Abstract
Background
Gomisin G, isolated from herb Schisandra chinensis, exhibits anti-tumor activities. Therefore, Gomisin G is a drug candidate for anti-liver cancer therapy.
Aims
To predict the metabolic behavior and metabolism-based drug-drug interaction of gomisin G.
Methods
Molecular docking method was used. The crystal structure of CYP3A4 with the ligand ketoconazole was chosen from protein data bank (http://www.rcsb.org/pdb). Chemdraw software was used to draw the two-dimensional structure of gomisin G with standard bond lengths and angles.
Results
Gomisin G can be well docked into the activity site of CYP3A4, and distance between gomisin G the heme active site was 2.75 Å. To evaluate whether the inhibitors of CYP3A4 can affect the metabolism of gomisin G, co-docking of gomisin G and ketoconazole was further performed. The distance between ketoconazole and activity center (2.10 Å) is closer than the distance between gomisin G and activity center of CYP3A4, indicating the easy influence of CYP3A4's strong inhibitor towards the metabolism of gomisin G.
Conclusion
Gomisin G is a good substrate of CYP3A4, and CYP3A4 inhibitors easily affect the metabolism of Gomisin G.
Keywords: Gomisin G, CYP3A4, molecular docking
Introduction
The liver plays an important role in filtering blood that circulates through the body. It can perform catalytic biotransformation process of nutrients and drugs into the ready-to-use chemicals. It can be affected by primary liver cancer, and by cancer which forms in other parts of the body and then spreads to the liver1. Searching efficient therapeutic drugs for liver cancers is very important and necessary.
Schisandra chinensis, also named wuweizi in Chinese, has wide application in clinic, including anti-tumor effects. Many efficient anti-tumor components have been isolated from Schisandra chinensis. For example, the lignans isolated from Schisandra chinensis showed anti-proliferative activity in human colorectal carcinoma2.
Schisandra chinensis polysaccharide exerts antitumor and antiangiogenic activity towards renal cell carcinoma model3. Schizandrin has been reported to exhibit anti-tumor activity4. Lignan component gomisin G is an important ingredient isolated from Schisandra chinensis, and is a potent drug candidate for treatment of liver cancer.
Lignan components have been reported to be good substrates of drug-metabolizing enzymes (DMEs). For example, drug-metabolizing enzyme cytochrome P450 3A catalyzes the biotransformation of major lignan component schizandrin4. Therefore, the potential drugdrug interaction between gomisin G and the inhibitor of CYP3A ketoconazole was predicted using molecular docking in the present study.
Materials and methods
The source of the crystal structure of CYP3A4 and molecular structure of gomisin G
Preparation of suitable crystal structure of protein and chemical structure of compound is the first key step for molecular docking. In the present study, the crystal structure of CYP3A4 with the ligand ketoconazole was chosen from protein data bank (http://www.rcsb.org/pdb). The structure was processed using the protein preparation wizard in the Schrödinger suite of programs, and the missing residues in the middle of the chain were added, and hydrogen atoms were assigned. Chemdraw software was used to draw the two-dimensional structure of gomisin G with standard bond lengths and angles.
Docking process
The gomisin G ligand docking and CYP450 3A4 protein-ligand complex studies were performed with Tripos molecular modeling packages according to previous literature5,6. Firstly, the three-dimensional structure of the gomisin G molecules was built and optimized by using the Tripos force field. The receptor-ligand binding geometry was optimized by using a flexible docking method with the Tripos FlexiDock program.
In this docking simulation, a CYP3A4 binding pocket was first defined to cover all residues within 4Å of the ligand in the initial CYP3A4-ketoconazole complex. During flexible docking by the FlexiDock module, all of the single bonds of residue side chains inside the defined 3A4 receptor binding pocket were regarded as rotatable or flexible bonds, and the ligand was allowed to rotate on all single bonds and move flexibly within the tentative binding pocket. The atomic charges were recalculated by using the Gasteiger-Huckel approach for the ligand. H-bonding site was marked for suitable atoms. The binding interaction energy was calculated to include van der Waals, electrostatic, and torsional energy terms defined in the Tripos force field. The structure optimization was performed for 20000-generations, using a Genetic Algorithm, and the 20 best-scoring ligand-protein complexes were kept for further analysis.
The Flexidock simulation indicated that the obtained 20 best scoring gomisin G-3A4 complex models have very similar 3D structures with little different energies.
Results
The inhibitor ketoconazole was first extracted from the activity cavity of CYP3A4, and then the structure of gomisin G was docked into the activity cavity of CYP3A4.
As shown in Figure 1, gomisin G can be well docked into the activity site of CYP3A4, and distance between gomisin G the heme active site was 2.75 Å. To evaluate whether the inhibitors of CYP3A4 can affect the metabolism of gomisin G, co-docking of gomisin G and ketoconazole was further performed.
Fig. 1.
Molecular docking of gomisin G into the activity cavity of CYP3A4. The crystal structure of CYP3A4 with ketoconazole in the activity cavity (PDB code 2V0M) was selected from protein data bank (http://www.rcsb.org/pdb). The structure of ketoconazole was firstly extracted from the cavity of CYP3A4 before the docking of gomisin G into the activity cavity.
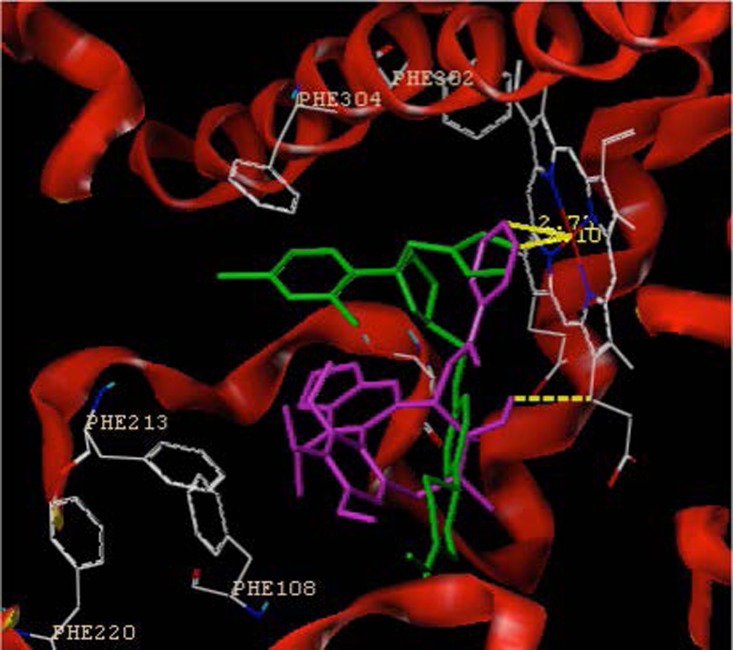
The distance between ketoconazole and activity center (2.10 Å) is closer than the distance between gomisin G and activity center of CYP3A4 (Figure 2), indicating the easy influence of CYP3A4's strong inhibitor towards the metabolism of gomisin G.
Fig. 2.
Co-docking of both gomisin G and ketoconazole into the activity cavity of CYP3A4. The green color represents the structure of ketoconazole, and the purple color represents the structure of gomisin G.
Discussion
The investigation of metabolic behavior and metabolism-based drug-drug interaction plays an important role in the R&D of new chemical entities towards new drugs. The development of many new chemical entities with poor metabolic properties was limited. For example, the anti-tumor drug candidate noscapine has been demonstrated to exhibit inhibition towards drug-metabolizing enzyme cytochrome P450 (CYP) 3A4 and 2C9, which strongly limited the R&D of noscapine7. Additionally, CYP3A4 and CYP2C9-catalyzed metabolic activation of noscapine also increase the risk of noscapine, limiting the development of noscapine8,9.
Molecular docking, predicting the preferred orientation of one molecule into the second one, is frequently used to predict the binding orientation of small drug candidates to their protein targets. Therefore, this method is suitable for the predicting the metabolic behavior through docking the compounds into the activity cavity of drug-metabolizing enzymes. For example, Kobayashi et al. used molecular docking method to determine the influence of single nucleotide polymorphisms in CYP2B6 on substrate recognition10. Liu et al. used molecular docking to deeply understand the metabolic behavior of GNF-351 by CYP3A4 and its potential drugdrug interaction with ketoconazole11.
The present study aims to predict the interaction between gomisin G and CYP3A4, and the well docking of gomisin G into the activity cavity of CYP3A4 indicated the good substrate of gomisin G for CYP3A4. Additionally, the inhibitors of CYP3A4 easily induce drugdrug interaction with Gomisin G through inhibiting the metabolism of gomisin G.
References
- 1.Szabo G, Saha B, Bukong TN. Alcohol and HCV: Implications for Liver Cancer. Adv Exp Med Biol. 2015;815:197–216. doi: 10.1007/978-3-319-09614-8_12. [DOI] [PubMed] [Google Scholar]
- 2.Gnabre J, Unlu I, Chang TC, Lisseck P, Bourne B, Scolnik R, Jacobsen NE, Bates R, Huang RC. Isolation of lignans from Schisandra chinensis with anti-proliferative activity in human colorectal carcinoma: structure-activity relationships. J Chromatogr B Analyt Technol Biomed Life Sci. 2010;878(28):2693–2700. doi: 10.1016/j.jchromb.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Qu HM, Liu SJ, Zhang CY. Antitumor and antiangiogenic activity of Schisandra chinensis polysaccharide in a renal cell carcinoma model. Int J Biol Macromol. 2014 doi: 10.1016/j.ijbiomac.2014.02.025. [DOI] [PubMed] [Google Scholar]
- 4.Cao YF, Zhang YY, Li J, Ge GB, Hu D, Liu HX, Huang T, Wang YC, Fang ZZ, Sun DX, Huo H, Yin J, Yang L. CYP3A catalyses schizandrin biotransformation in human, minipig, and rat liver microsomes. Xenobiotica. 2010;40(1):38–47. doi: 10.3109/00498250903366052. [DOI] [PubMed] [Google Scholar]
- 5.Jiang F, Jiang AF, Zhang LG, Li YZ, Yu SE. Molecular docking to predict the metabolic site of corynoline and the possible drug-drug interaction. Lat Am J Pharm. 2013;32(10):1581–1583. [Google Scholar]
- 6.Jiang J, Liu L, Wang W, Dong N, Fu X, Pang C, Yang H, Lv J, Ding L, Sun X. Metabolic behavior prediction of anti-glioma drug gefitinib by molecular docking. Lat Am J Pharm. 2014;33(7):1213–1215. [Google Scholar]
- 7.Fang ZZ, Zhang YY, Ge GB, Huo H, Liang SC, Yang L. Time-dependent inhibition (TDI) of CYP3A4 and CYP2C9 by noscapine potentially explains clinical noscapine-warfarin interaction. Br J Clin Pharmacol. 2010;69(2):193–199. doi: 10.1111/j.1365-2125.2009.03572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang ZZ, Krausz KW, Li F, Cheng J, Tanaka N, Gonzalez FJ. Metabolic map and bioactivation of the anti- tumor drug noscapine. Br J Pharmacol. 2012;167(6):1271–1286. doi: 10.1111/j.1476-5381.2012.02067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang ZZ, Gonzalez FJ. LC-MS-based metabolomics: an update. Arch Toxicol. 2014;88(8):1491–1502. doi: 10.1007/s00204-014-1234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi K, Takahashi O, Hiratsuka M, Yamaotsu N, Hirono S, Watanabe Y, Oda A. Evaluation of influence of single nucleotide polymorphisms in cytochrome P450 2B6 on substrate recognition using computational docking and molecular dynamics simulation. PLoS One. 2014;9(5):e96789. doi: 10.1371/journal.pone.0096789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu T, Qian G, Wang W, Zhang Y. Molecular docking to understand the metabolic behavior of GNF-351 by CYP3A4 and its potential drug-drug interaction with ketoconazole. Eur J Drug Metab Pharmacokinet. 2014 doi: 10.1007/s13318-014-0201-1. [DOI] [PubMed] [Google Scholar]