We validated the microarray experiments by 220 quantitative real-time PCR (RT PCR) tests using the same total RNA samples. Twenty genes were tested. Hint1 was selected as a reference gene for bone based on the stability of expression values across all samples obtained from the microarray experiments and then tested by RT PCR. All bear samples showed similar expression values for Hint1 with low standard deviation in multiple RT PCR tests. Total RNA concentrations were measured with a NanoDrop ND-1000 spectrophotometer, and cDNA was synthesized from 0.5 μg of total RNA from each sample. The reverse transcription was carried out with MiltiScribeTM reverse transcriptase (Applied Biosystems) with oligo d(T)16 primer in 25-μl reactions at 25°C for 10 min, 48°C for 30 min, and at 95°C for 5 min. The synthesized cDNA was diluted four times with RNase-free water, and 4 μl of diluted cDNA was used in the 20-μl volume real-time PCR. Primers were designed with the Primer3 software (http://frodo.wi.mit.edu/primer3/) using bear EST sequences (Table S1). Real-time PCR was performed in triplicates with Power SYBR Green PCR Master Mix (Applied Biosystems) on an ABI-7900 HT. Cycling parameters were 50°C for 2 min of incubation, 95°C for 10 min of Taq activation, and 40 cycles of 95°C for 15 s and 60°C for 1 min. Controls with no template were set to exclude contamination, and controls with no reverse transcriptase but all other components were taken to exclude nonspecific amplification from genomic DNA. Specificity of amplification was checked with the melting curve analysis and agarose gel electrophoresis. Four tenfold dilutions of a sample with mixed cDNA were used for a standard curve for each primer set for calculating RT PCR efficiency. We tested a difference in gene expression between hibernating and summer active black bears with P<0.10 as cutoff according to Pfaffl (2001). We calculated the fold change in level of expression of a target gene relative to a reference gene for each sample and then compared the values for each group using Student’s t test as described by Livak and Schmittgen (2001).
Results
Body temperature and metabolism
At the time of sampling between March 1 and 11, hibernating bears had core body temperatures of 34.5±0.5°C (mean ± SD, n=5) and minimum rates of oxygen consumption of 0.078±0.001 ml g−1 h−1 (n=4), when measured over at least a 0.5-h interval 2–9 h before euthanasia (Fedorov et al. 2009). In three bears anesthetized before euthanasia, body temperature had decreased to 33.6±1.0°C and metabolic rate had increased to 0.105±0.012 ml g−1 h−1. Ambient temperatures during the study period in winter were −10°C to −35°C with short periods with extremes of −43°C to 5°C. Temperatures within bear dens were about 10°C above the outside temperature. Bears lost 4.3±0.6% (n=5) of their body mass per month during the 4–5-month hibernation period. In two summer active bears, fasted for 24 h and anesthetized before euthanasia metabolic rates were 0.232 ml g−1 h−1 (range 0.252 to 0.213 ml g−1 h−1) and body temperatures averaged 37.2°C. Thus, immediately prior to tissue sampling, metabolic rate of anesthetized hibernating bears was 45.4% of that of anesthetized summer bears, with a 3.5°C lower body temperature.
Differentially expressed genes
Signals from 2,602 of 3,200 probes (81%) on bear array BA01 showed median intensities that were above background, whereas 9,473 of 9,600 (99%) probes showed significant signals on bear array BA02. To identify genes that were differentially expressed in hibernating compared to summer active bears, we used P<0.01 and |log2FC|>0.5, where FC is fold change (the mean expression value in the hibernating bears divided by the mean expression value in the summer active bears) as the criteria for differentially expressed genes as previously reported for other tissues (Fedorov et al. 2009, 2011). A total of 241 genes (2.6% of all genes with significant signals) were differentially expressed in bone during hibernation (Table S2). All but two differentially expressed genes demonstrated changes in expression that were less than fourfold differences (|log2FC|<2). Of the significantly differentially expressed genes, we identified 71 (29.5%) genes that were overexpressed and 170 (70.5%) genes that were underexpressed in bone during hibernation.
The estimated mean false discovery rate of 24% indicates that the expected proportion of false positives in the list of differentially expressed genes from array experiments is relatively high. To obtain an experimental estimate of the false discovery rate, we conducted quantitative real-time PCR tests for 20 randomly selected genes that showed differences in expression at P<0.05 in the array hybridization. Eighteen out of 20 genes (90%) tested showed significant changes in the same direction as the array results (Table 1, Fig. 2). The observed value of 10% for the false discovery rate is reasonable for this exploratory study. An important point of support for microarray results comes from the highly significant positive correlation (r=0.89, <0.0001) between fold changes for true positive in RT PCR and microarray experiments. Expression fold changes do not depend on significance level (P value) of individual genes, and thus, they are not sensitive to false discovery rate that results from multiple testing.
Table 1.
Gene expression differences obtained in microarray experiments and tested with real-time PCR in bear bone
| Gene symbol | Gene name | RT PCR
|
Microarray
|
||
|---|---|---|---|---|---|
| P | log2FC | P | log2FC | ||
| B4galt6 | UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 6 | 0.100 | −0.573 | 0.024 | −0.843 |
| Cav1 | Caveolin 1, caveolae protein | 0.039 | 1.395 | 0.023 | 1.055 |
| Cd47 | Leukocyte surface antigen CD47 precursor | 0.012 | −0.823 | 0.001 | −1.300 |
| Cfl1 | Cofilin-1 | 0.380 | n/a | 0.012 | −1.429 |
| Dcn | Decorin | 0.005 | 1.142 | 0.008 | 0.955 |
| Ect2 | Epithelial cell transforming sequence 2 oncogene | 0.040 | −1.392 | 0.047 | −0.792 |
| Fn1 | Fibronectin | 0.013 | 1.334 | 0.027 | 1.192 |
| Gpc3 | Glypican 3 | 0.015 | 1.847 | 0.012 | 1.922 |
| Hoxa10 | Homeobox A10 | 0.003 | 1.181 | 0.008 | 0.831 |
| Lnpep | Leucyl/cystinyl aminopeptidase | 0.080 | 0.561 | 0.041 | 0.631 |
| Mapk8 | Mitogen-activated protein kinase 8 | 0.180 | n/a | 0.006 | 0.837 |
| Mef2c | Myocyte enhancer factor 2 C | 0.011 | 1.196 | 0.013 | 1.055 |
| Omg | Oligodendrocyte myelin glycoprotein | 0.043 | −1.053 | 0.009 | −0.746 |
| Prrx1 | Paired related homeobox 1 | <0.001 | 1.515 | <0.001 | 1.488 |
| Sdc2 | Syndecan 2 | 0.050 | 0.422 | 0.013 | 0.889 |
| Srgn | Serglycin | <0.001 | −1.761 | 0.002 | −1.863 |
| Timp2 | Tissue inhibitor of metalloproteinase 2 | 0.005 | 0.992 | 0.021 | 1.138 |
| Ube2d3 | Ubiquitin-conjugating enzyme E2D 3 | <0.001 | −0.581 | 0.039 | −0.613 |
| Yap1 | Yes-associated protein 1 | 0.002 | 1.180 | 0.011 | 0.598 |
| Zeb1 | Zinc finger E-box binding homeobox 1 | 0.003 | 0.585 | <0.001 | 0.841 |
Inconsistent significance levels are in bold
Fig. 2.
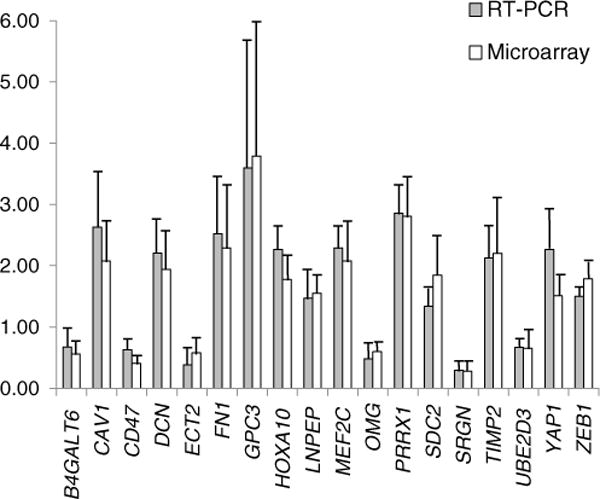
Differentially expressed genes in black bear bone tissue confirmed with real-time PCR. Solid and open bars show normalized expression values obtained in real-time PCR and microarray experiments, respectively; error bars represent SD. The values were normalized to the mean in summer active bears
Functional gene sets enriched by differentially expressed genes
Genes with a significant hybridization signal on the arrays were classified according to their GO categories of biological processes. GO categories with less than five differentially expressed genes detected were excluded from the analysis (Zeeberg et al. 2003). Significant enrichment of biological processes categories by differentially expressed genes was validated by the results of GSEA (Table 2; Fig. 1). GSEA ranks all genes with significant signals in the experiment; thus, its results do not depend on the selection of genes above cutoffs for significance of expression differences and false discovery (Subramanian et al. 2005).
Table 2.
Selected Gene Ontology categories of biological processes significantly enriched with differentially expressed genes in black bear bone
| GO category | Total genes on array | Changed genes | GO Miner
|
GSEA
|
||
|---|---|---|---|---|---|---|
| Enrichment | FDR | NES | FDR | |||
| Organ morphogenesis (GO:0009887) | 400 | 34↑ | 2.400 | <0.001 | 1.74 | 0.008 |
| Skeletal development (GO:0001501) | 119 | 14↑ | 3.268 | 0.008 | 1.59 | 0.077 |
| Central nervous system development (GO:0007417) | 218 | 18↑ | 2.305 | 0.030 | 1.35 | 0.189 |
| Muscle tissue development (GO:0060537) | 117 | 13↑ | 3.087 | 0.015 | 1.25 | 0.178 |
| Cartilage development (GO:0051216) | 33 | 6↑ | 5.051 | 0.038 | 1.57 | 0.074 |
| Ossification (GO:0001503) | 49 | 6↑ | 3.402 | 0.084 | 1.54 | 0.023 |
| Phagocytosis (GO:0006909) | 48 | 12↓ | 3.111 | 0.023 | −1.43 | 0.094 |
Arrows indicate direction of gene regulation in hibernating animals
FDR false discovery rate, NES normalized enrichment score
During hibernation, the proportion of overexpressed genes was significantly elevated for six gene sets involved in anabolic processes of tissues morphogenesis and development (Table 2). Among upregulated gene sets, there was the skeletal development category that includes genes involved in cartilage development and ossification (Table 3). Although the ossification category demonstrated enrichment by upregulated genes only at P=0.084 level in the GO miner and included six downregulated genes, mostly involved in bone mineralization, the GSEA showed overall significant (P=0.023) upregulation of bone biosynthesis genes (Fig. 1). Phagocytosis was the only biological processes category with significantly elevated proportion of downregulated genes (Table 2).
Table 3.
Genes in selected Gene Ontology categories of the biological processes significantly enriched with differentially expressed genes in black bear bone
| GO category | Gene name | Gene symbol | P value | Fold change (log2FC) |
|---|---|---|---|---|
| Skeletal development (GO:0001501) | Decorin | Dcn | 0.008 | 0.955 |
| Fibronectin 1 | Fn1 | 0.027 | 1.192 | |
| Homeobox A10 | Hoxa10 | 0.008 | 0.831 | |
| Insulin-like growth factor 1 receptor | Igf1r | 0.005 | 0.515 | |
| KIAA1217 | Kiaa1217 | 0.029 | 1.083 | |
| Kruppel-like factor 10 | Klf10 | 0.042 | 0.746 | |
| Mitogen-activated protein kinase 8 | Mapk8 | 0.006 | 0.837 | |
| Myocyte enhancer factor 2C | Mef2c | 0.013 | 1.055 | |
| Paired related homeobox 1 | Prrx1 | <0.001 | 1.488 | |
| Retinol binding protein 4, plasma | Rbp4 | 0.022 | 0.898 | |
| Schwannomin interacting protein 1 | Schip1 | 0.025 | 0.769 | |
| SRY (sex determining region Y)-box 9 | Sox9 | 0.010 | 0.568 | |
| Yes-associated protein 1 | Yap1 | 0.011 | 0.598 | |
| Zinc finger E-box binding homeobox 1 | Zeb1 | <0.001 | 0.841 | |
| Cartilage development (GO:0051216) | Decorin | Dcn | 0.008 | 0.955 |
| Myocyte enhancer factor 2 C | Mef2c | 0.013 | 1.055 | |
| Paired related homeobox 1 | Prrx1 | <0.001 | 1.488 | |
| SRY (sex determining region Y)-box 9 | Sox9 | 0.010 | 0.568 | |
| Yes-associated protein 1 | Yap1 | 0.011 | 0.598 | |
| Zinc finger E-box binding homeobox 1 | Zeb1 | <0.001 | 0.841 | |
| Ossification (GO:0001503) | Fibronectin 1 | Fn1 | 0.027 | 1.192 |
| Insulin-like growth factor 1 receptor | Igf1r | 0.005 | 0.515 | |
| Kruppel-like factor 10 | Klf10 | 0.042 | 0.746 | |
| Mitogen-activated protein kinase 8 | Mapk8 | 0.006 | 0.837 | |
| Myocyte enhancer factor 2C | Mef2c | 0.013 | 1.055 | |
| SRY (sex determining region Y)-box 9 | Sox9 | 0.010 | 0.568 |
Apart from validation of the GO miner result, we also used GSEA to test enrichment in selected gene sets known to be important for bone homeostasis. Similar to bone biosynthesis, bone morphogenetic protein signaling pathway (GO:0030509, 12 genes on the array) showed elevated proportion of upregulated genes (NES=1.39; FDR=0.13). In contrast, the bone mineralization category (GO:0030282) was represented by ten genes on the microarray and was enriched by downregulated genes (NES=−1.43; FDR=0.062) during hibernation. No significant directional changes (FDR=0.576) were detected in expression of seven genes involved in bone resorption (GO:0045453). However, two genes that induce osteoclast formation and bone resorption, small GTP-binding Rab9 protein (Rab9a) and osteoclast stimulating factor-1 (Ostf1), were both downregulated (Table S2) during hibernation. Receptor activation of NF-κb ligand (RANKL) signaling pathway (Biocarta: M2602, two genes on the array) that plays an important role in resorption during bone remodeling showed downregulation (NES=−1.32; FDR=0.085) during hibernation. In the RANKL gene set, c-Fos gene, an important mediator of osteoclast differentiation, was the most downregulated (Table S2). The apoptosis gene set including all apoptosis-related genes from Molecular Signatures Database was represented by 202 genes on the microarray and demonstrated a clear tendency for downregulation during hibernation with 38 genes being downregulated and overrepresented at the bottom of the GSEA ranked list of apoptosis genes (NES=−1.22; FDR=0.11).
Discussion
We sampled hibernating bears in early March after at least 4 months of continuous hibernation and when they were 4–6 weeks from emergence and their resumption of summer active levels of metabolism, body temperature, and feeding (Fedorov et al. 2009). All bears included as summer active or hibernating animals showed physiology and behavior typical for bears during summer and winter seasons (Tøien et al. 2011).
Of the genes that were differentially expressed in bone during hibernation, 70.5% were downregulated. This contrasts to the lower proportions of downregulated genes that were previously reported for the liver (48% underexpressed genes), skeletal muscle (46% underexpressed genes), and heart (25% downregulated genes) in hibernating bears (Fedorov et al. 2009, 2011). The decrease in the transcription levels for a number of genes may reflect a lower level of homeostatic activity in bone compared to other organs associated with prolonged period of immobility and skeleton unloading during hibernation.
Our study reveals coordinated induction in transcription of genes involved in anabolic processes of tissues development and formation including osteogenesis and cartilage development. This finding implies elevation of some forms of anabolic activity in bone during hibernation. In contrast, no coordinated directional changes were detected for genes involved in bone resorption, although some genes responsible for osteoclast formation and differentiation (Ostf1, Rab9a, and c-Fos) were significantly underexpressed in bone of hibernating bears. Pronounced underexpression of the c-Fos gene, member of RANKL signaling pathway, is remarkable as mice lacking this gene have overly dense bone and decreased bone resorption due to reduced osteoclast differentiation (Takayanagi et al. 2002). Transcriptional changes detected in our study may have implications for an adaptive mechanism preserving bone mass and structure through prolonged periods of immobility during hibernation.
Bears demonstrate a unique ability to prevent bone loss through prolonged periods of inactivity and skeleton disuse during 4–6 months of winter hibernation. In other mammalian species, mechanical unloading over periods of 4–17 weeks resulted in bone loss of 9–29% due to decreases in bone formation (McGee-Lawrence et al. 2008). Balanced remodeling preserves the architecture and strength of hibernating bear bone (Floyd et al. 1990; McGee-Lawrence et al. 2008, 2009; Pardy et al. 2004). Hibernating grizzly bears demonstrate decreases in the overall number of remodeling sites in the cortical bone (McGee et al. 2008), but no differences in the total number of remodeling sites in trabecular bone in the ilium (McGee-Lawrence et al. 2009). Ilium biopsies from hibernating black bears, albeit a small sample size, suggested some increase in trabecular bone remodeling (with balanced resorption and formation) during hibernation, which is believed to maintain bone structure and calcium homeostasis (Floyd et al. 1990). The molecular mechanisms that maintain constant and balanced trabecular bone remodeling during hibernation, despite the challenge of physical inactivity, are unknown.
Bone loss during immobility in mammals typically results from unbalanced remodeling due to decrease in bone formation alone or both decreased bone formation and increased resorption (McGee et al. 2008). Prolonged periods of mechanical unloading during hibernation potentially inhibit anabolic processes and promote reduction of bone formation. Coordinated induction in transcription of genes involved in anabolic processes implies elevation of anabolic activity in trabecular bone (ilium) that may counteract disuse-induced reductions in osteoanabolic activity, thus, preventing bone loss during hibernation. There was a lack of directional changes in expression of the group of genes involved in bone resorption. However, there was significant downregulation of three genes (Ostf1, Rab9a, and c-Fos) stimulating osteoclast differentiation. This decreased gene expression may counteract the mechanisms responsible for the increased osteoclastogenesis and bone resorption normally seen in disuse conditions. We also found a reduction in the transcription of genes responsible for bone mineralization, which is consistent with histological data showing decreased mineral apposition rate in the ilium (McGee-Lawrence et al. 2009). Decreased mineral apposition rate may be part of globally reduced metabolism for the conservation of metabolic energy during hibernation.
It has been suggested that reduction in disuse-induced osteoblast apoptosis contributes to the maintenance of bone formation during hibernation (Bradford et al. 2009). Coordinated suppression in transcription of apoptosis-related genes detected in our study supports decreased apoptotic activity in the ilium bone during hibernation. Underexpression of the two key apoptotic genes (Casp3, Casp7) during hibernation is consistent with reduction of caspase 3/7 activity detected in MC3T3-E1 osteoblasts cultured in hibernation serum as compared to osteoblasts treated with sera from fall and spring active black bears (Bradford et al. 2009). Seasonal changes in serum factors may maintain normal osteoblastic activity and bone formation during hibernation by reducing osteoblast loss due to apoptosis.
Our finding of increased expression of bone anabolic genes without an increase in transcription of bone resorption genes is similar to the elevated expression of anabolic genes involved in protein biosynthesis and unchanged transcriptional level of protein catabolism genes reported previously for skeletal muscles of hibernating bears (Fedorov et al. 2009). In addition to preventing disuse-induced bone loss, bears have a unique ability to preserve muscle mass and strength during hibernation (Harlow et al. 2001; Lundberg et al. 1976). Coordinated increase in transcriptional level of anabolic genes involved in protein biosynthesis implies induction of translation that may help prevent muscle atrophy during hibernation (Fedorov et al. 2009). Induction of anabolic processes without increased catabolism (inferred here from transcriptional changes) suggests that similar adaptive mechanisms contribute to the preservation of bone and muscle mass during prolonged periods of physical inactivity and starvation (i.e., hibernation). Elevation of energy expensive anabolic processes is generally unexpected because of the lack of dietary intake during hibernation, which is an adaptive strategy involving metabolic suppression to conserve energy during periods of low food availability. However, the energy cost of increased anabolism in bone and muscle may be an important trade-off with the adaptive mechanisms that allow bears to maintain full musculoskeletal function and preserve mobility during and immediately after hibernation, thus promoting survival.
In conclusion, this study represents the first research effort to elucidate transcriptional changes for thousands of genes in trabecular bone during hibernation in comparison to summer active black bears. Elevated expression of multiple anabolic genes without induction of bone resorption genes, as well as the downregulation of apoptosis-related genes, likely contribute to the adaptive mechanisms that preserve bone mass and structure through prolonged periods of immobility during hibernation. Future studies on the hibernating transcriptome in homogenous populations of different bone cell types (e.g., osteoblasts and osteoclasts) with gene probes more specific for bone will identify co-regulated functional groups of genes and provide new insight to the molecular basis of unusual bone homeostasis in hibernating bears. Understanding the molecular mechanism that prevents bone loss in hibernating bears may identify novel therapeutic targets to improve treatments for osteoporosis.
Supplementary Material
Acknowledgments
We thank the Alaska Department of Fish and Game for supplying bears. This work was supported by the National Science Foundation EPSCOR program and USAMRMC (05178001).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10142-012-0266-3) contains supplementary material, which is available to authorized users.
Contributor Information
Vadim B. Fedorov, Email: vfedorov@alaska.edu, Institute of Arctic Biology, University of Alaska Fairbanks, Fairbanks, AK 99775, USA
Anna V. Goropashnaya, Institute of Arctic Biology, University of Alaska Fairbanks, Fairbanks, AK 99775, USA
Øivind Tøien, Institute of Arctic Biology, University of Alaska Fairbanks, Fairbanks, AK 99775, USA.
Nathan C. Stewart, Institute of Arctic Biology, University of Alaska Fairbanks, Fairbanks, AK 99775, USA
Celia Chang, Systems and Computational Biology Center, the Wistar Institute, Philadelphia, PA 19104, USA.
Haifang Wang, CAS-MPG Partner Institute for Computational Biology, Shanghai Institutes of Biological Sciences, 320 Yue Yang Road, Shanghai 200031, China.
Jun Yan, CAS-MPG Partner Institute for Computational Biology, Shanghai Institutes of Biological Sciences, 320 Yue Yang Road, Shanghai 200031, China.
Louise C. Showe, Systems and Computational Biology Center, the Wistar Institute, Philadelphia, PA 19104, USA
Michael K. Showe, Systems and Computational Biology Center, the Wistar Institute, Philadelphia, PA 19104, USA
Seth W. Donahue, Department of Biomedical Engineering, Michigan Technological University, 309 Minerals and Materials Eng. Bldg., 1400 Townsend Drive, Houghton, MI 49931, USA
Brian M. Barnes, Institute of Arctic Biology, University of Alaska Fairbanks, Fairbanks, AK 99775, USA
References
- Bradford RM, Heiden DM, Gray SK, Buckendahl P, Vaughan MR, Tsai CJ, Donahue SW. Serum from hibernating bears exhibits increased osteocalcin and stimulates decreased apoptotic signaling in differentiating MC3T3-E1 osteoblasts [abstract]; ASBMR 31st Annual Meeting SU0205.2009. [Google Scholar]
- Donahue SW, Galley SA, Vaughan MR, Patterson-Buckendahl P, Demers LM, Vance JL, McGee ME. Parathyroid hormone may maintain bone formation in hibernating black bears (Ursus americanus) to prevent disuse osteoporosis. J Exp Biol. 2006;209:1630–1638. doi: 10.1242/jeb.02185. [DOI] [PubMed] [Google Scholar]
- Fedorov VB, Goropashnaya AV, Tøien Ø, Stewart NC, Gracey AY, Chang CL, Qin SZ, Pertea G, Quackenbush J, Showe LC, Showe MK, Boyer BB, Barnes BM. Elevated expression of protein biosynthesis genes in liver and muscle of hibernating black bears (Ursus americanus) Physiol Genomics. 2009;37:108–118. doi: 10.1152/physiolgenomics.90398.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov VB, Goropashnaya AV, Tøien Ø, Stewart NC, Chang C, Wang H, Yan J, Showe LC, Showe MK, Barnes BM. Modulation of gene expression in heart and liver of hibernating black bears (Ursus americanus) BMC Genomics. 2011;12:171. doi: 10.1186/1471-2164-12-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink T, Rasmussen JG, Emmersen J, Pilgaard L, Fahlman A, Brunberg S, Josefsson J, Arnemo JM, Zachar V, Swenson JE, Frobert O. Adipose-derived stem cells from the brown bear (Ursus arctos) spontaneously undergo chondrogenic and osteogenic differentiation in vitro. Stem Cell Res. 2011;7:89–95. doi: 10.1016/j.scr.2011.03.003. [DOI] [PubMed] [Google Scholar]
- Floyd T, Nelson RA, Wynne GF. Calcium and bone metabolic homeostasis in active and denning black bears (Ursus americanus) Clin Orthop Relat Res. 1990;255:301–309. [PubMed] [Google Scholar]
- Harlow HJ, Lohuis T, Beck TD, Iaizzo PA. Muscle strength in overwintering bears. Nature. 2001;409:997. doi: 10.1038/35059165. [DOI] [PubMed] [Google Scholar]
- Kaneps AJ, Stover SM, Lane NE. Changes in canine cortical and cancellous bone mechanical properties following immobilization and remobilization with exercise. Bone. 1997;21:419–423. doi: 10.1016/s8756-3282(97)00167-1. [DOI] [PubMed] [Google Scholar]
- Kari L, Loboda A, Nebozhyn M, Rook AH, Vonderheid EC, Nichols C, Virok D, Chang C, Horng WH, Johnston J, Wysocka M, Showe MK, Showe LC. Classification and prediction of survival in patients with the leukemic phase of cutaneous T cell lymphoma. J Exp Med. 2003;197:1477–1488. doi: 10.1084/jem.20021726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lundberg DA, Nelson RA, Wahner HW, Jones JD. Protein metabolism in the black bear before and during hibernation. Mayo Clin Proc. 1976;51:716–722. [PubMed] [Google Scholar]
- McGee ME, Maki AJ, Johnson SE, Nelson OL, Robbins CT, Donahue SW. Decreased bone turnover with balanced resorption and formation prevent cortical bone loss during disuse (hibernation) in grizzly bears (Ursus arctos horribilis) Bone. 2008;42:396–404. doi: 10.1016/j.bone.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee-Lawrence ME, Carey HV, Donahue SW. Mammalian hibernation as a model of disuse osteoporosis: the effects of physical inactivity on bone metabolism, structure, and strength. Am J Physiol Regul Integr Comp Physiol. 2008;295:R1999–R2014. doi: 10.1152/ajpregu.90648.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee-Lawrence ME, Wojda SJ, Barlow LN, Drummer TD, Castillo AB, Kennedy O, Condon KW, Auger J, Black HL, Nelson OL, Robbins CT, Donahue SW. Grizzly bears (Ursus arctos horribilis) and black bears (Ursus americanus) prevent trabecular bone loss during disuse (hibernation) Bone. 2009;6:1186–1191. doi: 10.1016/j.bone.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RA. Protein and fat metabolism in hibernating bears. Fed Proc. 1980;39:2955–2958. [PubMed] [Google Scholar]
- Nelson RA, Jones JD, Wahner HW, McGill DB, Code CF. Nitrogen metabolism in bears: urea metabolism in summer starvation and in winter sleep and role of urinary bladder in water and nitrogen conservation. Mayo Clin Proc. 1975;50:141–146. [PubMed] [Google Scholar]
- Pardy CK, Wohl GR, Ukrainetz PJ, Sawers A, Boyd SK, Zernicke RF. Maintenance of bone mass and architecture in denning black bears (Ursus americanus) J Zool. 2004;263:359–364. [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:2001–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayanagi H, Kim S, Matsuo K, Suzuki H, Suzuki T, Sato K, Yokochi T, Oda H, Nakamura K, Ida N, Wagner EF, Taniguchi T. RANKL maintains bone homeostasis through c-Fos-dependent induction of interferon-beta. Nature. 2002;416:744–749. doi: 10.1038/416744a. [DOI] [PubMed] [Google Scholar]
- Tøien Ø, Blake J, Edgar DM, Grahn DA, Heller HC, Barnes BM. Hibernation in black bears: independence of metabolic suppression from body temperature. Science. 2011;331:906–909. doi: 10.1126/science.1199435. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Ohshima H, Mizuno K, Sekiguchi C, Fukunaga M, Kohri K, Rittweger J, Felsenberg D, Matsumoto T, Nakamura T. Intravenous pamidronate prevents femoral bone loss and renal stone formation during 90-day bed rest. J Bone Miner Res. 2004;19:1771–1778. doi: 10.1359/JBMR.040811. [DOI] [PubMed] [Google Scholar]
- Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, Narasimhan S, Kane DW, Reinhold WC, Lababidi S, Bussey KJ, Riss J, Barrett JC, Weinstein JN. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeberg BR, Qin H, Narasimhan S, Sunshine M, Cao H, Kane DW, Reimers M, Stephens RM, Bryant D, Burt SK, Elnekave E, Hari DM, Wynn TA, Cunningham-Rundles C, Stewart DM, Nelson D, Weinstein JN. High-Throughput GoMiner, an ‘industrial-strength’ integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of common variable immune deficiency (CVID) BMC Bioinforma. 2005;6:168. doi: 10.1186/1471-2105-6-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerwekh JE, Ruml LA, Gottschalk F, Pak CYC. The effects of twelve weeks of bed rest on bone histology, biochemical markers of bone turnover, and calcium homeostasis in eleven normal subjects. J Bone Miner Res. 1998;13:1594–1601. doi: 10.1359/jbmr.1998.13.10.1594. [DOI] [PubMed] [Google Scholar]
- Zhao S, Shao CX, Goropashnaya AV, Stewart NC, Xu YC, Tøien Ø, Barnes BM, Fedorov VB, Yan J. Genomic analysis of expressed sequence tags in American black bear Ursus americanus. BMC Genomics. 2010;11:201. doi: 10.1186/1471-2164-11-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


