Abstract
Background
Breast cancer remains the leading reason of cancer death among women worldwide, and gefitinib is the efficient drug for breast cancer.
Aims
To use targeted metabolomics method to elucidate the therapeutic mechanism of gefitinib through profiling the amino acids.
Methods
Healthy women (n=56) and women with breast cancer (n=60) were enrolled in Affiliated Yuhuangding hospital, medical college of Qingdao University from 2012–2014. API 3200 triple quadrupole mass spectrometer was used to analyze the serum samples.
Results
The concentration of amino acids was compared between healthy women and women with breast cancers. Compared with the healthy women, the concentration of arginine in breast cancer women significantly decreased (p<0.0001). To show the representative capability of arginine towards the pathogenesis of breast cancers, the receiver operating characteristic (ROC) curve was drawn, and the area under the curve (AUC) was calculated to be 0.96 ± 0.02, indicating the high predictive capability of arginine for breast cancer . The reversing ability of gefitinib towards the level of arginine was further determined, and 1 month treatment of gefitinib (500 mg/day) significantly reversed the arginine level of breast cancer patients (p<0.0001)
Conclusion
The therapy of gefitinib towards breast cancer through reversing breast cancer biomarker arginine was demonstrated.
Keywords: Breast cancer, arginine, metabolomics, gefitinib
Introduction
Breast cancer remains the leading reason of cancer death among women worldwide1. There are many risk factors for breast cancers, such as obesity, lack of physical exercise, and drinking alcohol. Many drugs have been used, are being developed, or exert the therapeutic potential towards breast cancers, such as paclitaxel, doxorubicin, fluorouracil, Everolimus, and anastrozole (http://www.cancer.gov/cancertopics/druginfo/breastcancer).
Gefitinib, under the trade name Iressa, marketed by AstraZeneca and Teva, is a drug employed to treat breast cancer2. Additionally, gefitinib can also be employed to treat many other tumors, such as lung cancer. Gefitinib inhibits EGFR tyrosine kinase by binding to the adenosine triphosphate (ATP)-binding site of the enzyme. Therefore, EGFR tyrosine kinase has been widely accepted as the major target for gefitinib in breast cancer treatment. However, a drug always has multiple therapeutic targets and mechanisms, and combination of multiple target pathways can facilitate the explanation of therapeutic mechanism. Therefore, searching new therapeutic mechanism of gefitinib towards breast cancer will provide the opportunity to completely explain the therapeutic mechanism of gefitinib towards breast cancer.
Metabolomics is the scientific study of chemical processes involving small molecules metabolites3. Metabolomics study has played a key role in elucidating the mechanism of various cancers. Metabolomics study of colon cancers showed that the elevation of level of taurine, isoglutamine, choline, lactate, phenylalanine and tyrosine, and the reduction of lipids and triglycerides is closely related with the pathogenesis of colon cancer 4. For breast cancer, the alteration of histidine, acetoacetate, glycerol, pyruvate, glycoproteins (N-acetyl), mannose, glutamate and phenylalanine has been demonstrated to be the key factor5.
The present study aims to determine the therapeutic mechanism of gefitinib using targeted metabolomics.
Materials and methods
The statement of Ethics
Ethical approval for this study was granted from affiliated Yuhuangding hospital, and all enrolled women in this study gave the written informed consent.
Study Population
Healthy women (n=56) and women with breast cancer (n=60) were enrolled in Affiliated Yuhuangding hospital, medical college of Qingdao University from 2012–2014. The serum was taken before and after treatment with 500 mg/day for 1 months through centrifugation (the speed 8,000 g) of blood.
Targeted metabolomics analysis of amino acids components
The aliquots (10 uL) of serum were dropped on the filter paper, and then filter paper was placed into 96-well polypropylene micro titer plates. 100 µL of methanol solution containing isotope labeled amino acid internal standards (NSK-A, NSK-B, Cambridge Isotope Laboratories, Andover, USA) was employed to extract the samples. Gentle shaking was performed at room temperature for 30 min. After evaporation at 70°C for 40 min, 60 µL of 3n butanolic-HCL was added for derivatisation at 65°C. After a second evaporation step the samples were reconstituted with 150 µL of the mobile phase (1/1 v/v isopropanol/water) and analyzed by flow injection analysis (FIA)-MS/MS using a API 3200 triple quadrupole mass spectrometer (AB SCIEX, Darmstadt, Germany). The mobile phase is 70/30 (v/v) acetonitrile/water, and changed flow rate was used as followed: 0.01–0.1 min, 0.26 mL/min; 0.11–0.45 min, 0.02 mL/min; 1–1.2 min, 0.1–0.6 mL/min; 1.2–1.4 min, 0.6 mL/min; 1.41–2 min, 0.26 mL/min. The following mass conditions were used: Curtain gas, 20 psi; Collision gas, 8 psi; Ionspray voltage, 4500 V; Temperature, 350°C; Ion source gas (GS) 1 and 2, 35 psi. Neutral loss (NL) of 102.1 Da and precursor ion monitoring of 85.1 Da were used to monitoring the amino acids and their corresponding internal standard.
Statistical analysis
The data was given using Prism software (version 4.0; GraphPad). Statistical difference was performed using one student two-tailed t-test.
Results
The concentration of amino acids was firstly compared between healthy women and women with breast cancers. As shown in Figure 1, the concentration of arginine was 14.7 ± 0.9 uM.
Figure 1.
Arginine significantly decreased in pre-treatment patients with breast cancer. The concentration of arginine in health volunteers (n=56) and patients (n=60) was given in Figure. ****, p<0.0001.
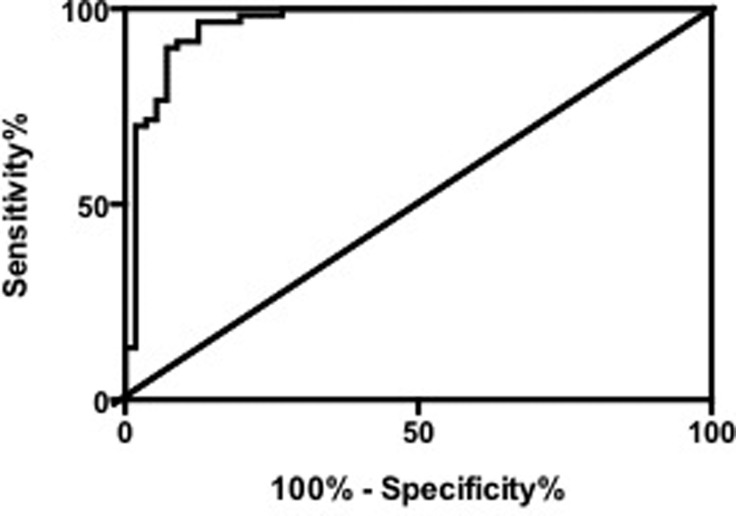
Compared with the healthy women, the concentration of arginine in breast cancer women significantly decreased (value= 5.4 ± 0.3, p<0.0001). To show the representative capability of arginine towards the pathogenesis of breast cancers, the receiver operating characteristic (ROC) curve was drawn, and the area under the curve (AUC) was calculated to be 0.96 ± 0.02, indicating the high predictive capability of arginine for breast cancer (Figure 2)
Figure 2.
Receiver operating characteristic (ROC) curve to demonstrate the sensitivity and specificity for Arginine as the prediction biomarker.
The reversing ability of gefitinib towards the level of arginine was further determined, and 1 month treatment of gefitinib (500 mg/day) significantly reversed the arginine level of breast cancer patients (p<0.0001) (Figure 3). Other amino acids did not changed among these three groups (data not shown).
Figure 3.
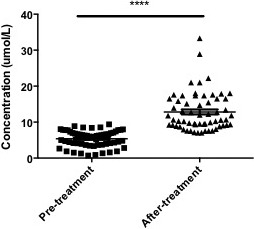
Comparison of arginine level in serum from pre-treatment and after-treatment patients (n=60). ****, p<0.0001.
Discussion
Amino acids are organic compounds that combine to form proteins, and amino acids and proteins are the building blocks of life. Additionally, amino acids are important energy source for the body6. The present study showed that arginine significantly decreased in breast cancer patients in comparison with healthy volunteers. The decrease of arginine in serum indicated possible elevation of arginine in tumor tissues. Previous literature showed that arginine starvation could protect breast cancer cells through impairing mitochondrial respiratory function7. Therefore, gefitinib-induced elevation of arginine in serum for gefitinib-treated patients might indicate the possible therapy role through decreasing the arginine level in tumor tissues.
It should be noted previous literature showed that tryptophan is the key native marker in cells to determine the level of metastasis competence in breast cell lines using native fluorescence spectroscopy8. However, the present study did not detect the alteration of tryptophan in breast cancers, and the reversing effect of gefitinib towards tryptophan. The reason might be the number of patients is not enough, and maybe the in vitro results are not easily translated into in vivo situation.
In conclusion, the therapy of Gefitinib towards breast cancer through reversing breast cancer biomarker arginine was demonstrated.
References
- 1.Sandholm J, Selander KS. Toll-like receptor 9 in breast cancer. Front Immunol. 2014;5:330. doi: 10.3389/fimmu.2014.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalykaki A, Agelaki S, Kallergi G, Xyrafas A, Mavroudis D, Georgoulias V. Elimination of EGFR-expressing circulating tumor cells in patients with metastatic breast cancer treated with gefitinib. Cancer Chemother Pharmacol. 2014;73(4):685–693. doi: 10.1007/s00280-014-2387-y. [DOI] [PubMed] [Google Scholar]
- 3.Fang ZZ, Gonzalez FJ. LC-MS-based metabolomics: an update. Arch Toxicol. 2014;88(8):1491–1502. doi: 10.1007/s00204-014-1234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jimenez B, Mirnezami R, Kinross J, Cloarec O, Keun HC, Holmes E, et al. 1H HR-MAS NMR spectroscopy of tumor-induced local metabolic “field-effects” enables colorectal cancer staging and prognostication. J Proteome Res. 2013;12(2):959–968. doi: 10.1021/pr3010106. [DOI] [PubMed] [Google Scholar]
- 5.Bezabeh T, Ijare OB, Nikulin AE, Somorjai RL, Smith IC. MRS-based Metabolomics in Cancer Research. Magn Reson Insights. 2014;7:1–14. doi: 10.4137/MRI.S13755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou J, Yan W, Hu G, Shen B. Amino Acid network for the discrimination of native protein structures from decoys. Curr Protein Pept Sci. 2014;15(6):522–528. doi: 10.2174/1389203715666140724084709. [DOI] [PubMed] [Google Scholar]
- 7.Qiu F, Chen YR, Liu X, Chu CY, Shen LJ, Xu J, et al. Arginine starvation impairs mitochondrial respiratory function in ASS1-deficient breast cancer cells. Sci Signal. 2014;7(319):ra31. doi: 10.1126/scisignal.2004761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, Pu Y, Xue J, Pratavieira S, Xu B, Achilefu S, et al. Tryptophan as the fingerprint for distinguishing aggressiveness among breast cancer cell lines using native fluorescence spectroscopy. J Biomed Opt. 2014;19(3):37005. doi: 10.1117/1.JBO.19.3.037005. [DOI] [PubMed] [Google Scholar]