Abstract
Proteinaceous components of the biofilm matrix include secreted extracellular proteins, cell surface adhesins and protein subunits of cell appendages such as flagella and pili. Biofilm matrix proteins play diverse roles in biofilm formation and dissolution. They are involved in attaching cells to surfaces, stabilizing the biofilm matrix via interactions with exopolysaccharide and nucleic acid components, developing three-dimensional biofilm architectures, and dissolving biofilm matrix via enzymatic degradation of polysaccharides, proteins, and nucleic acids. In this chapter, we will review functions of matrix proteins in a selected set of microorganisms, studies of the matrix proteomes of Vibrio cholerae and Pseudomonas aeruginosa, and roles of outer membrane vesicles and of nucleoid-binding proteins in biofilm formation.
Introduction
Microorganisms in the natural environment typically live on or in close association with surfaces and predominantly exist as biofilms, surface attached microbial communities composed of cells and extracellular matrix (1–4). The exact compositions of biofilm matrixes differ greatly between different microorganisms and growth conditions under which biofilms are formed, but generally consist of exopolysaccharides, proteins, and nucleic acids. Proteinaceous components include cell surface adhesins, protein subunits of flagella and pili, secreted extracellular proteins, and proteins of outer membrane vesicles.
Cell surface proteins, pili and flagella participate in initial attachment to surfaces and, in some microorganisms, are also involved in migration along the surfaces, thereby facilitating surface colonization. Matrix proteins contribute to biofilm structure and stability. Such proteins were identified mostly by mutational studies, which showed that the absence of matrix proteins results in reduced biofilm formation and stability, and altered biofilm architectures (5–14). Structural analysis and biofilm localization studies have provided further insights into the functions and mechanisms of action of matrix proteins. Some matrix proteins exhibit enzymatic properties towards matrix components, such as the glycosyl hydrolase dispersin B that hydrolyzes polysaccharides (15); proteases that target matrix proteins (16); and DNases that degrade extracellular nucleic acids (17, 18), thus facilitating either biofilm matrix reorganization or biofilm matrix degradation and dispersal.
Several studies have been carried out to identify the matrix proteome of several microorganisms, including Vibrio cholerae (19), Pseudomonas aeruginosa (20), Myxococcus xanthus (21) and natural biofilm communities of acid mine drainage (22). These studies revealed that, in addition to secreted proteins, the biofilm matrix also contains large numbers of periplasmic, cytoplasmic, inner, and outer membrane proteins. These results implicate the involvement of cell lysis and/or outer membrane vesicles (OMVs) in modulating biofilm proteome composition.
In this chapter, we focus on the matrix proteins that play structural roles in the biofilm formation. We will discuss the functions and mechanisms of action of matrix proteins and lectins produced by V. cholerae and P. aeruginosa, the biofilm-associated proteins from Staphylococcus aureus and the hydrophobin from Bacillus subtilis in biofilm formation. Finally, we will review matrix proteome studies of V. cholerae and P. aeruginosa and the roles of OMVs and nucleoid-binding proteins in biofilm formation.
V. cholerae Matrix Proteins
V. cholerae is a facultative human pathogen that colonizes the human intestine and survives for extended periods in natural aquatic environments. Both pathogenesis and environmental survival are closely linked to the microbe’s ability to form biofilms. Mature biofilm formation in V. cholerae depends on the production of Vibrio exopolysaccharides (VPS) (23, 24). V. cholerae produces two different types of VPS. The repeating unit of the major variant consists of -4)-α-GulNAcAGly3OAc-(1-4)-β-D-Glc-(1-4)-α-Glc-(1-4)-α-D-Gal-(1-. In the minor variant, α-D-Glc is replaced with α-D-GlcNAc (25). Three major biofilm matrix proteins (RbmA, Bap1 and RbmC) (5, 6) are important for biofilm formation on abiotic surfaces, and the extracellular chitin-binding protein GbpA mediates attachment to chitinous surfaces of zooplankton (26). The structure, function, and mechanistic roles of these matrix proteins in V. cholerae surface adhesion and biofilm formation are reviewed below.
Rugosity and biofilm structure modulator A (RbmA)
RbmA is a 26-kDa matrix protein involved in facilitating intercellular adhesion during biofilm formation (5, 27). Studies carried out using a rugose variant of V. cholerae, which exhibits enhanced biofilm formation and colony corrugation due to increased production of VPS and matrix proteins, revealed that an rbmA mutant exhibits a decrease in colony corrugation (Figure 1), forms a biofilm with altered biofilm architectures and disperses easily by shear force (5). Similarly, pellicles, which are biofilms formed at the air-liquid interface, formed by the rbmA mutant are less wrinkled and more fragile, and disintegrate upon force (5). Addition of exogenous purified RbmA rescues the altered pellicle phenotype of an rbmA mutant strain (19), indicating that extracellular provision of RbmA enhances intercellular interactions. Taken together, these studies point out the importance of RbmA in development of mature biofilm architecture and in stabilization of biofilms.
Figure 1.
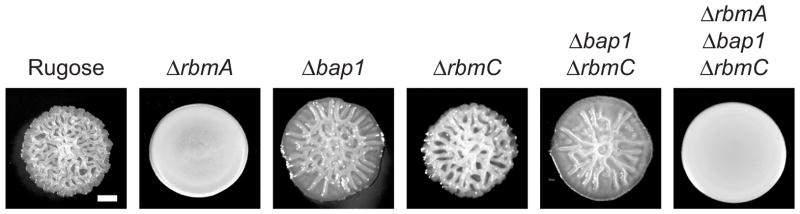
Colony morphology of V. cholerae rugose variant and mutant strains unable to produce RbmA, RbmC and Bap1 matrix proteins. Bar = 0.5mm.
The crystal structure of RbmA revealed that it consists of two tandem fibronectin type III (FnIII) domains and functions as a 49-kDa dimer (28). The approximately 100 aa FnIII domain is found widely in many proteins, including eukaryotic cell surface receptors and prokaryotic carbohydrate-binding proteins (29), suggesting a possible role of RbmA in binding carbohydrates and in cell adhesion. The two tandem FnIII domains (Figure 2) are not identical in peptide sequence, but share 24% identity and 44% similarity (30). The FnIII domains of RbmA fold as a seven-strand β-sandwich, with the N-terminal of the FnIII domain of one monomer interacting tightly with a second monomer of the asymmetric unit (28). The crystal structure of RbmA also revealed a positively-charged groove formed by the two adjacent FnIII domains (28). Three arginine residues (R116, R219 and R234) located within this groove, which are predicted to be involved in ligand binding, were found to be critical for RbmA function. Strains that produce mutated versions of RbmA, containing point mutations in these positively-charged residues, exhibit a decrease in colony corrugation and/or pellicle formation when compared to the parental strain (28). RbmA also contains a negatively-charged groove, formed between the two FnIII domains of the same monomer (28). However, site-directed mutagenesis resulting in either removing (E84A) or reversing (E84R) the negative charges did not affect RbmA function, suggesting that this negatively-charged groove does not play a major role in RbmA-mediated biofilm formation under the conditions studied (28). The biological role of the negatively-charged groove remains to be determined.
Figure 2.
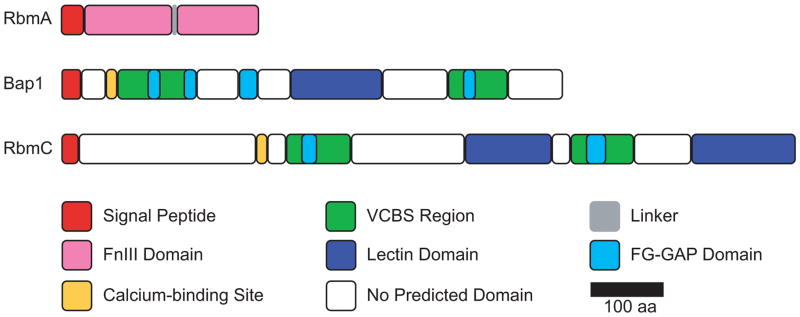
Domain organization of V. cholerae RbmA, Bap1 and RbmC. FnIII, fibronectin type III; VCBS, Vibrio-Colwellia-Bradyrhizobium-Shewanella repeat; FG-GAP, phenyl-alanyl-glycyl (FG) and glycyl-alanyl-prolyl (GAP).
Possible ligands of RbmA were identified by glycan array studies (30). RbmA exhibits saccharide-binding specificity and multivalency towards sialic acid and fucose, and has a lower binding preference to galactose, rhamnose, and N-acetylgalactosamine (GalNAc) (30). The ability of RbmA to bind to galactose, a component of VPS (23), suggests that RbmA-mediated biofilm formation may be, in part, due to RbmA-VPS interactions. The significance of RbmA binding to other sugars remains to be determined. It has also been speculated that RbmA may bind the lipopolysaccharide (LPS) found on the cell surface via interaction with sialic acid derivatives (30). Therefore, the positively-charged groove may interact with negatively-charged ligands such as carbohydrates on bacterial cell surfaces (30), VPS (28), and/or LPS (30). The unique flexibility of the FnIII domains (31, 32), together with the predicted interactions of RbmA with VPS, LPS and/or cell surfaces, support a model where RbmA can act as an elastic scaffold in the biofilm matrix. RbmA may mediate flexible contacts with other matrix components and bacterial cell surfaces, thus increasing shear resistance and integrity of the biofilm matrix.
RbmA is secreted and contains a secretion signal sequence in its N-terminal region (Figure 2) (5), indicating involvement of the general secretion (Sec) pathway in its secretion. However, the mechanism of RbmA secretion outside of the cell is currently unknown. Localization studies revealed that RbmA is distributed throughout the biofilm (19, 27), specifically surrounding the bacterial cells (27), suggesting that RbmA facilitates cell-cell adhesion. In fact, RbmA was detected on cell surfaces 30 minutes after surface attachment and aids in the retention of newly divided daughter cells (27). Furthermore, retention of RbmA on cell surfaces is dependent on the presence of VPS. This indicates that interaction with VPS is essential for RbmA spatial distribution within the biofilm matrix and further reinforces the notion that RbmA binds VPS. The spatial and temporal localization of RbmA, as well as the dependency on VPS for retention on cell surfaces, strongly supports the role of RbmA as a scaffold that mediates cell-cell and cell-matrix interactions.
Biofilm-associated protein 1 (Bap1) and rugosity and biofilm structure modulator C (RbmC)
The 75-kDa Bap1 and 104-kDa RbmC matrix proteins share 47% peptide sequence similarity and are involved in biofilm formation in V. cholerae (6, 33). Despite their sequence similarity, Bap1 and RbmC affect biofilm formation differently. While lack of either protein affects colony corrugation (Figure 1), pellicle formation and biofilm formation; the defects differ in magnitude (6). A mutant unable to produce both RbmC and Bap1 exhibits marked decreases in biofilm formation and the ability to stably attach to surfaces. Complementation analysis of rbmC bap1 double mutant showed that Bap1 and RbmC can partially complement each other, but they are not functionally redundant (6, 27).
Sequence similarity database (SSDB) and Pfam motif searches, as well as sequence alignment analysis, showed that Bap1 and RbmC contain four overlapping Vibrio-Colwellia-Bradyrhizobium-Shewanella repeat (VCBS) domains that form two VCBS regions (Figure 2). In addition, Bap1 contains four FG-GAP domains, while RbmC contains two FG-GAP domains. Although the VCBS domain (PF13517) is commonly found in multiple copies in proteins from several species of Vibrio, Colwellia, Bradyrhizobium, and Shewanella, very little is known about its function, except that it may be involved in cell adhesion. FG-GAP repeats (PF01839) are found in the N-terminal region of the eukaryotic integrin α-chain, which is important for ligand recognition and binding to the extracellular matrix or cell surface proteins (34–36). NCBI conserved domain searches also revealed that Bap1 and RbmC contain one and two Jacalin-like lectin domains, respectively. The Jacalin-like lectin domains (PF01419), or β-prism domains, have binding specificities to galactose, mannose and/or glucose and their derivatives (37, 38). Since VPS contains these sugars (23), it is possible that Bap1 and RbmC may bind to the galactose, mannose and/or glucose residues of VPS. The presence of VCBS, FG-GAP and lectin domains suggests that Bap1 and RbmC likely mediate biofilm formation by facilitating adhesion and carbohydrate binding. Both Bap1 and RbmC contain a predicted EF-hand calcium-binding motif (Figure 2), indicative of calcium binding. However, it remains to be determined whether these predicted sites bind calcium and if binding to calcium regulates protein function.
Both Bap1 and RbmC contain predicted secretion signal sequences at the N-terminal regions (Figure 2), indicating that they are secreted from the cytoplasm via the Sec-dependent pathway. V. cholerae type II secretion system (T2SS) proteome analysis showed that RbmC is secreted by the T2SS (39), while the mechanism of secretion of Bap1 is currently unknown. Biofilm localization studies revealed that RbmC and Bap1 form envelopes around microcolonies/cell clusters, and that Bap1, but not RbmC, localizes to the biofilm-surface interface (27). The localization of Bap1 and RbmC within the biofilm corroborates their functions and the biofilm phenotypes observed. While the presence of both Bap1 and RbmC in the cell envelope likely allows partial functional redundancy, the additional role of Bap1 in anchoring developing biofilms onto surfaces cannot be fulfilled by RbmC. It is noteworthy that the specific spatial localization of Bap1 and RbmC within the biofilm is not due to localized transcription of bap1 and rbmC within the cell population (19). The mechanisms by which Bap1 and RbmC are targeted to their specific location during biofilm formation are currently unknown. Retention of Bap1 and RbmC on cell surfaces is dependent on the presence of VPS, while the localization of Bap1 at the biofilm-substratum interface is VPS-independent (27). These observations indicate that Bap1 and RbmC could bind VPS, and these interactions may influence the spatial localization of these matrix proteins. The second lectin domain at the C-terminal region of RbmC has been reported to be non-essential for protein function, as a bap1 rbmC double mutant strain, harboring a rbmC allele with a truncation of the C-terminal lectin domain, is able to form biofilms (19). However, it is yet to be determined if the complemented strain can form biofilms with wild-type architectures.
It has been reported that Bap1 could associate with outer membrane vesicles (OMVs) by binding to OmpT (40). Bap1-OmpT interaction requires the presence of the integrin-binding domain (leucine-aspartic acid-valine LDV peptide) of OmpT and the FG-GAP domains of Bap1 (Figure 2). It was also reported that RbmC does not interact with OmpT in the OMVs, although it is currently unclear why Bap1, but not RbmC, binds to OmpT. It is possible that Bap1 may exhibit higher binding affinity to OmpT as Bap1 contains four FG-GAP domains, while RbmC only contains two FG-GAP domains. Although it is yet to be determined if OMVs are part of the V. cholerae biofilm matrix, this observation highlights possible bifunctional roles of Bap1 in biofilm formation: facilitating cell attachment to surfaces and interactions among different biofilm components, i.e. VPS and OMVs. While the localization of Bap1 and RbmC in the biofilm matrix and their involvement in biofilm formation have been demonstrated, the importance of the various predicted adhesion and carbohydrate-binding domains is unknown. Further studies in determining the roles of these domains and critical residues for protein function will help gain better insights into the mechanistic functions of these matrix proteins.
V. cholerae GlcNAc-binding protein A (GbpA)
In the natural aquatic environment, V. cholerae adheres to chitinous surfaces of phytoplankton and zooplankton, including exoskeletons of copepods, and colonization of these surfaces enhances V. cholerae survival in the rapidly changing environment (41, 42). Chitin is a polymer of N-acetyl-D-glucosamine (GlcNAc) and is one of the most abundant polysaccharides in nature that can be used as a carbon and energy source by V. cholerae (43). The V. cholerae GbpA is a chitin-binding protein (26, 44) predicted to be critical for V. cholerae environmental survival by facilitating adhesion to chitinous surfaces. Indeed, gbpA mutant strains exhibited decreased attachment to zooplankton chitinous exoskeletons and egg sacs (26, 45). In addition, GbpA-deficient mutants exhibited reduced adherence to chitin and GlcNAc-coated beads (26, 44). Similarly, GbpA-chitin interaction was also observed in GbpA-containing cell lysates and the interaction was abolished upon the addition of GlcNAc in a concentration-dependent manner, indicating that GbpA specifically binds to the chitin monomer GlcNAc (26). Although GbpA mediates adhesion to chitinous surfaces, it was reported that GbpA does not play a role in attachment and biofilm formation on abiotic surfaces. A gbpA mutant strain exhibited wild-type levels of attachment efficiency on inorganic quartz, quartzite and calcium carbonate (marine sediment) surfaces (45) and was able to form biofilms on polyvinyl chloride surfaces (46), suggesting that the main role of GbpA is adhesion to GlcNAc/chitin-containing biotic surfaces. Furthermore, GbpA has also been reported to bind to intestinal mucin, which also contains GlcNAc (47). As such, GbpA functions as a virulence factor (26, 47, 48) and an adhesin in mediating intestinal attachment and adhesion on chitinous surfaces.
Crystal structures of GbpA revealed that it is an unusual, elongated protein. It consists of four domains (D1 to D4) that do not interact, and are completely surface-exposed (48). The two terminal domains D1 and D4 exhibit structural similarity to chitin-binding protein 21 (CBP21) and sequence similarity to the C-terminal chitin-binding domain of chitinase B (ChiB) from Serratia marcescens, respectively (48). Domains D1 and D4 were shown to bind chitin in glycan array binding assays. However, in contrast to previous reports that showed binding of GbpA to GlcNAc using V. cholerae cells and cell lysates (26, 44), the purified full-length and various truncated versions of GbpA did not exhibit GlcNAc binding ability (48), suggesting that additional V. cholerae factors may be required for GbpA-GlcNAc interaction. Domains D2 and D3 are essential for cell-surface interactions (48). Domain D2 shows distant structural similarity to the β-domain of protein p5 of Sphingomonas sp. A1 (48), which is predicted to be involved in cell-surface interactions (49, 50). Domain D3 exhibits an immunoglobulin fold that is associated with cell adhesion (48, 51, 52). Furthermore, D1 is essential for mucin binding and D1 to D3 are required for intestinal colonization in infant mice (48). GbpA contains an N-terminal secretion signal sequence and has been reported to be secreted by the type II secretion system (26, 39). It appears that the unique elongated structure of GbpA results in modular binding, in which the two terminal domains D1 and D4 bind to chitin, while the two central domains D2 and D3 interact with cell surfaces. Although the binding targets of each domain have been elucidated, the essential residues for their specific binding are still unknown; such studies would provide mechanistic insights into GbpA-mediated adhesion.
Matrix proteome of V. cholerae
The proteome of the V. cholerae biofilm matrix from biofilms grown in nutrient broth under static biofilm conditions has been reported (19). A total of 74 proteins with predicted extracytoplasmic localization were identified. These include known biofilm matrix proteins (RbmA and RbmC), several outer membrane proteins (OmpA, OmpU, OmpT, OmH, OmpK, OmpW, OmpS, and TolC), periplasmic proteins, flagellar proteins, mannose-sensitive hemagglutinin (MSHA) pili proteins, enzymes (hemagglutinin/protease HapA and chitinase ChiA-2), and the hemolysin HlyA. Protein localization studies, using immunofluorescence microscopy, further confirmed biofilm matrix localization of RbmA, Bap1, MshA, and HlyA (19). The cell-associated MSHA type IV pili, which facilitates attachment to abiotic and chitinous biotic surfaces (53, 54), and the matrix proteins RbmA and Bap1, containing adhesion and carbohydrate-binding lectin domains, are expected to be retained within the biofilm matrix. HlyA contains a Jacalin-like lectin domain, suggesting that HlyA may be binding to the biofilm matrix via the lectin domain. The role of outer membrane proteins (OMPs) in biofilm formation is yet to be determined. Several hypothetical proteins were also identified in the study, and determining their functions may lead to the identification of new biofilm matrix proteins of V. cholerae.
P. aeruginosa Matrix Proteins
P. aeruginosa is an opportunistic human pathogen capable of causing diverse infections in humans. The capacity of P. aeruginosa to form biofilms on various surfaces in the human body is critical for its pathogenicity. The P. aeruginosa biofilm matrix consists of exopolysaccharides (55–57), extracellular DNA (eDNA) (58, 59) and proteins (7, 8, 14, 60). In P. aeruginosa, three exopolysaccharides (alginate, Pel, and Psl) have been shown to be involved in biofilm formation in a strain-specific manner. Alginate is a high molecular weight acetylated exopolysaccharide that consists of uronic acids (mannuronic and guluronic acids) (55, 61) and is not essential for biofilm formation, as shown in the clinical alginate-overproducing mucoid strain (FRD1) and the laboratory non-mucoid (PAO1 and PA14) strains (62–64). However, overproduction of alginate affects the development of mature biofilm architectures (62, 64, 65). Pel is a glucose-rich polysaccharide and is required for pellicle and biofilm formation in P. aeruginosa PA14, while Psl is a mannose- and galactose-rich polysaccharide that is critical for pellicle and biofilm formation in PAO1 (56, 57, 66–69). eDNA has also been reported to be important in the initial stages of biofilm formation. The presence of DNase I prevents biofilm formation, and addition of DNase I on pre-formed biofilms at early stages of biofilm formation leads to dissolution of the biofilm matrix (58). While a wealth of knowledge is available on the exopolysaccharides produced by different strains of P. aeruginosa, only a few P. aeruginosa biofilm matrix proteins have been described. These include the lectins LecA and LecB, and the Psl-binding matrix protein CdrA.
LecA and LecB lectins
P. aeruginosa produces two lectins, LecA and LecB (formerly known as PA-IL and PA-IIL, respectively) (70–72) that are involved in biofilm formation (7, 8, 60) and play a role during infections (73–75). Lectins are carbohydrate-binding proteins that exhibit sugar binding specificity (76, 77). LecA is required for P. aeruginosa biofilm formation on polystyrene and stainless steel surfaces. A lecA mutant of PAO1 exhibits reduced substratum coverage while a LecA-overproducing strain exhibits increased biofilm formation (7). LecA binds to galactose, N-acetyl-D-galactosamine and glucose (60, 78). It is yet to be determined if LecA binds to the galactose-rich Psl and glucose-rich Pel; such an interaction would contribute to biofilm formation. LecB is involved in biofilm formation on glass surfaces. A lecB mutant of PAO1 forms a biofilm with reduced thickness and surface coverage than that formed by wild type (8). LecB exhibits high affinity for fucose and binds to several other monosaccharaides with the following preference: L-fucose > L-galactose > D-arabinose > D-fructose > D-mannose (79). LecB may mediate biofilm formation of P. aeruginosa via interactions with the galactose and mannose residues in Psl. Orthologs of LecB have also been identified in other bacteria (80–83) but their role in biofilm formation is unknown.
Crystal structures of LecA and LecB have been reported both in the native form and in complex with their binding saccharides (79, 84–86). LecA (51 kDa) is a tetrameric protein consisting of four 12.8-kDa subunits (84, 85, 87). In the LecA structure, each monomer adopts a small jelly-roll β-sandwich fold, consisting of two curved sheets, with a calcium-dependent ligand binding site at the apex that binds one galactose ligand and one calcium ion (85). LecA also contains a secondary glucose-binding site in close proximity to the primary galactose-binding site, but the bound glucose residue does not interact with the amino acid residues of the galactose-binding site (78). LecA-mediated biofilm formation involves the galactose binding site. Addition of galactosides that have high affinities to LecA, such as IPTG (isopropyl-β-D-thiogalactoside) and NPG (p-nitrophenyl-α-D-galactoside), reduce wild-type P. aeruginosa PAO1 biofilm formation and induce dispersion in mature biofilms (7).
LecB (47 kDa) is a tetrameric protein consisting of four 11.7-kDa subunits (88). In the LecB structure, each monomer is arranged as a nine-stranded antiparallel β-sandwich (84, 86). The amino acid residues N95 to D104 form a single loop, and together with S22, S23 and G114 make up the ligand binding site in LecB that interacts with the two calcium ions. The two calcium ions not only directly interact with the ligands but are required to stabilize the N95-D104 loop, which forms the core ligand-binding site.
LecB is localized to the outer membrane (8). A mutation in the calcium binding site (D104A) abolishes binding of LecB to the outer membrane. Furthermore, treatment of outer membrane fractions with NPF (p-nitrophenyl-α-L-fucose), which has a high affinity for LecB, resulted in dissociation of LecB from the outer membrane; in addition, pre-incubation of a fluorescently-labeled LecB with L-fucose inhibited interaction of the lectin with cell surfaces (8). Collectively, these findings suggest that LecB likely interacts with a ligand on the cell surface in a calcium-dependent manner.
In a study designed to identify the cell surface ligand for LecB, direct interaction between LecB and the outer membrane protein OprF was demonstrated (89). Western blot analysis revealed that OprF was necessary for proper LecB localization, as LecB was released into the culture supernatant of the oprF mutant, instead of being retained on the outer membrane as observed in the wild-type strain. Specific LecB-OprF interaction was further demonstrated with co-purification of OprF and LecB. In the same study, a far-western blot analysis using purified LecB detected several positive protein bands from the membrane fraction of P. aeruginosa (89), suggesting that LecB may interact with other ligands on the membrane in addition to OprF. The mechanism of LecB-OprF interaction remains to be determined. It is also currently unclear if LecA interacts with any outer membrane proteins in P. aeruginosa. Although both LecA and LecB are localized in the cytosolic and outer membrane fractions in PAO1 (7, 8), they do not contain predicted signal sequence as determined by SignalP 4.1. However, using SecretomeP 2.0, both LecA and LecB are predicted to be secreted via a non-classical pathway (7). The exact mechanism of LecA and LecB secretion is currently unknown.
Cyclic diguanylate-regulated two-partner secretion partner A (CdrA)
P. aeruginosa produces a matrix protein, CdrA, which binds to the exopolysaccharide Psl and mediates biofilm formation on abiotic surfaces (14). cdrA is in an operon with cdrB, predicted to encode a putative outer membrane transporter. CdrA and CdrB are predicted to be members of the two-partner secretion (TPS) systems. Western blot analysis demonstrated that CdrA exists as a full-length cell-associated protein that can be processed into a smaller fragment, which is released into culture supernatant. The exact mechanism of this proteolytic processing is not clear. A cdrA mutant forms biofilms that are thinner and less structured than biofilms formed by wild type (14). Overproduction of CdrA leads to an increase in cell auto-aggregation in liquid cultures and CdrA-mediated auto-aggregation is dependent on Psl, but not Pel: a Psl-deficient strain is unable to auto-aggregate while a Pel-deficient strain still exhibits auto-aggregation phenotype when CdrA is overproduced. Addition of mannose reduces the auto-aggregation phenotype, suggesting that CdrA binds to the mannose residues in Psl. CdrA also exhibits multivalency as the addition of fucose, fructose, or GlcNAc also reduces the auto-aggregation phenotype. Direct binding of CdrA to Psl exopolysaccharide was demonstrated with co-immunoprecipitation of CdrA with Psl. Therefore, CdrA mediates biofilm formation and cell auto-aggregation in P. aeruginosa PAO1 by binding to the Psl exopolysaccharide, leading to either cross-linking the exopolysaccharide polymers and/or tethering Psl to cells.
The secondary structure of CdrA is predicted to be dominated mainly by β-strands, and tertiary structure prediction revealed that CdrA forms a long, rod-shape structure with a β-helix structural motif (14). CdrA contains several putative adhesion domains (14), including a carbohydrate-dependent hemagglutination activity domain, a glycine-rich sugar-binding domain, and a RGD (Arg-Gly-Asp) sequence motif that may function as an integrin recognition site (51). Although the functions of these domains and motifs remain to be tested, it is likely that CdrA mediates biofilm formation via these putative adhesion domains.
Matrix proteome of P. aeruginosa
A study designed to identify proteins associated with P. aeruginosa PAO1 biofilm matrix (matrix proteome) was recently carried out (20). Forty-five proteins were identified exclusively in the matrix proteome that were not present in the whole cell samples, including the Psl-binding CdrA and the cognate transporter CdrB. Overall, the matrix proteome of P. aeruginosa contains secreted proteins (13.3%), cytoplasmic proteins (28.9%), periplasmic proteins (11.1%), cytoplasmic membrane proteins (2.2%), and most abundantly outer membrane proteins (OMPs) (35.6%). The cellular locations of the rest of the proteins (8.9%) could not be identified with confidence. The presence of OMPs and cytoplasmic proteins in the matrix proteome could be due to cell lysis as well as the presence of outer membrane vesicles (OMVs) in the biofilm matrix. When compared to proteins identified from biofilm OMVs, 53% of predicted OMPs were found in both matrix and OMV samples, indicating that a large portion of the matrix proteins are associated with OMVs. A large 362-kDa protein, predicted to be localized in the outer membrane, was also identified that may function as a surface protein for adhesion and biofilm formation. Several enzymes were also found, although not exclusively, in the biofilm matrix of P. aeruginosa, including alkaline protease, chitinase, protease IV, and a putative magnesium-dependent DNase (20), which are likely retained in the matrix by interaction with matrix components such as exopolysaccharides, eDNA or proteins. The function of these proteins in biofilm formation is yet to be determined.
Biofilm-associated Protein (Bap)
The Bap protein family represents one of the most studied groups of matrix proteins involved in biofilm formation (Table 1). Members of the Bap family are usually very large secreted proteins of up to several hundred kDa in molecular mass. The most unique feature of these proteins is the presence of multiple repeats of identical or near-identical amino acid residues in the core region. Members of the Bap family exhibit the following unique domain features (11). They contain an N-terminal secretion signal sequence, followed by a non-repetitive N-terminal Region B, which may be absent in some Bap orthologs (9). The central region, which makes up most of the protein, consists of multiple identical or near-identical repeats that may contain amyloid-like peptide sequences (90). The number of repeats in the central region can vary in different species and isolates, resulting in different extended structures and protein variants that likely aid in the evasion of host immune responses (91, 92). In Gram-positive bacteria, the protein ends with the C-terminal region carrying an LPxTG cell-wall anchoring motif (11). Most of these Bap proteins can function both as virulence factors involved in pathogenesis and as matrix proteins mediating abiotic surface adhesion and subsequent biofilm formation. In the following section, we will discuss representative Bap proteins from Staphylococcus aureus, Staphylococcus epidermidis and Salmonella enterica.
Table 1.
Members of the Bap family.
| Microorganism1 | Strain | Protein (residues)2 | GenBank/UniProtKB | Size (kDa) | A repeats | C repeats | D repeats | Amyloid peptide3 | References |
|---|---|---|---|---|---|---|---|---|---|
| Staphylococcus aureus | V329 | Bap (2,276 aa) | Q79LN3 | 239 | 2 | 13 (2 partial) | 3 (1 partial) | 1 (STVTVTD) | (11, 90, 91) |
| Staphylococcus epidermidis | C533 | Bap (2,742 aa) | AAY28519.1 | 284 | 2 | 16 | 6 | 1 (STVTVTD) 17 (STVTVTF) |
(90, 91) |
| Staphylococcus chromogenes | C483 | Bap (1,530 aa) | Q4ZHU4 | 162 | 1 | 5 | 4 | 1 (STVTVTD) 5 (STVTVTF) |
(90, 91) |
| Staphylococcus hyicus | 12 | Bap (3,278 aa) | Q4ZHU0 | 338 | 2 | 25 | 3 | 1 (STVTVTD) 26 (STVTVTF) |
(90, 91) |
| Staphylococcus xylosus | C482 | Bap (3,271 aa) | AAY28517.1 | 337 | 0 | 24 | 9 | 23 (STVTVTF) | (91) |
| Staphylococcus simulans | ATCC1362 | Bap (1,674 aa) | Q4ZHU2 | 177 | 2 | 6 | 4 | 1 (STVTVTD) 5 (STVTVTF) |
(90, 91) |
| Salmonella enterica Typhimurium | 3938 | BapA (3,824 aa) | A9LS56 | 386 | ND | 29 | ND | 1 (STVTVTL) | (9, 90) |
Selected members of Bap family proteins.
Protein lengths are in parentheses.
Number of repetitions of the amyloid peptide within the protein were a combination of data reported by Lembre et. al. (90) and from manual scanning of the protein sequences. The amyloid peptide sequences are in parentheses.
aa – amino acids, ND – no data or no domains identified.
S. aureus Bap
Bap, a 239-kDa cell surface protein (Table 1), was first identified in a study designed to identify genes involved in Staphylococcal biofilm formation. Mutants unable to produce Bap exhibit decreased colony corrugation, reduced intercellular adhesion (cell aggregation), and impaired biofilm formation on abiotic surfaces (11, 92–94).
Bap exhibits a unique domain organization (Figure 3) containing repeats that can be grouped into four regions: A to D (11). The N-terminal region of Bap contains a putative secretion signal sequence and thus likely involves the general Sec-dependent pathway for its secretion. Following the N-terminal signal peptide is Region A, which consists of two 32 amino acids repeats (A repeats) separated by 26 amino acids (11). Region B does not contain any repeats. A dimerization domain is predicted in Region A and Region B, suggesting that Bap may form homodimers or heterodimers with other Bap orthologs (94). Formation of heterodimers of S. aureus Bap and Bap orthologs from other bacteria could facilitate mixed-species biofilm formation. The most distinctive region of Bap is the core region (Region C), which is comprised of thirteen near-identical repeats (C repeats) of 86 amino acids and two partial repeats at each end of Region C. Region D consists of three short repeats of eighteen amino acids (D repeats) in a stretch of sequence rich in serine and aspartic acid residues, and the cell-wall anchoring LPxTG motif at the C-terminal region (11). The repeats in Region C are predicted to fold as a seven-strand β-sandwich and exhibit similarity to members of HYR family that contain extracellular adhesion modules (51, 94). Thus, it is possible that the C repeats could mediate adhesion and be involved in biofilm formation. However, differences in the number of repeats in Region C do not appear to be critical for biofilm formation on abiotic surfaces or colony corrugation (92, 93): a mutant producing a shorter variant of Bap, which contains one C repeat forms biofilms and exhibits colony morphology indistinguishable from those formed by the parental strain, which produces the wild-type Bap with thirteen C repeats (93). This notion was further supported by the observation that there is no association between biofilm formation and natural bap-positive isolates containing a varying number of repeats in Region C (92). Although the number of C repeats does not appear to affect biofilm formation, the presence of a single C repeat may be sufficient to mediate adherence and biofilm formation in S. aureus. Alternatively, Region A and/or Region B may be involved in biofilm formation. In fact, the N-terminal region of Esp, a Bap ortholog in Enterococcus faecalis, which exhibits 33% sequence identity to Region B of S. aureus Bap, was found to be essential for biofilm formation (10, 95). Therefore, a more in-depth domain analysis would be critical to determine which region of Bap is required for biofilm formation on abiotic surfaces.
Figure 3.
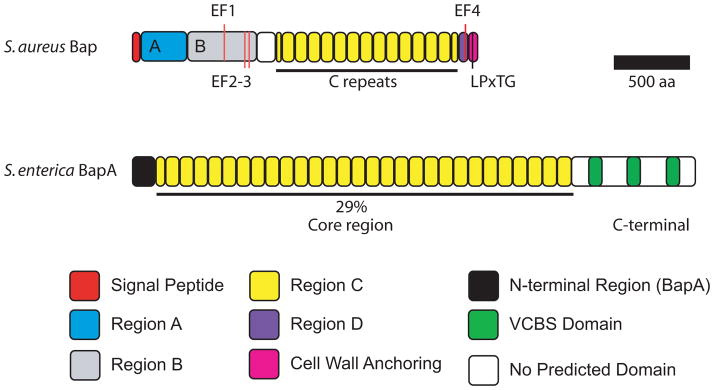
Domain organization of S. aureus Bap and S. enterica BapA. EF-hand calcium-binding motifs EF1 to 4 in Bap are indicated. LPxTG is the cell-wall anchoring motif. The repeats in the core regions of S. enterica BapA shares 29% identity with the C repeats of S. aureus Bap.
Post-translational regulation of Bap function by calcium has been reported (96, 97). Calcium inhibits Bap-mediated biofilm formation, likely through induction of a conformational change (96, 97). Bap contains four predicted Ca2+-binding EF-hand motifs (PS00018) that exhibit ≥80% similarity to the loop consensus of EF-hand motifs (96). Three are found within Region B while the fourth one is located in Region D (Figure 3). Addition of calcium in the millimolar range reduced wild-type S. aureus biofilm formation on polystyrene plates and cell aggregation in liquid cultures, while no inhibitory effect on biofilm formation was observed with natural Bap-deficient S. aureus isolates (96). In addition, calcium also decreases S. aureus biofilm thickness (97). Mutations in two of the predicted EF-hand motifs abolish the biofilm-inhibitory effect of calcium (96). Importantly, western blot analysis of surface protein samples revealed that wild-type Bap harvested from cells grown in the presence of calcium is more resistant to protease degradation than that from cells grown in the absence of calcium. This suggests that binding to calcium, while inhibiting Bap function, also renders the protein more resistant to proteolytic degradation (96), possibly due to conformational changes.
Bap orthologs in other Gram-positive and Gram-negative bacteria
Bap orthologs have also been identified in other Staphylococcus species, including S. epidermidis. A S. epidermidis mutant lacking Bap is incapable of forming wild-type biofilms, while complementation with bap in trans increases the biofilm forming capacity (91). In the Region C of Bap proteins, amyloid-like peptide sequences was identified (90). Amyloid proteins, such as Escherichia coli curli (98) and B. subtilis TasA (99), form extracellular fibers that are involved in biofilm formation. The amyloid consensus peptide sequences TVTVTF, STVTVT and STVTVTF are found in S. aureus and S. epidermidis Bap (Table 1) (90) and Bap orthologs from other Gram-negative bacteria, including BapA from S. enterica Typhimurium (Table 1). While the STVTVTF peptides form stiff fibers of several microns long in vitro, as observed with atomic force microscopy (90), formation of amyloid-like fibers by Bap proteins has not been reported. It is predicted that the amyloid sequence repeats contribute to the adhesive properties of Bap in promoting cell-cell adhesion rather than participating in amyloid-like fiber formation. Site-directed mutagenesis or STVTVTF motif deletion studies are likely to provide more insights into the role of these repeats in surface adhesion and biofilm formation by Bap and Bap orthologs.
In S. enterica Enteritidis, BapA was identified through sequence homology searches using S. aureus Bap (9). Phenotypic analyses using bapA deletion and overexpression strains revealed that BapA is involved in pellicle and biofilm formation (9). The core region of BapA contains 29 imperfect tandem repeats of 86 to 106 amino acid residues and shares 29% identity with C repeats of S. aureus Bap (Figure 3, Table 1). Despite the absence of calcium-binding motifs, BapA-dependent biofilm formation was found to be mediated by calcium in the millimolar range, similar to that observed for S. aureus Bap. The mechanism of calcium regulation of BapA-mediated biofilm formation is currently unknown. Nonetheless, modulation of protein conformation by calcium may be a conserved regulatory mechanism in Bap-mediated biofilm formation in both Gram-positive and Gram-negative bacteria. BapA also contains three VCBS repeats at the C-terminal region (9), which have been implicated in cell adhesion, further corroborating the involvement of BapA in biofilm formation. BapA is secreted and loosely associated with the bacterial cell surface (9). Secretion of BapA is likely through a type I secretion system, as BapA contains three non-interacting α-helices at the C-terminal region that resemble the C-terminal secretion signal recognized by the type I secretion ABC transporter (9). In addition, deletion of downstream coding regions, including a putative ABC-type exporter, abolished BapA secretion and resulted in a strain unable to form pellicles (9). The presence of Bap orthologs with similar domain organization in both Gram-positive and Gram-negative bacteria highlights the importance of Bap-family proteins in biofilm formation in a diverse group of bacteria.
B. subtilis BslA (biofilm surface layer protein)
The amphiphilic protein BslA, formerly known as YuaB, has been reported to be a major factor in contributing to surface repellency and colony corrugation of biofilms formed by B. subtilis, a Gram-positive soil-dwelling bacterium. BslA plays a synergistic role with other matrix components, specifically TasA and exopolysaccharides, in the late stages of biofilm formation and confers enhanced repellency in the resultant biofilm with the formation of a unique hydrophobic layer on the biofilm surface (12, 13, 100, 101). Such increased repellency in biofilm surfaces may enhance B. subtilis survival in a natural soil habitat by repelling environmental pollutants and toxic compounds such as heavy metals and anti-microbial agents.
Strains lacking BslA form colonies and pellicles with decreased corrugation, altered surface microstructures and loss of surface repellency (12, 13, 100, 101). Increased expression of bslA from an IPTG-inducible promoter in a BslA-deficient strain results in increased colony and pellicle corrugation (12) and exogenous addition of purified BslA complements the altered pellicle and colony phenotypes of a bslA mutant strain (13). Importantly, overproduction of exopolysaccharide or the amyloid protein TasA, the other two major biofilm matrix components, cannot complement the loss of colony and pellicle corrugation phenotypes due to bslA mutation (12), indicating that BslA functions synergistically with TasA and the exopolysaccharide in mediating biofilm formation in B. subtilis. Co-culturing a BslA-deficient strain and a strain that is unable to produce TasA and exopolysaccharide complements the colony-biofilm phenotype (12). These observations reinforce the notion that the biofilm matrix is composed of secreted matrix components and these extracellular components contribute collectively to biofilm formation by serving as communal resources. While strains unable to produce BslA, TasA or exopolysaccharide exhibit altered pellicle structures, only TasA- and exopolysaccharide-deficient strains are incapable of forming cell clusters and aggregates (13), suggesting that BslA likely plays a role in biofilm formation after TasA- and exopolysaccharide-dependent cell clusters are formed.
As a bacterial hydrophobin, BslA also functions synergistically with TasA and the exopolysaccharide in mediating surface repellency, via its ability to form rough surface microstructures (13). BslA exhibits self-polymerization (13), similar to other hydrophobins, and forms a hydrophobic layer at the bottom liquid-cell interface of pellicles (101), and on both the top and bottom surfaces of colony biofilms (13, 101). The BslA hydrophobic layer localized at the bottom of the floating pellicle biofilm may form a protein raft carrying the biofilm mass. Using a bslA promoter-green fluorescent protein construct, it was shown that spatial localization of BslA at the surfaces of biofilms is not due to localized transcription of bslA (101). BslA therefore migrates, via an unknown mechanism, to the biofilm surface. BslA exhibits unique dual functions in biofilm formation: it functions as a hydrophobin in increasing biofilm repellency and a matrix protein critical for biofilm architecture formation.
BslA contains a secretion signal sequence (13) and has been reported to be part of the secretome of B. subtilis grown in liquid cultures (102); however, the mechanism of secretion is unclear. Using western blot analyses (12, 13) and transmission electron microscopy (12), BslA was shown to be secreted and retained within the biofilm matrix. Although secretion is required for BslA-mediated biofilm formation, it is currently unclear if BslA is associated with the exopolysaccharides or cell wall, since BslA was found in different cellular fractions in different studies and was detected in the matrix only in standing, but not in shaking cultures (12, 13).
Crystal structures reveal that BslA exhibits an Ig-like fold and contains a hydrophobic cap (101). The presence of the Ig-like fold corroborated the hypothesis that BslA plays an adhesive role in biofilm formation. Site-directed mutagenesis of the residues located in the hydrophobic cap (L76K, L77K, L79K, L121K, L123K, L153K and I155K) resulted in mutant strains exhibiting altered colony biofilm phenotypes but only two of these (L77K and L79K) show a loss of surface repellency, indicating that loss of hydrophobicity is not merely due to a decrease in colony corrugation and complexity (101). However, the loss of surface repellency due to site-directed mutagenesis is always accompanied by loss of complex colony architectures. As such, BslA-mediated surface repellency in B. subtilis biofilms is dependent on the hydrophobic cap, which in turn affects biofilm formation, but loss of BslA function in biofilm formation does not necessarily affect biofilm repellency. Currently, the mechanism by which BslA affects biofilm formation and surface repellency is unknown. In addition, it is not clear how individual BslA proteins interact during the self-assembling process, how BslA localizes to the surface of the biofilm, and what type of interaction occurs between the BslA hydrophilic layer and the exopolysaccharide.
Outer Membrane Vesicles (OMVs)
OMVs are small spherical structures produced by Gram-negative bacteria through pinching off or blebbing from the outer membrane. They range from 10 to 300 nm in diameter, and generally contain cytoplasmic and periplasmic contents such as proteases, alkaline phosphatase, lipases, and toxins, as well as outer membrane proteins, and LPS. OMVs are involved in many biological processes, including biofilm formation, pathogenesis, quorum signaling, nutrient acquisition and horizontal gene transfer (103–105). OMVs are produced by several Gram-negative bacteria, including P. aeruginosa (20, 106), Helicobacter pylori (107), V. cholerae (40, 108), and Vibrio fischeri (112), as observed with transmission electron microscopy of thin-section biofilms or extracted OMVs. A growing number of studies have been focused on investigating the possible role of OMVs in biofilm formation.
P. aeruginosa OMVs are a component of the biofilm matrix (106). Using transmission electron microscopy, OMVs were observed to be present within the biofilm matrix formed by P. aeruginosa PAO1 (106). OMVs extracted from biofilm cultures of P. aeruginosa PAO1 differ in quantity, quality, and in protein identity compared to those isolated from planktonic cultures (20, 106), suggesting that OMVs may play different roles in these different physiological growth states (i.e. biofilm versus planktonic). Importantly, there are more OMVs isolated from biofilms than from planktonic cultures (106). However, a direct role of OMVs in P. aeruginosa biofilm formation has not been demonstrated. Nonetheless, interactions of OMVs with secreted matrix proteins, through binding to outer membrane proteins (OMPs) present in the OMVs, have been reported, thus implicating OMVs in biofilm formation. The OMP OprF was identified in both the biofilm matrix and the OMVs (20). Since OprF interacts with the lectin LecB (89), it is possible that OMVs may be localized in the biofilm matrix via interaction of OMV-associated OprF and the potentially Psl exopolysaccharide-binding LecB, thus forming an OMV-OprF-LecB-Psl interaction. It has been documented that eDNA, a biofilm matrix component of P. aeruginosa PAO1, associates with OMVs in P. aeruginosa PAO1 (58, 109, 110), which suggests that OMVs may be involved in the process of biofilm formation by interacting with different biofilm matrix components, including eDNA.
A direct role of OMVs in biofilm formation has been demonstrated in H. pylori. Addition of purified OMV-fraction induces biofilm formation in a dose-dependent manner (107). OMV production in H. pylori, similar to that observed in other bacteria, depends on culture conditions and the physiological state of the cells, as more OMV production and enhanced biofilm formation were observed in growth medium containing fetal calf serum in a dose-dependent manner (107). Although the protein compositions of OMVs produced by H. pylori have not been determined, protein profiles, as determined by SDS-PAGE, indicate that OMVs isolated at different stages of biofilm formation exhibit differences (111), suggesting that these OMVs may have distinct cargo.
Production of OMVs has also been reported in V. cholerae (40, 108); however, direct involvement of OMVs in V. cholerae biofilm formation has not been demonstrated. Nonetheless, OMVs isolated from planktonic V. cholerae cultures grown in the presence of the anti-microbial peptide polymyxin B, which induces cell envelope stress, have been shown to bind Bap1 (40), a matrix protein that is involved in biofilm formation (6, 27). As mentioned earlier, Bap1 associates with OMVs by binding to OmpT, and since Bap1 also functions as an adhesive protein, it is likely that Bap1 binding to OMVs results in adherence and localization of OMVs onto surfaces and/or exopolysaccharides, implicating OMVs in biofilm formation.
There is a clear connection between OMV production and biofilm formation in V. fischeri (112). Transmission electron microscopy analysis of colony biofilms showed that OMVs are present within the extracellular matrix. A V. fischeri strain that exhibits increased biofilm formation, due to overproduction of the RscS sensor kinase, produces approximately 2.5-fold more OMVs when compared to the parental strain. Furthermore, the increase in OMV production is dependent on the production of the symbiosis polysaccharide.
OMV involvement in biofilm formation may be multifaceted, as OMVs could interact with various biofilm matrix components, including proteins, exopolysaccharides and eDNA. These interactions likely involve proteins and/or LPS found on the surfaces of OMVs, and therefore could reinforce structural integrity of the biofilm matrix. OMVs may also be deposited onto surfaces, thus conditioning the substratum for subsequent bacterial attachment. Further investigations will help to gain greater insights into the role of OMVs in biofilm formation.
Bacterial Nucleoid-Binding Proteins
Recent studies revealed that nucleoid-binding proteins, besides being involved in the maintenance of DNA supercoiling and compaction, also play a “moonlighting” role in biofilm formation (113–115). DNABII family proteins, members of the nucleoid-associated protein superfamily, can be grouped into two subtypes: HU (histone-like protein from E. coli strain U39), which is ubiquitous in Eubacteria, and IHF (integration host factor), which is only found in bacteria within the α- and γ-proteobacteria genera (113, 116). Extracellular-localized nucleoid-binding proteins have been found in association with eDNA in the biofilm matrix from sputum samples of cystic fibrosis patients (114, 115). eDNA contributes to biofilm stability in many bacterial species, including P. aeruginosa PAO1 (58). It has also been reported that addition of anti-IHF serum, which exhibits avidity to both IHF and HU, to pre-formed biofilms results in dissolution of E. coli, Haemophilus influenza and Burkholderia cenocepacia biofilms (113, 114). Similar dissolutions of pre-formed biofilm with the addition of anti-IHF serum were also observed with other bacterial species, including S. aureus, S. epidermidis, uropathogenic E. coli, Neisseria gonorrhoeae, and P. aeruginosa (113). A study carried out to identify proteins found in the biofilm matrix of P. aeruginosa PAO1 identified the nucleoid-binding protein HU in the biofilm matrix (20), further suggesting that nucleoid-binding protein may be involved in biofilm formation. While several other nucleoid-binding proteins, including H-NS and DPS, are produced in different bacteria, anti-sera directed against these proteins did not result in alteration of biofilm structures when added to the biofilms (113), suggesting that H-NS and DPS are unlikely to be involved in biofilm formation. Currently, it is unclear how nucleoid-binding proteins are localized outside of the cells. Since they do not contain a secretion signal sequence, it is likely that they are released into the biofilm matrix via cell lysis or through a similar unknown mechanism involved in eDNA release. Since OMVs may be involved in eDNA release into the biofilm matrix, it is possible that the extracellular localization of these nucleoid-binding proteins may be OMV-mediated. Little is known about the identity of nucleoid-binding proteins in different bacterial biofilms and the role they play in biofilm formation.
Concluding Remarks
Matrix proteins play diverse and important roles in biofilm formation by mediating initial surface attachment, cell cluster and aggregate formation, and stabilization and establishment of elaborate three-dimensional biofilm architectures. Matrix proteins exhibit unique characteristics and function synergistically with each other and with other matrix components, such as exopolysaccharides and eDNA, in biofilm formation. A common feature of these matrix proteins is that they are organized into modules or domains. These include FnIII, FG-GAP, VCBS, lectin, glycine-rich sugar-binding domains, and RGD sequence motifs that participate in cell-cell adhesion and/or binding to extracellular matrix, cell surface proteins, or carbohydrates. Identification of residues within the domains that are critical for ligand interactions as well as identification of the target ligands will be crucial in deciphering the mechanisms of these matrix proteins in biofilm formation. The regulation of matrix proteins is complex, involving multiple positive and negative transcriptional regulators, alternative sigma factors and small-regulatory RNAs (5, 100, 117–125). Matrix protein production is commonly coordinated with the production of other matrix components, such as exopolysaccharides, leading to optimal biofilm structure and function. Better characterization of the biofilm matrix proteome, structure/function relationships of matrix proteins, and regulatory circuits controlling biofilm matrix production will provide further mechanistic insights into biofilm formation and facilitate development of anti-biofilm therapeutics.
Acknowledgments
We thank Karen Visick and Holger Sondermann as well as members of Yildiz laboratory for their valuable comments on the manuscript. This work is supported by NIH R01 AI055987.
References
- 1.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 2.O’Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 3.Branda SS, Vik S, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 5.Fong JC, Karplus K, Schoolnik GK, Yildiz FH. Identification and characterization of RbmA, a novel protein required for the development of rugose colony morphology and biofilm structure in Vibrio cholerae. J bacteriol. 2006;188:1049–1059. doi: 10.1128/JB.188.3.1049-1059.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fong JC, Yildiz FH. The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J bacteriol. 2007;189:2319–2330. doi: 10.1128/JB.01569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diggle SP, Stacey RE, Dodd C, Camara M, Williams P, Winzer K. The galactophilic lectin, LecA, contributes to biofilm development in Pseudomonas aeruginosa. Environ Microbiol. 2006;8:1095–1104. doi: 10.1111/j.1462-2920.2006.001001.x. [DOI] [PubMed] [Google Scholar]
- 8.Tielker D, Hacker S, Loris R, Strathmann M, Wingender J, Wilhelm S, Rosenau F, Jaeger KE. Pseudomonas aeruginosa lectin LecB is located in the outer membrane and is involved in biofilm formation. Microbiology. 2005;151:1313–1323. doi: 10.1099/mic.0.27701-0. [DOI] [PubMed] [Google Scholar]
- 9.Latasa C, Roux A, Toledo-Arana A, Ghigo JM, Gamazo C, Penades JR, Lasa I. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol Microbiol. 2005;58:1322–1339. doi: 10.1111/j.1365-2958.2005.04907.x. [DOI] [PubMed] [Google Scholar]
- 10.Toledo-Arana A, Valle J, Solano C, Arrizubieta MJ, Cucarella C, Lamata M, Amorena B, Leiva J, Penades JR, Lasa I. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl Environ Microbiol. 2001;67:4538–4545. doi: 10.1128/AEM.67.10.4538-4545.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cucarella C, Solano C, Valle J, Amorena B, Lasa I, Penades JR. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J bacteriol. 2001;183:2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ostrowski A, Mehert A, Prescott A, Kiley TB, Stanley-Wall NR. YuaB functions synergistically with the exopolysaccharide and TasA amyloid fibers to allow biofilm formation by Bacillus subtilis. J bacteriol. 2011;193:4821–4831. doi: 10.1128/JB.00223-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi K, Iwano M. BslA(YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms. Mol Microbiol. 2012;85:51–66. doi: 10.1111/j.1365-2958.2012.08094.x. [DOI] [PubMed] [Google Scholar]
- 14.Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol. 2010;75:827–842. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaplan JB, Ragunath C, Ramasubbu N, Fine DH. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous beta-hexosaminidase activity. J bacteriol. 2003;185:4693–4698. doi: 10.1128/JB.185.16.4693-4698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marti M, Trotonda MP, Tormo-Mas MA, Vergara-Irigaray M, Cheung AL, Lasa I, Penades JR. Extracellular proteases inhibit protein-dependent biofilm formation in Staphylococcus aureus. Microbes Infect. 2010;12:55–64. doi: 10.1016/j.micinf.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR, Bayles KW. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One. 2009;4:e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nijland R, Hall MJ, Burgess JG. Dispersal of biofilms by secreted, matrix degrading, bacterial DNase. PLoS One. 2010;5:e15668. doi: 10.1371/journal.pone.0015668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Absalon C, Van Dellen K, Watnick PI. A communal bacterial adhesin anchors biofilm and bystander cells to surfaces. PLoS Pathog. 2011;7:e1002210. doi: 10.1371/journal.ppat.1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toyofuku M, Roschitzki B, Riedel K, Eberl L. Identification of proteins associated with the Pseudomonas aeruginosa biofilm extracellular matrix. J Proteome Res. 2012;11:4906–4915. doi: 10.1021/pr300395j. [DOI] [PubMed] [Google Scholar]
- 21.Curtis PD, Atwood J, 3rd, Orlando R, Shimkets LJ. Proteins associated with the Myxococcus xanthus extracellular matrix. J bacteriol. 2007;189:7634–7642. doi: 10.1128/JB.01007-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiao Y, D’Haeseleer P, Dill BD, Shah M, Verberkmoes NC, Hettich RL, Banfield JF, Thelen MP. Identification of biofilm matrix-associated proteins from an acid mine drainage microbial community. Appl Environ Microbiol. 2011;77:5230–5237. doi: 10.1128/AEM.03005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yildiz FH, Schoolnik GK. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci U S A. 1999;96:4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fong JC, Syed KA, Klose KE, Yildiz FH. Role of Vibrio polysaccharide (vps) genes in VPS production, biofilm formation and Vibrio cholerae pathogenesis. Microbiology. 2010;156:2757–2769. doi: 10.1099/mic.0.040196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yildiz F, Fong J, Sadovskaya I, Grard T, Vinogradov E. Structural Characterization of the Extracellular Polysaccharide from Vibrio cholerae O1 El-Tor. PLoS One. 2014;9:e86751. doi: 10.1371/journal.pone.0086751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirn TJ, Jude BA, Taylor RK. A colonization factor links Vibrio cholerae environmental survival and human infection. Nature. 2005;438:863–866. doi: 10.1038/nature04249. [DOI] [PubMed] [Google Scholar]
- 27.Berk V, Fong JC, Dempsey GT, Develioglu ON, Zhuang X, Liphardt J, Yildiz FH, Chu S. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science. 2012;337:236–239. doi: 10.1126/science.1222981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giglio KM, Fong JC, Yildiz FH, Sondermann H. Structural basis for biofilm formation via the Vibrio cholerae matrix protein RbmA. J bacteriol. 2013;195:3277–3286. doi: 10.1128/JB.00374-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bork P, Doolittle RF. Proposed acquisition of an animal protein domain by bacteria. Proc Natl Acad Sci U S A. 1992;89:8990–8994. doi: 10.1073/pnas.89.19.8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maestre-Reyna M, Wu WJ, Wang AH. Structural insights into RbmA, a biofilm scaffolding protein of Vibrio Cholerae. PLoS One. 2013;8:e82458. doi: 10.1371/journal.pone.0082458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oberhauser AF, Marszalek PE, Erickson HP, Fernandez JM. The molecular elasticity of the extracellular matrix protein tenascin. Nature. 1998;393:181–185. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
- 32.Craig D, Gao M, Schulten K, Vogel V. Tuning the mechanical stability of fibronectin type III modules through sequence variations. Structure. 2004;12:21–30. doi: 10.1016/j.str.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 33.Moorthy S, Watnick PI. Identification of novel stage-specific genetic requirements through whole genome transcription profiling of Vibrio cholerae biofilm development. Mol Microbiol. 2005;57:1623–1635. doi: 10.1111/j.1365-2958.2005.04797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baneres JL, Roquet F, Martin A, Parello J. A minimized human integrin alpha(5)beta(1) that retains ligand recognition. J Biol Chem. 2000;275:5888–5903. doi: 10.1074/jbc.275.8.5888. [DOI] [PubMed] [Google Scholar]
- 35.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 36.Hynes RO. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 37.Raval S, Gowda SB, Singh DD, Chandra NR. A database analysis of jacalin-like lectins: sequence-structure-function relationships. Glycobiology. 2004;14:1247–1263. doi: 10.1093/glycob/cwh140. [DOI] [PubMed] [Google Scholar]
- 38.Sankaranarayanan R, Sekar K, Banerjee R, Sharma V, Surolia A, Vijayan M. A novel mode of carbohydrate recognition in jacalin, a Moraceae plant lectin with a beta-prism fold. Nat Struct Biol. 1996;3:596–603. doi: 10.1038/nsb0796-596. [DOI] [PubMed] [Google Scholar]
- 39.Sikora AE, Zielke RA, Lawrence DA, Andrews PC, Sandkvist M. Proteomic analysis of the Vibrio cholerae type II secretome reveals new proteins, including three related serine proteases. J Biol Chem. 2011;286:16555–16566. doi: 10.1074/jbc.M110.211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duperthuy M, Sjostrom AE, Sabharwal D, Damghani F, Uhlin BE, Wai SN. Role of the Vibrio cholerae matrix protein Bap1 in cross-resistance to antimicrobial peptides. PLoS Pathog. 2013;9:e1003620. doi: 10.1371/journal.ppat.1003620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huq A, Small EB, West PA, Huq MI, Rahman R, Colwell RR. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl Environ Microbiol. 1983;45:275–283. doi: 10.1128/aem.45.1.275-283.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chowdhury MA, Huq A, Xu B, Madeira FJ, Colwell RR. Effect of alum on free-living and copepod-associated Vibrio cholerae O1 and O139. Appl Environ Microbiol. 1997;63:3323–3326. doi: 10.1128/aem.63.8.3323-3326.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hunt DE, Gevers D, Vahora NM, Polz MF. Conservation of the chitin utilization pathway in the Vibrionaceae. Appl Environ Microbiol. 2008;74:44–51. doi: 10.1128/AEM.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meibom KL, Li XB, Nielsen AT, Wu CY, Roseman S, Schoolnik GK. The Vibrio cholerae chitin utilization program. Proc Natl Acad Sci U S A. 2004;101:2524–2529. doi: 10.1073/pnas.0308707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stauder M, Huq A, Pezzati E, Grim CJ, Ramoino P, Pane L, Colwell RR, Pruzzo C, Vezzulli L. Role of GbpA protein, an important virulence-related colonization factor, for Vibrio cholerae’s survival in the aquatic environment. Environ Microbiol Rep. 2012;4:439–445. doi: 10.1111/j.1758-2229.2012.00356.x. [DOI] [PubMed] [Google Scholar]
- 46.Stauder M, Vezzulli L, Pezzati E, Repetto B, Pruzzo C. Temperature affects Vibrio cholerae O1 El Tor persistence in the aquatic environment via an enhanced expression of GbpA and MSHA adhesins. Environ Microbiol Rep. 2010;2:140–144. doi: 10.1111/j.1758-2229.2009.00121.x. [DOI] [PubMed] [Google Scholar]
- 47.Bhowmick R, Ghosal A, Das B, Koley H, Saha DR, Ganguly S, Nandy RK, Bhadra RK, Chatterjee NS. Intestinal adherence of Vibrio cholerae involves a coordinated interaction between colonization factor GbpA and mucin. Infect Immun. 2008;76:4968–4977. doi: 10.1128/IAI.01615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wong E, Vaaje-Kolstad G, Ghosh A, Hurtado-Guerrero R, Konarev PV, Ibrahim AF, Svergun DI, Eijsink VG, Chatterjee NS, van Aalten DM. The Vibrio cholerae colonization factor GbpA possesses a modular structure that governs binding to different host surfaces. PLoS Pathog. 2012;8:e1002373. doi: 10.1371/journal.ppat.1002373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maruyama Y, Momma M, Mikami B, Hashimoto W, Murata K. Crystal structure of a novel bacterial cell-surface flagellin binding to a polysaccharide. Biochemistry. 2008;47:1393–1402. doi: 10.1021/bi701872x. [DOI] [PubMed] [Google Scholar]
- 50.Hashimoto W, He J, Wada Y, Nankai H, Mikami B, Murata K. Proteomics-based identification of outer-membrane proteins responsible for import of macromolecules in Sphingomonas sp. A1: alginate-binding flagellin on the cell surface. Biochemistry. 2005;44:13783–13794. doi: 10.1021/bi050873b. [DOI] [PubMed] [Google Scholar]
- 51.Callebaut I, Gilges D, Vigon I, Mornon JP. HYR, an extracellular module involved in cellular adhesion and related to the immunoglobulin-like fold. Protein Sci. 2000;9:1382–1390. doi: 10.1110/ps.9.7.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kelly G, Prasannan S, Daniell S, Fleming K, Frankel G, Dougan G, Connerton I, Matthews S. Structure of the cell-adhesion fragment of intimin from enteropathogenic Escherichia coli. Nat Struct Biol. 1999;6:313–318. doi: 10.1038/7545. [DOI] [PubMed] [Google Scholar]
- 53.Chiavelli DA, Marsh JW, Taylor RK. The mannose-sensitive hemagglutinin of Vibrio cholerae promotes adherence to zooplankton. Appl Environ Microbiol. 2001;67:3220–3225. doi: 10.1128/AEM.67.7.3220-3225.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watnick PI, Fullner KJ, Kolter R. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J bacteriol. 1999;181:3606–3609. doi: 10.1128/jb.181.11.3606-3609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Evans LR, Linker A. Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J bacteriol. 1973;116:915–924. doi: 10.1128/jb.116.2.915-924.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Friedman L, Kolter R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J bacteriol. 2004;186:4457–4465. doi: 10.1128/JB.186.14.4457-4465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Friedman L, Kolter R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol. 2004;51:675–690. doi: 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- 58.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 59.Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, Tolker-Nielsen T. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol. 2006;59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- 60.Garber N, Guempel U, Belz A, Gilboa-Garber N, Doyle RJ. On the specificity of the D-galactose-binding lectin (PA-I) of Pseudomonas aeruginosa and its strong binding to hydrophobic derivatives of D-galactose and thiogalactose. Biochim Biophys Acta. 1992;1116:331–333. doi: 10.1016/0304-4165(92)90048-y. [DOI] [PubMed] [Google Scholar]
- 61.Schurks N, Wingender J, Flemming HC, Mayer C. Monomer composition and sequence of alginates from Pseudomonas aeruginosa. Int J Biol Macromol. 2002;30:105–111. doi: 10.1016/s0141-8130(02)00002-8. [DOI] [PubMed] [Google Scholar]
- 62.Nivens DE, Ohman DE, Williams J, Franklin MJ. Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J bacteriol. 2001;183:1047–1057. doi: 10.1128/JB.183.3.1047-1057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stapper AP, Narasimhan G, Ohman DE, Barakat J, Hentzer M, Molin S, Kharazmi A, Hoiby N, Mathee K. Alginate production affects Pseudomonas aeruginosa biofilm development and architecture, but is not essential for biofilm formation. J Med Microbiol. 2004;53:679–690. doi: 10.1099/jmm.0.45539-0. [DOI] [PubMed] [Google Scholar]
- 64.Wozniak DJ, Wyckoff TJ, Starkey M, Keyser R, Azadi P, O’Toole GA, Parsek MR. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc Natl Acad Sci U S A. 2003;100:7907–7912. doi: 10.1073/pnas.1231792100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hentzer M, Teitzel GM, Balzer GJ, Heydorn A, Molin S, Givskov M, Parsek MR. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J bacteriol. 2001;183:5395–5401. doi: 10.1128/JB.183.18.5395-5401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ma L, Lu H, Sprinkle A, Parsek MR, Wozniak DJ. Pseudomonas aeruginosa Psl is a galactose- and mannose-rich exopolysaccharide. J bacteriol. 2007;189:8353–8356. doi: 10.1128/JB.00620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Colvin KM, Gordon VD, Murakami K, Borlee BR, Wozniak DJ, Wong GC, Parsek MR. The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 2011;7:e1001264. doi: 10.1371/journal.ppat.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jackson KD, Starkey M, Kremer S, Parsek MR, Wozniak DJ. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J bacteriol. 2004;186:4466–4475. doi: 10.1128/JB.186.14.4466-4475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ma L, Jackson KD, Landry RM, Parsek MR, Wozniak DJ. Analysis of Pseudomonas aeruginosa conditional Psl variants reveals roles for the Psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J bacteriol. 2006;188:8213–8221. doi: 10.1128/JB.01202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gilboa-Garber N. Purification and properties of hemagglutinin from Pseudomonas aeruginosa and its reaction with human blood cells. Biochim Biophys Acta. 1972;273:165–173. [PubMed] [Google Scholar]
- 71.Gilboa-Garber N, Mizrahi L, Garber N. Mannose-binding hemagglutinins in extracts of Pseudomonas aeruginosa. Can J Biochem. 1977;55:975–981. doi: 10.1139/o77-145. [DOI] [PubMed] [Google Scholar]
- 72.Gilboa-Garber N. Pseudomonas aeruginosa lectins. Methods Enzymol. 1982;83:378–385. doi: 10.1016/0076-6879(82)83034-6. [DOI] [PubMed] [Google Scholar]
- 73.Adam EC, Mitchell BS, Schumacher DU, Grant G, Schumacher U. Pseudomonas aeruginosa PA-II lectin stops human ciliary beating: therapeutic implications of fucose. Am J Respir Crit Care Med. 1997;155:2102–2104. doi: 10.1164/ajrccm.155.6.9196121. [DOI] [PubMed] [Google Scholar]
- 74.Avichezer D, Gilboa-Garber N. Antitumoral effects of Pseudomonas aeruginosa lectins on Lewis lung carcinoma cells cultured in vitro without and with murine splenocytes. Toxicon. 1991;29:1305–1313. doi: 10.1016/0041-0101(91)90117-a. [DOI] [PubMed] [Google Scholar]
- 75.Bajolet-Laudinat O, Girod-de Bentzmann S, Tournier JM, Madoulet C, Plotkowski MC, Chippaux C, Puchelle E. Cytotoxicity of Pseudomonas aeruginosa internal lectin PA-I to respiratory epithelial cells in primary culture. Infect Immun. 1994;62:4481–4487. doi: 10.1128/iai.62.10.4481-4487.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gabius HJ, Andre S, Kaltner H, Siebert HC. The sugar code: functional lectinomics. Biochim Biophys Acta. 2002;1572:165–177. doi: 10.1016/s0304-4165(02)00306-9. [DOI] [PubMed] [Google Scholar]
- 77.Barondes SH, Gitt MA, Leffler H, Cooper DN. Multiple soluble vertebrate galactoside-binding lectins. Biochimie. 1988;70:1627–1632. doi: 10.1016/0300-9084(88)90298-2. [DOI] [PubMed] [Google Scholar]
- 78.Blanchard B, Imberty A, Varrot A. Secondary sugar binding site identified for LecA lectin from Pseudomonas aeruginosa. Proteins. 2013 doi: 10.1002/prot.24430. [DOI] [PubMed] [Google Scholar]
- 79.Imberty A, wimmerova M, Mitchell EP, Gilboa-Garber N. Structures of the lectins from Pseudomonas aeruginosa: insight into the molecular basis for host glycan recognition. Microbes Infect. 2004;6:221–228. doi: 10.1016/j.micinf.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 80.Sudakevitz D, Kostlanova N, Blatman-Jan G, Mitchell EP, Lerrer B, Wimmerova M, Katcoff DJ, Imberty A, Gilboa-Garber N. A new Ralstonia solanacearum high-affinity mannose-binding lectin RS-IIL structurally resembling the Pseudomonas aeruginosa fucose-specific lectin PA-IIL. Mol Microbiol. 2004;52:691–700. doi: 10.1111/j.1365-2958.2004.04020.x. [DOI] [PubMed] [Google Scholar]
- 81.Zinger-Yosovich K, Sudakevitz D, Imberty A, Garber NC, Gilboa-Garber N. Production and properties of the native Chromobacterium violaceum fucose-binding lectin (CV-IIL) compared to homologous lectins of Pseudomonas aeruginosa (PA-IIL) and Ralstonia solanacearum (RS-IIL) Microbiology. 2006;152:457–463. doi: 10.1099/mic.0.28500-0. [DOI] [PubMed] [Google Scholar]
- 82.Lameignere E, Malinovska L, Slavikova M, Duchaud E, Mitchell EP, Varrot A, Sedo O, Imberty A, Wimmerova M. Structural basis for mannose recognition by a lectin from opportunistic bacteria Burkholderia cenocepacia. Biochem J. 2008;411:307–318. doi: 10.1042/bj20071276. [DOI] [PubMed] [Google Scholar]
- 83.Pokorna M, Cioci G, Perret S, Rebuffet E, Kostlanova N, Adam J, Gilboa-Garber N, Mitchell EP, Imberty A, Wimmerova M. Unusual entropy-driven affinity of Chromobacterium violaceum lectin CV-IIL toward fucose and mannose. Biochemistry. 2006;45:7501–7510. doi: 10.1021/bi060214e. [DOI] [PubMed] [Google Scholar]
- 84.Mitchell E, Houles C, Sudakevitz D, Wimmerova M, Gautier C, Perez S, Wu AM, Gilboa-Garber N, Imberty A. Structural basis for oligosaccharide-mediated adhesion of Pseudomonas aeruginosa in the lungs of cystic fibrosis patients. Nat Struct Biol. 2002;9:918–921. doi: 10.1038/nsb865. [DOI] [PubMed] [Google Scholar]
- 85.Cioci G, Mitchell EP, Gautier C, Wimmerova M, Sudakevitz D, Perez S, Gilboa-Garber N, Imberty A. Structural basis of calcium and galactose recognition by the lectin PA-IL of Pseudomonas aeruginosa. FEBS Lett. 2003;555:297–301. doi: 10.1016/s0014-5793(03)01249-3. [DOI] [PubMed] [Google Scholar]
- 86.Loris R, Tielker D, Jaeger KE, Wyns L. Structural basis of carbohydrate recognition by the lectin LecB from Pseudomonas aeruginosa. J Mol Biol. 2003;331:861–870. doi: 10.1016/s0022-2836(03)00754-x. [DOI] [PubMed] [Google Scholar]
- 87.Avichezer D, Katcoff DJ, Garber NC, Gilboa-Garber N. Analysis of the amino acid sequence of the Pseudomonas aeruginosa galactophilic PA-I lectin. J Biol Chem. 1992;267:23023–23027. [PubMed] [Google Scholar]
- 88.Gilboa-Garber N, Katcoff DJ, Garber NC. Identification and characterization of Pseudomonas aeruginosa PA-IIL lectin gene and protein compared to PA-IL. FEMS Immunol Med Microbiol. 2000;29:53–57. doi: 10.1111/j.1574-695X.2000.tb01505.x. [DOI] [PubMed] [Google Scholar]
- 89.Funken H, Bartels KM, Wilhelm S, Brocker M, Bott M, Bains M, Hancock RE, Rosenau F, Jaeger KE. Specific association of lectin LecB with the surface of Pseudomonas aeruginosa: role of outer membrane protein OprF. PLoS One. 2012;7:e46857. doi: 10.1371/journal.pone.0046857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lembre P, Vendrely C, Martino PD. Identification of an amyloidogenic peptide from the Bap protein of Staphylococcus epidermidis. Protein Pept Lett. 2014;21:75–79. doi: 10.2174/09298665113209990072. [DOI] [PubMed] [Google Scholar]
- 91.Tormo MA, Knecht E, Gotz F, Lasa I, Penades JR. Bap-dependent biofilm formation by pathogenic species of Staphylococcus: evidence of horizontal gene transfer? Microbiology. 2005;151:2465–2475. doi: 10.1099/mic.0.27865-0. [DOI] [PubMed] [Google Scholar]
- 92.Cucarella C, Tormo MA, Ubeda C, Trotonda MP, Monzon M, Peris C, Amorena B, Lasa I, Penades JR. Role of biofilm-associated protein Bap in the pathogenesis of bovine Staphylococcus aureus. Infect Immun. 2004;72:2177–2185. doi: 10.1128/IAI.72.4.2177-2185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Valle J, Latasa C, Gil C, Toledo-Arana A, Solano C, Penades JR, Lasa I. Bap, a biofilm matrix protein of Staphylococcus aureus prevents cellular internalization through binding to GP96 host receptor. PLoS Pathog. 2012;8:e1002843. doi: 10.1371/journal.ppat.1002843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cucarella C, Tormo MA, Knecht E, Amorena B, Lasa I, Foster TJ, Penades JR. Expression of the biofilm-associated protein interferes with host protein receptors of Staphylococcus aureus and alters the infective process. Infect Immun. 2002;70:3180–3186. doi: 10.1128/IAI.70.6.3180-3186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tendolkar PM, Baghdayan AS, Shankar N. The N-terminal domain of enterococcal surface protein, Esp, is sufficient for Esp-mediated biofilm enhancement in Enterococcus faecalis. Journal of bacteriology. 2005;187:6213–6222. doi: 10.1128/JB.187.17.6213-6222.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Arrizubieta MJ, Toledo-Arana A, Amorena B, Penades JR, Lasa I. Calcium inhibits bap-dependent multicellular behavior in Staphylococcus aureus. J bacteriol. 2004;186:7490–7498. doi: 10.1128/JB.186.22.7490-7498.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shukla SK, Rao TS. Effect of calcium on Staphylococcus aureus biofilm architecture: a confocal laser scanning microscopic study. Colloids Surf B Biointerfaces. 2013;103:448–454. doi: 10.1016/j.colsurfb.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 98.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Romero D, Aguilar C, Losick R, Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci U S A. 2010;107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Verhamme DT, Murray EJ, Stanley-Wall NR. DegU and Spo0A jointly control transcription of two loci required for complex colony development by Bacillus subtilis. J bacteriol. 2009;191:100–108. doi: 10.1128/JB.01236-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hobley L, Ostrowski A, Rao FV, Bromley KM, Porter M, Prescott AR, MacPhee CE, van Aalten DM, Stanley-Wall NR. BslA is a self-assembling bacterial hydrophobin that coats the Bacillus subtilis biofilm. Proc Natl Acad Sci U S A. 2013;110:13600–13605. doi: 10.1073/pnas.1306390110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Voigt B, Antelmann H, Albrecht D, Ehrenreich A, Maurer KH, Evers S, Gottschalk G, van Dijl JM, Schweder T, Hecker M. Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis. J Mol Microbiol Biotechnol. 2009;16:53–68. doi: 10.1159/000142894. [DOI] [PubMed] [Google Scholar]
- 103.Beveridge TJ. Structures of gram-negative cell walls and their derived membrane vesicles. J bacteriol. 1999;181:4725–4733. doi: 10.1128/jb.181.16.4725-4733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol. 2010;64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bielig H, Dongre M, Zurek B, Wai SN, Kufer TA. A role for quorum sensing in regulating innate immune responses mediated by Vibrio cholerae outer membrane vesicles (OMVs) Gut Microbes. 2011;2:274–279. doi: 10.4161/gmic.2.5.18091. [DOI] [PubMed] [Google Scholar]
- 106.Schooling SR, Beveridge TJ. Membrane vesicles: an overlooked component of the matrices of biofilms. J bacteriol. 2006;188:5945–5957. doi: 10.1128/JB.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yonezawa H, Osaki T, Kurata S, Fukuda M, Kawakami H, Ochiai K, Hanawa T, Kamiya S. Outer membrane vesicles of Helicobacter pylori TK1402 are involved in biofilm formation. BMC Microbiol. 2009;9:197. doi: 10.1186/1471-2180-9-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kondo K, Takade A, Amako K. Release of the outer membrane vesicles from Vibrio cholerae and Vibrio parahaemolyticus. Microbiol Immunol. 1993;37:149–152. doi: 10.1111/j.1348-0421.1993.tb03192.x. [DOI] [PubMed] [Google Scholar]
- 109.Schooling SR, Hubley A, Beveridge TJ. Interactions of DNA with biofilm-derived membrane vesicles. Journal of bacteriology. 2009;191:4097–4102. doi: 10.1128/JB.00717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Renelli M, Matias V, Lo RY, Beveridge TJ. DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology. 2004;150:2161–2169. doi: 10.1099/mic.0.26841-0. [DOI] [PubMed] [Google Scholar]
- 111.Yonezawa H, Osaki T, Woo T, Kurata S, Zaman C, Hojo F, Hanawa T, Kato S, Kamiya S. Analysis of outer membrane vesicle protein involved in biofilm formation of Helicobacter pylori. Anaerobe. 2011;17:388–390. doi: 10.1016/j.anaerobe.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 112.Shibata S, Visick KL. Sensor kinase RscS induces the production of antigenically distinct outer membrane vesicles that depend on the symbiosis polysaccharide locus in Vibrio fischeri. J bacteriol. 2012;194:185–194. doi: 10.1128/JB.05926-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Goodman SD, Obergfell KP, Jurcisek JA, Novotny LA, Downey JS, Ayala EA, Tjokro N, Li B, Justice SS, Bakaletz LO. Biofilms can be dispersed by focusing the immune system on a common family of bacterial nucleoid-associated proteins. Mucosal Immunol. 2011;4:625–637. doi: 10.1038/mi.2011.27. [DOI] [PubMed] [Google Scholar]
- 114.Novotny LA, Amer AO, Brockson ME, Goodman SD, Bakaletz LO. Structural stability of Burkholderia cenocepacia biofilms is reliant on eDNA structure and presence of a bacterial nucleic acid binding protein. PLoS One. 2013;8:e67629. doi: 10.1371/journal.pone.0067629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gustave JE, Jurcisek JA, McCoy KS, Goodman SD, Bakaletz LO. Targeting bacterial integration host factor to disrupt biofilms associated with cystic fibrosis. J Cyst Fibros. 2013;12:384–389. doi: 10.1016/j.jcf.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Swinger KK, Rice PA. IHF and HU: flexible architects of bent DNA. Curr Opin Struct Biol. 2004;14:28–35. doi: 10.1016/j.sbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 117.Yildiz FH, Liu XS, Heydorn A, Schoolnik GK. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol Microbiol. 2004;53:497–515. doi: 10.1111/j.1365-2958.2004.04154.x. [DOI] [PubMed] [Google Scholar]
- 118.Beyhan S, Bilecen K, Salama SR, Casper-Lindley C, Yildiz FH. Regulation of rugosity and biofilm formation in Vibrio cholerae: comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR. J bacteriol. 2007;189:388–402. doi: 10.1128/JB.00981-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fong JC, Yildiz FH. Interplay between cyclic AMP-cyclic AMP receptor protein and cyclic di-GMP signaling in Vibrio cholerae biofilm formation. J bacteriol. 2008;190:6646–6659. doi: 10.1128/JB.00466-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kovacs AT, Kuipers OP. Rok regulates yuaB expression during architecturally complex colony development of Bacillus subtilis 168. J bacteriol. 2011;193:998–1002. doi: 10.1128/JB.01170-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 122.Jones CJ, Newsom D, Kelly B, Irie Y, Jennings LK, Xu B, Limoli DH, Harrison JJ, Parsek MR, White P, Wozniak DJ. ChIP-Seq and RNA-Seq Reveal an AmrZ-Mediated Mechanism for Cyclic di-GMP Synthesis and Biofilm Development by Pseudomonas aeruginosa. PLoS Pathog. 2014;10:e1003984. doi: 10.1371/journal.ppat.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol. 2010;78:158–172. doi: 10.1111/j.1365-2958.2010.07320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Burrowes E, Baysse C, Adams C, O’Gara F. Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosa PAO1, as revealed by transcriptome analysis. Microbiology. 2006;152:405–418. doi: 10.1099/mic.0.28324-0. [DOI] [PubMed] [Google Scholar]
- 125.Moscoso JA, Mikkelsen H, Heeb S, Williams P, Filloux A. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ Microbiol. 2011;13:3128–3138. doi: 10.1111/j.1462-2920.2011.02595.x. [DOI] [PubMed] [Google Scholar]


