Abstract
Plasmids are important vehicles for rapid adaptation of bacterial populations to changing environmental conditions. To reduce the cost of plasmid carriage, it is thought that only a fraction of a local population carries plasmids or is permissive to plasmid uptake. Plasmids provide various accessory traits which might be beneficial under particular conditions. The genetic variation generated by plasmid carriage within populations ensures the robustness towards environmental change. Plasmid-mediated gene transfer plays an important role not only in the mobilization and dissemination of antibiotic resistance genes but also in the spread of degradative pathways and pathogenicity determinants of pathogens. Here we summarize the state-of-the-art methods to study the occurrence, abundance and diversity of plasmids in environmental bacteria. Increasingly, cultivation independent total community DNA methods are being used to characterize and quantify the diversity and abundance of plasmids in relation to various biotic and abiotic factors. An improved understanding of the ecology of plasmids and their hosts is crucial in the development of intervention strategies for antibiotic resistance gene spread. We discuss the potentials and limitations of methods used to determine the host range of plasmids as the ecology of plasmids is tightly linked to their hosts. The recent advances in sequencing technologies provide an enormous potential for plasmid classification, diversity and evolution studies but numerous challenges still exist.
Keywords: Plasmid ecology, horizontal gene transfer, exogenous plasmid isolation, metamobilome, host range, plasmidome
1. Plasmids - ancient means of bacterial adaptation and diversification
To date plasmid-mediated horizontal gene transfer is recognized as a major driving force for bacterial adaptation and diversification. Different environmental settings have their distinct bacterial community composition, which determines - possibly with the exception of broad-host range plasmids - the type of dominant plasmids that can be found. It is assumed that only a fraction of a population carries plasmids, which will ensure a rapid adaptation of the population to changing environmental conditions [1]. Without any doubt plasmid-mediated spread of antibiotic resistance genes among bacteria of different taxa is one of the most impressive examples of bacterial plasticity in response to various selective pressures [2,3]. While the molecular biology of plasmid-encoded replication, maintenance and transfer processes of some plasmids has been studied for decades, little attention has been paid to their dissemination in the environment, their ecology and the factors that drive their spread and diversification. In an overwhelming number of studies, the investigated plasmid-carrying strains originate from clinical specimen or diseased plant material, mostly human or plant pathogens. The reason for the lack of studies regarding the ecology of plasmids in natural settings was mainly the lack of tools to detect and quantify plasmids and to successfully culture their hosts.
Thanks to the rapidly advancing sequencing technologies, the number of completely sequenced plasmids increased in the last decades. Comparative plasmid sequence analysis has provided insights into the evolution of plasmids and their relatedness, their modular structure and the existence of hot spots for the insertion of accessory genes [4–6]. The growing plasmid sequence data base is also the prerequisite for the development of primers and probes to detect or classify plasmids. With the development of cultivation-independent DNA-based methods it became possible to study the occurrence of plasmids in various environmental samples and to quantify their abundance. Here we emphasize that studying plasmid ecology is without any doubt a prerequisite for a better understanding of the role of plasmids and their contribution to bacterial diversification and adaptation. In particular, the disentanglement of factors that foster the proliferation of plasmid carrying strains, horizontal gene exchange, and determine the cost and benefits of plasmid carriage to their hosts needs further consideration. This chapter aims to provide an overview on state-of-the-art methods being used to detect, isolate and characterize plasmids and to study various aspects of their ecology. Examples from recent studies will be given to illustrate the potentials and limitations of the methods employed to study plasmid occurrence and diversity, as well as some insights obtained. Although the methods described are applicable to Gram-negative and Gram-positive bacteria from any kind of environment, the major focus of the chapter is on plasmids in Gram-negative bacteria.
2. Detection and quantification of plasmid-specific sequences in total community DNA
Back in the 1980’s, Staley and Konopka [7] described the great plate count anomaly as the discrepancy between microscopic and colony forming counts, the latter being often much lower than the microscopic counts observed for the same sample. In general, the more oligotrophic an environment, the smaller is the proportion of bacteria able to form colonies on plates [8]. In addition, many bacteria that are well known to form colonies on plates can, under environmental conditions, enter a state called viable but nonculturable (vbnc) [9]. The vbnc status was reported for many human and plant pathogens.
In the last two decades, the development and application of cultivation-independent methods provided a better and more comprehensive picture of the occurrence and distribution of plasmids in different environmental settings. In particular the use of total community DNA (TC-DNA), extracted directly from environmental samples, became a more and more widely used approach for the detection and quantification of plasmid occurrence and abundance. For most types of environmental samples PCR-amplification with primers targeting replication or transfer related segments of the backbone of particular plasmid groups is required because of low plasmid abundance. When TC-DNA is analyzed for the presence of particular plasmid-specific sequences, it is important that the absence of PCR-inhibiting substances is confirmed, e.g. by the amplification of the 16S rRNA gene fragments and sample dilutions. Furthermore, PCR-amplicons from TC-DNA should be analyzed either by cloning and sequencing, amplicon sequencing or by Southern blot hybridization with labeled probes generated from PCR-amplicons obtained from reference strains to exclude false positive detection. This strategy has recently been used to screen TC-DNA from various types of environments originating from different geographic origins for the presence of IncP-1, IncP-7 and IncP-9 plasmids [10]. These plasmid groups are known to carry degradative genes or complete operons encoding degradative pathways [11,12]. The study by Dealtry et al. [10] showed a remarkably wide distribution of these plasmids and enabled the authors to identify so-called hot spots of plasmid occurrence. For example, samples from various pesticide bio-purification systems (BPS) and river sediments were identified as environments with high abundance of bacterial populations carrying IncP-1, IncP-7 and IncP-9 plasmids. Most remarkable was the presence of all IncP-1 subgroups in samples from BPS, except for the ζ subgroup that was recently described by Norberg et al. [5]. The strength of the hybridization signal obtained with probes targeting particular IncP-1 subgroups clearly differed in intensity indicating differences in their abundances, although different amplification efficiency of the different primer systems used could not be excluded. Cloning and sequencing of amplicons obtained with a newly designed IncP-9 primer system from TC-DNA of BPS samples revealed the presence not only of known plasmid groups but also the presence of unknown, yet to be isolated IncP-9 plasmids in these samples [10]. In many studies, cloning of PCR-amplicons provided insights into the sequence diversity of the amplicons. Bahl et al. [13] were the first to show the presence of the different IncP-1 subgroups in the inflow of a wastewater treatment plant. These authors had designed primers targeting the plasmid replication initiation gene trfA of the different IncP-1 groups, also including the γ, ε and δ subgroups that were amplified by the primers used in the study by Götz et al. [14]. Due to the sequence divergence of the trfA gene, three different primer systems had to be mixed in order to guarantee the parallel detection of all IncP-1 plasmid subgroups discovered at the time.
In another approach, 454 pyrosequencing of amplicons was employed to study IncP-1 plasmid diversity. In contrast to the traditional cloning and sequencing approaches, the number of sequences achieved by amplicon pyrosequencing at relatively low costs is impressive but many of the limitations, such as limitations of available sequences in databases, remain [15,16]. The 454 amplicon sequencing of trfA genes amplified from TC-DNA of samples taken from an on-farm BPS showed changes in the composition of the relative abundance of different IncP-1 subgroups over the agricultural season. While the relative abundance of IncP-1ε plasmids decreased over the season, the relative abundance of IncP-1β plasmids increased [15]. These results strongly point to an enrichment of populations carrying IncP-1β plasmids that code for enzymes likely involved in the degradation of pesticides under field conditions. Other studies showed a high abundance of plasmids in different types of environments such as manure, manure-treated soils, river water sediments, and sea sediments by PCR Southern blot hybridization [17–19] or quantitative real-time PCR [20,21].
The use of quantitative real-time PCR (qPCR) became an important tool in plasmid ecology such as for the elucidation of environmental factors influencing plasmid relative abundance in microbial communities. Although the enormous diversity of plasmid groups out there is certainly not covered by the primer systems available, recent examples have demonstrated the applicability of this tool in ecological studies [22–24]. Different biotic and abiotic factors can influence the relative abundance of plasmids and their hosts, and these factors can now be investigated in experiments comprising sufficient replicates and proper controls. This type of experiments are needed in order to test hypotheses and confirm correlations. For example, in a study by Smalla et al. [18] effects of mercury pollution on the abundance of IncP-1 plasmids in river sediments were investigated. Replicated river sediment samples were taken along a gradient of mercury pollution and, although different hybridization signal intensities were only semi-quantitative, PCR Southern blot analysis indicated that the abundance of the mercury resistance gene merRTΔP and the IncP-1 specific trfA gene correlated with the concentration of mercury pollution in the sediment samples. However, the authors could not exclude that additional factors could have influenced the bacterial community composition and the relative abundance of IncP-1 carrying bacterial cells. Another example is the study by Jechalke et al. [23] investigating plasmids in on-farm BPS over the agricultural season. In this study it was shown that the relative abundance of IncP-1 plasmids, which was quantified by qPCR with primers targeting IncP-1 korB, increased and that this effect was correlated with an increased concentration of a wide diversity of pesticides. The korB gene encodes a protein involved in the partitioning system and the regulatory network of the plasmid [25,26]. As no controls were available, no causal relationship could be demonstrated. However, in microcosm experiments in which linuron was added to BPS material the relative abundance of IncP-1 plasmids was significantly increased compared to the controls, providing evidence for the previously assumed correlation (Dealtry et al., submitted). It was also shown by qPCR that the application of manure containing antibiotics to arable soils increased the relative abundance of plasmids and class 1 integrons in the rhizosphere and in bulk soil [27,28].
Furthermore, it is important to note that the abundance of plasmid-carrying populations can increase in response to various triggers, e.g. root exudates, as recently shown by Jechalke et al. [22]. In the rhizosphere of lettuce grown in three different soil types a significantly increased relative abundance of the IncP-1 korB gene was found, while in the rhizosphere of potato plants grown in the same type of soils no enrichment of IncP-1 plasmids was observed by PCR Southern blot. The 454 pyrosequencing of 16S rRNA gene fragments amplified from TC-DNA of lettuce rhizosphere samples revealed that several genera were enriched that are known to include strains with the potential to degrade aromatic compounds [29], and the detection of aromatic compounds in the root exudates of lettuce supported the assumption that degraders of these molecules were enriched [30]. Most likely, the bacterial populations that were enriched in response to the lettuce root exudates carried IncP-1 plasmids with degradative genes. To test this hypothesis it will be necessary to isolate the IncP-1 plasmids either by a traditional cultivation approach or by exogenous isolation.
In summary, the detection and quantification of plasmid-related sequences in TC-DNA allows for the first time surveys of the dissemination of plasmid-specific sequences and, moreover, to link plasmid abundance with environmental factors and pollutant concentrations. However, experiments with treatments and controls performed in a sufficient number of independent replicates are needed in order to unequivocally test interdependencies.
3. Plasmid genome sequencing
Comparative analysis of whole plasmid genome sequences is rapidly becoming a standard approach to increase our understanding of the genetic diversity and evolutionary history of plasmids. Today, in November 2014, a total of 4,638 complete plasmid genome sequences are available in Genbank. Just over six years earlier, their number was only 1,490. Many of them have been sequenced as part of entire bacterial genomes, but an almost equal amount was submitted as plasmid genomes. The data not only help to define the molecular events that took place during the evolution of these plasmids, but also give us a more complete overview of the enormous collection of accessory genes encoded on plasmids. Future work will have to focus on annotating many of these genes that still have unknown or hypothetical functions.
Methods to determine the complete genome sequence of plasmids have evolved very rapidly over the last decade. To illustrate with an example, in a joint project to determine the genome sequence of 100 broad-host range plasmids with the U.S. Department of Energy Joint Genome Institute (JGI, Walnut Creek, CA), the institute used three different methods during the lifetime of the project. While in 2008 the plasmids were still being sequenced using Sanger sequencing of ~3 kb clone libraries, the following year a switch was made to the Roche/454 platform with GS FLX Titanium Sequencing chemistry, and soon after Illumina sequencing technology [6,31]. As a consequence of the novel sequence technology the cost of DNA sequencing per nucleotide has drastically decreased in recent years. Particularly for plasmids which have smaller genomes than bacterial chromosomes but require the same library preparation procedure, library preparation is relative expensive compared to the actual sequencing run. Moreover, improved bioinformatics pipelines are now available to separate chromosomal from plasmid DNA sequences. This has allowed us and others to simply determine the entire bacterial genome sequence and bioinformatically extract the plasmid genome sequence rather than invest a lot of labor time and cost in purifying the plasmid DNA. Previously, for many bacteria other than E. coli labor-intensive large-scale plasmid extraction methods were required to generate sufficient plasmid DNA low in contaminating chromosomal DNA. Today we can perform rapid total genomic DNA extractions instead. This approach works especially well in re-sequencing projects where the wild-type bacterial strain with its plasmid(s) has been completely sequenced previously, and the researcher wants to learn the genetic changes after experimental evolution or other genetic manipulations. The method can also work well in case of de novo plasmid sequence analysis when some information on the plasmids of interest is already available, and especially when the plasmid has first been transferred out of its native host into a previously sequenced laboratory strain.
For de novo sequencing projects of truly novel plasmids in their native host where the research question requires a correct assembly, we still recommend to at least enrich if not purify the plasmid DNA. Very often manually closing the sequence gaps between contigs by PCR-amplification and subsequent Sanger sequencing of the PCR-product is required. It is also highly advisable to compare experimental restriction fragment length patterns of the plasmid DNA with in silico digests, to confirm correct assembly. In our experience, with plasmids that have a well-known backbone structure, we have encountered a few cases where automated assembly was incorrect. This was even in a case of a single IncP-1 plasmid, where plasmid DNA had been purified, but a large duplication impeded the correct assembly of the sequence without additional experimental work [32]. In general, large duplications make it nearly impossible to correctly determine a plasmid genome sequence by short read sequencing like Illumina HiSeq and MiSeq. Methods such as SMRT (single-molecule real-time sequencing) applied by Pacific Biosciences (http://www.pacificbiosciences.com/) are very promising for plasmid genome sequencing, as fragments as long as 40 kb can be sequenced in one read. For many plasmids, 1–3 reads could thus be sufficient and assembly is no longer an issue. A recent example is the complete genome sequencing in a single run of a Klebsiella pneumoniae strain with four plasmids that encode NDM-1 and oxa-232 carbapenemases and other drug resistances [12].
When plasmids do not have a marker gene that can be used in the laboratory to select for the presence of the plasmid, we and others have marked the plasmid with a mini-transposon. One of the first examples was described by Tauch et al. [33] and Gstalder et al. [34] for plasmids pIPO2 and pMOL98. The drawback of the marking is that the obtained genome sequence does not represent the native, unmarked plasmid. In more recent work, the DNA sequences of the mini-transposons were removed from each plasmid sequence to reconstruct the genome sequences of the originally captured plasmids. By confirming the presence of repeats at each side of the transposon, we knew that the transposition had not caused deletions of flanking regions [31].
Annotation of plasmids is still a great challenge due to the confusing nomenclature of backbone genes, which is different for different plasmid incompatibility groups, and due to the large numbers of accessory genes with unknown or hypothetical functions. Automatic annotation is a first step but always requires a follow-up by careful manual annotation. In our previous plasmid sequencing projects, automatic annotation was carried out by the J. Craig Venter Institute Annotation Service (www.jcvi.org) or the Institute for Genome Sciences (IGS) (http://www.igs.umaryland.edu). Problems with annotation of plasmid encoded genes have been recently discussed by plasmid biology experts [35] but the debate is going on.
4. Detection of plasmid-specific sequences in metagenomes and metamobilomes
With the rapidly advancing sequencing technologies, deep sequencing of metagenomes or plasmidomes has opened a new path to discover mobile genetic elements by means of bioinformatics tools. Recently, first studies have been published attempting to describe plasmid genome diversity within a natural microbial community based on metagenomic approaches. In short, genomic or plasmid DNA is extracted directly from the sample lysates, for example by CsCl density gradient centrifugation, or bacterial populations are first isolated from the samples, which can be followed by removal of linear chromosomal DNA and a non-specific amplification step to increase the amount of circular plasmid DNA [36–38].
Plasmid extraction and purification directly from the microbial cell fraction of a sample by alkaline lysis, followed by size-dependent DNA separation methods such as ultracentrifugation or column-based binding assays was proposed by Li et al. [39] as a straight-forward approach to study plasmids in environmental samples. To minimize contamination with chromosomal fragments, sheared genomic and linear plasmid DNA is degraded by a plasmid-safe ATP-dependent DNase, amplified by φ29 DNA polymerase to ensure optimal amounts of plasmid DNA (micrograms), and subsequently sequenced. Although traditional cultivation biases were avoided and a large richness of previously unknown replicons was revealed, the method was found to favor small circular plasmids most likely due to the employed multiple displacement amplification. The finding was confirmed using pKJK10 and pBR322 as model plasmids [38]. The inclusion of an additional electroelution step provided an improved coverage of larger sized plasmids and thus could make the method more suitable to study the plasmid content of environmental bacteria. However, this method will obviously exclude the detection of linear plasmids.
Metagenomics will definitely shed more light on the diversity of plasmids and the accessory genes they carry, but due to the mosaic nature of plasmids with similar or identical sequences of considerable length, obtaining correctly closed genomes of large plasmids remains a challenge for complex microbial communities (see also previous section).
Another drawback of metagenomics methods is the lack of information on plasmid - host associations, just as for the plasmid capture methods described in the following section. Although an enormous diversity of novel and known resistance genes was revealed, based on metagenomic information it is currently impossible to determine which plasmid belongs to which host. Interestingly, promising techniques are being tested to physically link plasmid DNA in one cell to parts of chromosomal DNA, based on Hi-C. This is a method that relies on cross-linking molecules in close physical proximity and consequently identifies both, thus reflecting the spatial arrangement of DNA at the time of cross-linking within cells [40,41] (Figure 1). Proof of principle was recently shown by Beitel et al. [42].
Figure 1.
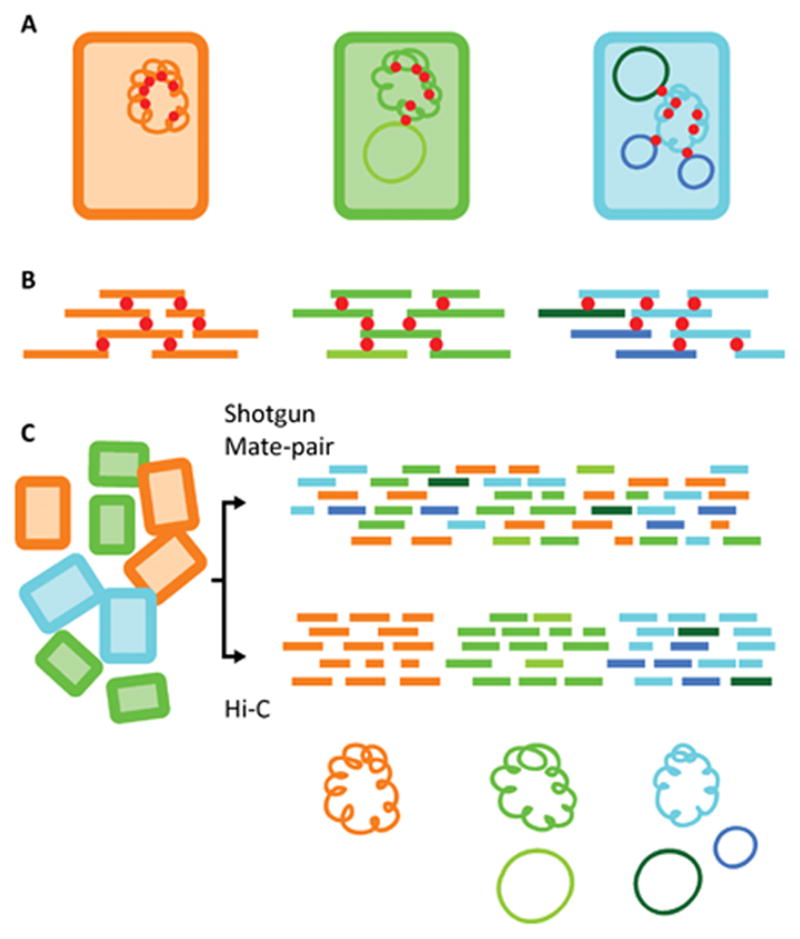
Overview of applying Hi-C technology to a mixed bacterial population to reliably associate plasmids with the chromosomes of their hosts (modified from Burton et al. [41]). (A) Rectangles indicate different cells carrying plasmids or not. Plasmids are cross-linked with bacterial chromosomes in close proximity (red circles). (B) The DNA in the cross-linked protein complexes is digested with HindIII endonuclease following cell lysis and free DNA ends are tagged with biotin. After ligation of blunt-ended DNA fragments under highly dilute conditions, which preferentially ligates fragments that are within the same cross-linked DNA/protein complex, cross-links are removed, DNA is purified, biotin is eliminated from un-ligated ends, DNA is size-selected, and ligation products are selected for through a biotin pull-down. The resulting Hi-C library is further analyzed by sequencing. (C) Workflow to create individual species/plasmid assemblies from a metagenome sample by combining shotgun, Hi-C, and (optionally) mate-pair libraries.
Another culture-independent method to isolate novel plasmids from microbial communities is the transposon-aided capture method (TRACA) [43]. In brief, this method uses purified plasmid DNA extracted from bacterial cells, cell cultures or environmental samples, which is then amended with an EZ-Tn5 OriV Kan2 transposon and a transposase for the in vitro transposition reaction. Following the transposition reaction, the transposition reaction mix is diluted, purified and electroporated into E. coli EPI300 cells. Transformants are plated on Luria Bertani broth containing 50 μg/ml kanamycin to select for EZ-Tn5 and captured plasmids can be further investigated or sequenced. Using this method, plasmids were successfully captured and characterized from different environments such as the human gut, activated sludge from a sewage treatment plant, and human dental plaque associated with periodontal disease [43–45]. The advantage of the TRACA method is that plasmids from Gram-positive and Gram-negative species can be captured without the requirement for selectable markers, mobilization or conjugative functions. However, the results suggest that small plasmids (<10 kb) are preferentially isolated, which might be due to a lower copy number and lower transformation frequency of larger plasmids [43–45].
In the future a combination of different plasmid isolation, sequencing, and bioinformatics methods will be needed to improve our view of the diversity and accessory gene content of these important mobile genetic elements in the horizontal gene pool.
5. Exogenous capturing of plasmids by means of biparental and triparental matings
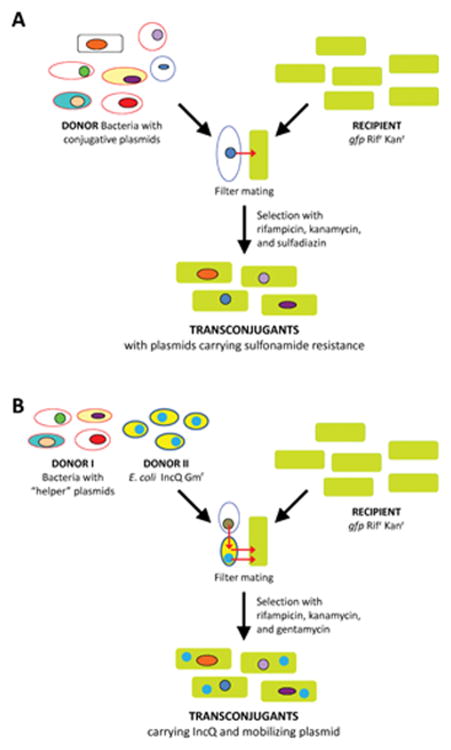
This method allows capturing conjugative as well as mobilizable plasmids from environmental bacteria without the need to culture their original host. The bacterial communities associated with an environmental sample are being mixed with recipient cells and after a filter mating on non-selective agar the cells are being re-suspended and plated on media containing rifampicin, kanamycin (to select for the recipient) and antibiotics or heavy metals to which the recipient is sensitive (Figure 2A). However, the few cells which received a mobile genetic element from the indigenous donor bacteria which confer the resistances required will form colonies on plates, and the transfer frequencies are in general given as the quotient of transconjugant and recipient numbers. The successful detection of transconjugants depends on the transfer and replication range as well as the presence and expression of respective resistance or degradative functions. In contrast to the biparental matings, the so-called triparental matings involve a second donor carrying a small mobilizable IncQ plasmid and the plasmid capturing is exclusively based on their plasmid mobilizing capacity (Figure 2B). Thus both methods can be seen as cultivation-independent methods although a successful transfer event requires cells of sufficient metabolical activity as conjugative type IV secretion (T4S) to translocate from a donor to a recipient bacterium is an energetically costly process [46]. A disadvantage of both capturing methods is that the original host remains unknown. But both methods allow capturing novel types of plasmids as shown for plasmids belonging to the PromA group [33,47,48] or the so-called LowG+C plasmid family [49,50].
Figure 2.
Exogenous capturing of plasmids by means of (A) biparental and (B) triparental mating. For biparental mating environmental bacteria are mixed with recipient cells and after a filter mating the cells are being re-suspended and plated on media containing rifampicin (Rif), kanamycin (Kan) (to select for the recipient) and antibiotics or heavy metals to which the recipient is sensitive. Triparental matings involve a second donor carrying a small mobilizable IncQ plasmid and the plasmid capturing is exclusively based on their plasmid-mobilizing capacity. To facilitate the identification of transconjugants the recipient is labeled with the green fluorescent protein (gfp).
A range of Gram-negative recipients have been successfully used in exogenous plasmid capturing experiments. Very often, independent from the environmental sample analyzed exogenously captured plasmids belonged to different types of IncP-1 plasmids. The frequent isolation of IncP-1 plasmids is remarkable as the relative abundance of IncP-1 plasmids is typically low, with the exception of some hot spots (e.g. sewage or BPS). However, also many other broad and narrow host range plasmids have been isolated with this method and subsequently characterized. Biparental matings with E. coli gfp as recipient strain were used to monitor the effect of the presence of antibiotics in manure added to soil on the frequency of capturing transferable antibiotic resistances [51]. Transfer frequencies from soils that did not receive manure were very low or below detection. In the presence of manure the transfer frequencies were several orders of magnitude higher compared to mating with control soil or the piggery manure. Interestingly, two months after the treatments from the sandy soil transconjugants were only captured when the manure was spiked with SDZ [51]. Thus from this type of experiments, we could once again show the importance of nutrient availability and selective pressure. Obviously, the latter depends on the physico-chemical properties of the antibiotic as these determine the interaction with the different anorganic and organic components of a soil or sediment sample [24]. It remains open whether the higher plasmid capturing frequencies were observed because of indeed higher transfer rates in the presence of antibiotics or because of the higher abundance of potential plasmid donors. In soil microcosms and in mesocosms planted with maize, or in field experiments with maize and grass, the majority of the plasmids, which were captured based on the sulfadiazine resistance conferred, belonged to the Low G+C plasmids and the IncP-1ε group [28,49,50,52].
When different recipient strains were used to exogenously capture plasmids, typically distinct sets of plasmids were obtained [53,54]. An exception was the capturing of similar IncP-1 plasmids in recipient strains that belonged to three different classes of the Proteobacteria (Agrobacterium tumefaciens, Cupriavidus necator, Pseudomonas putida, E. coli) from sewage sludge in Belgium [17]. Thus the type of plasmids captured into recipients obviously depends on the type and abundance of plasmid donor strains, the plasmid transfer and replication range but might be also influenced by the mating conditions (liquid or surface mating, duration, temperature, O2 availability, pH, chemotaxis). Variation of these factors might favor transfer/capture of distinct sets of plasmids essentially as they influence metabolic activity of the plasmid donor and host and the plasmid encoded transfer apparatus. Thus, exogenous plasmid isolation done in surface mating will obviously miss IncF and IncI types of plasmids that transfer better in broth matings. These plasmids seem to be highly important for antibiotic resistance and pathogenicity in E. coli.
6. Analyses of the plasmid content in bacterial isolates
Several chapters of this book discuss the in-depth characterization of plasmids from bacterial isolates such as enteropathogenic E. coli strains. That is why this chapter will focus on some aspects of the epidemiology and ecology of plasmids in cultivable bacteria. There is no doubt that plasmid ecology is tightly linked to the ecology of the host which remains unknown when plasmid occurrence and abundance is studied by total community DNA extraction. Here we picked some very recent studies in order to discuss the tools used for plasmid typing and diversity studies. Isolates were obtained after selective plating on nutrient media supplemented with a range of antibiotics. In other studies the plasmids originated from human, animal or plant pathogenic strains or from isolates with degradative functions. Many of the studies for obvious reasons focus on the detection of antibiotic resistance genes and their genetic context either by using a functional genomics approach or by investigating single isolates. While the fraction of bacteria forming colonies on plates likely does not represent the complete plasmid content of a given sample, the great advantage of the cultivation-based approach is that the localization of antibiotic resistance genes on the chromosome (integrons, ICE) or on plasmids and the genetic context of the acquired gene load can be analyzed. The presence and diversity of plasmids is traditionally assessed by plasmid DNA extraction and restriction analysis. To date the initial plasmid typing is done by PCR with primers targeting either plasmid replication [10,13,55] or transfer regions [56]. More details on the relaxase-based typing are given in the chapter by de Toro et al. [57] (this book). Plasmid multilocus sequencing typing is used for few plasmid families within the Enterobacteriaceae, e.g. IncI1 plasmids (http://pubmlst.org/plasmid/).
Prices for sequencing further decreasing it is likely that future plasmid typing will be done exclusively by sequencing. Moreover, the reconstruction of the plasmid content from whole genome sequencing data sets recently became feasible thanks to tools such as PLACNET (plasmid constellation networks). This tool was recently employed to analyze the plasmidome of 10 Escherichia coli lineage ST131 strains from whole genome sequencing (WGS) data sets [58]. The study showed that E. coli ST131 strains, which are practically identical in their core genomes, contain a striking variety of different plasmids including IncF and IncI.
Amos et al. [59] isolated Enterobacteriaceae resistant to extended spectrum beta-lactams from river sediments, taken at two time points upstream and downstream a sewage treatment. Third generation cephalosporin resistant Enterobacteria were enumerated and screened by PCR for blaTEM, blaSHV and blaCTX-M. Isolates which were positive for CTX-M15 were subjected to conjugation to a new E. coli background. These authors reported for the first time the occurrence of CTX-M-15 localized on IncF and IncI type plasmids in enterobacterial isolates from sediments. Several novel genetic contexts flanking the CTX-M-15 gene were discovered with a bunch of mobile genetic elements such as ISEcp1 and IS26 [59]. Although the number of samples investigated was limited, the data suggested the sewage treatment process increased the abundance of blaCTX-M-15 carrying Enterobacteriaceae in river sediments.
Many Gram-negative plant pathogenic bacteria carry genes involved in the interaction with plants, localized on pathogenicity islands or plasmids. We have recently characterized plant pathogenic Pseudomonas savastanoi strains isolated from different host plants and noticed that these strains could not be differentiated based on their 16S rRNA gene sequence and their BOX fingerprints. However, the isolates could clearly be distinguished based on their plasmid content. Based on their repA sequence the plasmids were assigned to the pAT family [60].
As described in the chapter by de Toro et al. [57] in this book, more and more plasmids are discovered and assembled from whole genome sequences of isolates. The assembly of plasmid replicons is particularly challenging due to the short read length of Illumina sequences and the high abundance of IS, ISCR, transposons, and truncated sequences due to multiple insertion processes. Bioinformatic tools are presently developed which will assist not only in the assembly of novel plasmids and comparative analyses, but also in their visualization. Publically available web-based tools are indeed urgently needed to manage the enormous plasmid-related sequence data sets. A few recently mentioned tools in addition to PLACNET introduced in the chapter by de Torro et al. [57] (this book) are Seqfindr and plasmid barcodes which were recently used for rapid plasmid profiling and comparison of plasmids encoding CTX-M-15 beta-lactamase genes from globally disseminated E. coli ST131 strains.
7. Plasmid host range determination and plasmid cost and benefits
The host range of a plasmid is thought to be a key parameter that determines plasmid ecology [61]. While traditionally plasmid host range studies were done by mating with a few selected recipient strains under optimal conditions, a comparative plasmid host range determination in the rhizosphere of grass was undertaken by Pukall et al. [62]. In this study E. coli donor strains carrying IncP-1 (pTH10, pTH16, RP4), IncQ (pIE639), IncN (pIE1037), IncW (pIE1056), IncI1 (pIE1040) or IncFII (pIE1055) plasmids, all carrying the Tn7-like transposon Tn1826 were applied together with an E. coli recipient strain into soil. The highest transfer frequencies were observed for IncP-1, followed by IncN and IncW, whereas under the same conditions no transfer was detected for IncI1 and IncFII. Fifty days after introducing the E. coli donor pTH16 and recipients into non-sterile soil, rhizosphere bacteria that captured the nourseothricin resistance plasmid were isolated and identified by BIOLOG as Agrobacterium, Pseudomonas and Flavobacterium. A similar approach was also taken to identify the host range of the IncP-1ε pHH3414 in soil microcosms planted with Acacia caven [63]. In this experiment soil bacteria that received pHH3414 were identified by 16S rRNA gene sequencing as Gammaproteobacteria (Enterobacter amnigenus, Xanthomonas codiaei) and Betaproteobacteria (Cupriavidus campinensis, Alcaligenes sp.). Soil microcosm experiments were also used by Goris et al. [36] to determine the host range of the catabolic plasmids pJP4 and pEMT1, conferring the ability to degrade the herbicide 2,4 dichlorophenoxyacetic acid. Recipients were mainly identified as Burkholderia species when no additional nutrients were added, while the amendment of the soils with nutrients resulted in additional transconjugants identified as Stenotrophomonas species. Thus again the plasmid hosts of both plasmids comprised Beta- and Gammaproteobacteria.
Fluorescent marker tagged plasmids have been used in different studies to elucidate the host range of plasmids. De Gelder et al. [64] could show for another rfp marked IncP-1 plasmid (pB10) that the recipients among a sewage sludge obtaining the plasmid depended on the donor (Ensifer meliloti, Cupriavidus necator, Pseudomonas putida) and the mating conditions (liquid vs plate matings). Recently, a molecular approach based on qPCR was presented to detect rare plasmid transfer events following a low initial pB10-donor inoculation to environmental samples, indicating that eukaryotic predation can affect plasmid transfer events [65]. Surprisingly, in the study by De Gelder et al. [64] the recipients of pB10 were affiliated to Alphaproteobacteria and Gammaproteobacteria. Also in the studies by Musovic et al. [66] Pseudomonas putida KT2440::lacIq1, harboring the IncP-ε plasmid conjugative plasmid pKJK10 was used to determine its host range in soil-barley microcosms. Plasmid pKJK10 had the gfp(mut3b) gene inserted downstream of the lac-repressible promoter PA1-04/03. Because this promoter is LacI repressed, the inserted gfp gene is silent in the donor strain. After seven days the bacterial fraction, collected by a Nycodenz centrifugation step, was subjected to cell sorting by flow cytometry. Transconjugants were obtained after cell sorting. 16S rRNA gene fragments amplified from gfp-positive cells revealed a broad diversity of transconjugants affiliated to Alpha-, Beta-, Gammaproteobacteria and, most remarkably, to Actinobacteria (Arthrobacter) as well. In a follow-up study Musovic et al. [67] developed a minimal-cultivation approach in combination with zygotic fluorescence expression and microscopy to quantify the recipient fraction of a soil bacterial community permissive to the gfp-tagged IncP-1α plasmid RP4. The authors showed that the host range which again comprised Alpha-, Beta-, and Gammaproteobacteria varied strongly dependent on the nutrient media used.
More recently cultivation-independent methods have been used to assess the in situ host range of plasmids. Klümper et al. [61] combined high throughput cell sorting of donor and transconjugant and used 454 sequencing of 16S rRNA gene fragments amplified from the transconjugant pools to determine the diversity of plasmid recipients in soil bacterial communities under conditions optimized for cell-to-cell contact. The plasmids RP4 (IncP-1α), pKJK5 (IncP-1ε) and pIPO2 (pPromA) were shown to have an unexpectedly broad transfer range, and more than 300 transconjugant OTUs were detected. Transconjugants did not only comprise all proteobacterial classes but also included diverse members of 10 additional phyla including Verrucomicrobia, Bacteriodetes and Actinobacteria. Thus several studies showed often a transfer range beyond the Gram-negative bacteria. It remains to be seen whether all these transconjugants also replicate the plasmid as separate mobile element or if integration in the chromosome allowed some or all of the genes to persist in these novel diverse hosts. This finding was confirmed by subsequent studies using the same technology but Shintani et al. [68] investigated the conjugative transfer ranges of three different gfp-tagged plasmids of the incompatibility groups IncP-1 (pBP136), IncP-7 (pCAR1), and IncP-9 (NAH7) in soil bacterial communities using both cultivation-dependent and cultivation-independent methods. Gfp-expressing transconjugants sorted by flow cytometry were characterized by sequencing of 16S rRNA genes following either whole-genome amplification or cultivation. In accordance with other studies the recipients of pBP136 belonged to diverse species within the phylum Proteobacteria as revealed by both culture-dependent and culture-independent methods. Transconjugants belonging to the phyla Actinobacteria, Bacteroidetes, and Firmicutes were detected only by the culture-independent method. Furthermore, the transconjugants of pCAR1 and NAH7 were identified by both methods as Pseudomonas indicating a rather narrow host range of the plasmids. The cultivation-independent methods indicated that Delftia species (class Betaproteobacteria) were “transient” hosts of pCAR1. Thus several studies showed that the transfer range of different plasmids studied might be far broader than previously expected which has implications for the spread of plasmid-encoded accessory genes. This was already known from matings in the laboratory [69], and is now confirmed for in situ plasmid spread. Additionally, it was shown that the permissiveness of a bacterial community and even of isolates to receive and maintain plasmids can change by several orders of magnitude in response to irregular environmental change such as manure fertilization [63,70].
It is well known that most plasmids impose a cost on their host when their accessory genes are not providing a benefit to that host, such as antibiotic resistance plasmids in the absence of antibiotics. In addition to the more obvious cases of fitness due to expression of costly proteins, recent studies are also trying to understand the more subtle interactions between plasmids and their hosts and the molecular level. For example, the impact of plasmid pCAR1 carriage on the expression of chromosomal genes of three different Pseudomonas hosts was investigated by transcriptome analyses of plasmid-free and pCAR1 carrying strains [11,71]. The transcriptomic studies revealed that carriage of the IncP-7 plasmid pCAR1 altered the gene expression of the iron acquisition in all strains likely due to the expression of the carbazole degradative genes carried on pCAR1. In P. putida KT2440 the expression of the pyoverdine gene was higher in the plasmid-carrying strain and also genes involved SOS response (lexA, recA), genes on a putative prophage were upregulated due to plasmid carriage. Furthermore an increased chloramphenicol resistance phenotype was observed. In vitro evolution analyses showed that the fitness cost of pCAR1 might be partially due to the pCAR1-induced constitutive expression of chromosomal genes and iron shortage induced due to car gene expression (Nojiri et al., in preparation).
In addition to the many competition studies performed in liquid cultures, recent work has shown the effect of antibiotic presence on the competitiveness of plasmid-bearing strains in soil. In two independent competition experiments with plasmid-free and plasmid-carrying Acinetobacter baylyi BD413 performed in soil microcosms, Jechalke et al. [72] demonstrated that the LowGC-type plasmid pHHV216 conferred a fitness advantage to Acinetobacter baylyi BD413 in soils that had received manure spiked with sulfonamide antibiotic sulfadiazine. Plasmid carriage had a disadvantage when the selective pressure by SDZ was absent. A very interesting study recently showed that even sublethal levels of antibiotics, far under the MIC, selected for plasmid-bearing strains in laboratory competitions with plasmid-free isogenic strains [73]. The findings suggest that even when growth inhibition is not detected by the traditional MIC assays, the antibiotic impedes growth enough to allow the drug resistance plasmid-bearing strain to outcompete its plasmid-free counterpart. To determine the true cost and benefits of plasmids to their hosts more studies need to be performed in the hosts’ natural environments and at various concentrations of selecting agents.
At the rate at which new plasmid sequence information is being released, it is no longer possible to empirically test the host range of all newly described plasmids. Therefore, genomics-based methods may be used to predict the likely host and host range of a specific plasmid based on its DNA sequence. Bacteria clearly differ in the relative abundance of di-, tri-, or tetranucleotides (also referred to as their genomic sequence signature), and it appears that plasmids that have a long-term association with hosts of a similar signature tend to acquire that signature [74]. In contrast, broad-host-range plasmids rather thought to move between distantly related bacteria, show distinct genomic signatures [75]. Therefore we can now easily examine the genomic signature of uncharacterized plasmids and infer their likely host or host range. While the method often accurately predicts a plasmid’s likely hosts in the case of well-characterized plasmids, more experimental validation is needed for some novel key plasmids that are being sequenced as part of genome and metagenome projects.
8. Conclusions
During the past two decades, significant progress has been made in our ability to detect and quantify specific groups of plasmids in environmental and clinical samples, and to determine and compare their entire genome sequences. This expansion in studies and methods has greatly increased our understanding of the diversity of plasmids that exist, even within already well-studied groups such as the IncP-1 plasmids. While until 2004 only two IncP-1 subgroups had been described, there are now, just a decade later, likely seven phylogenetically distinct clades [5,31]. We expect that the high pace of development of new genome sequencing methods and creative approaches such as applying Hi-C to metagenomic studies will further enhance our insight into the important role of plasmids in the rapid adaptation of microbial communities to man-made changes in their environment. Obvious examples of such environmental changes are the increased presence of antibiotics, heavy metals and xenobiotics, but also increased world-wide traveling, high densities of food animals and people, exploration of new territories, and climate change, which likely affect spread of bacterial pathogens and their often mobile virulence genes. There are plenty of challenges ahead of us, but given the progress made in the past decades these are exciting times for plasmid ecology.
Acknowledgments
S.J. was funded by the Federal Environment Agency (Umweltbundesamt) (FKZ 3713 63 402). We are also grateful for funding from NIH grant no. R01 AI084918 from the National Institute of Allergy and Infectious Diseases (NIAID) and DOD grant DM110149 to E.M.T. We thank Wesley Loftie-Eaton for drawing the figure 1.
References
- 1.Heuer H, Abdo Z, Smalla K. Patchy distribution of flexible genetic elements in bacterial populations mediates robustness to environmental uncertainty. FEMS Microbiol Ecol. 2008;65:361–371. doi: 10.1111/j.1574-6941.2008.00539.x. [DOI] [PubMed] [Google Scholar]
- 2.Heuer H, Smalla K. Plasmids foster diversification and adaptation of bacterial populations in soil. FEMS Microbiol Rev. 2012;36:1083–1104. doi: 10.1111/j.1574-6976.2012.00337.x. [DOI] [PubMed] [Google Scholar]
- 3.Djordjevic SP, Stokes HW, Roy Chowdhury P. Mobile elements, zoonotic pathogens and commensal bacteria: Conduits for the delivery of resistance genes into humans, production animals and soil microbiotia. Front Microbiol. 2013;4 doi: 10.3389/fmicb.2013.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schlüter A, Szczepanowski R, Pühler A, Top EM. Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol Rev. 2007;31:449–477. doi: 10.1111/j.1574-6976.2007.00074.x. [DOI] [PubMed] [Google Scholar]
- 5.Norberg P, Bergstrom M, Jethava V, Dubhashi D, Hermansson M. The IncP-1 plasmid backbone adapts to different host bacterial species and evolves through homologous recombination. Nat Commun. 2011;2 doi: 10.1038/ncomms1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sen D, Brown CJ, Top EM, Sullivan J. Inferring the evolutionary history of IncP-1 plasmids despite incongruence among backbone gene trees. Mol Biol Evol. 2012 doi: 10.1093/molbev/mss210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Staley JT, Konopka A. Measurement of in-situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Ornston, L N. 1985:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- 8.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliver JD. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol Rev. 2010;34:415–425. doi: 10.1111/j.1574-6976.2009.00200.x. [DOI] [PubMed] [Google Scholar]
- 10.Dealtry S, Ding GC, Weichelt V, Dunon V, Schlüter A, Martini MC, Del Papa MF, Lagares A, Amos GCA, Wellington EMH, Gaze WH, Sipkema D, Sjöling S, Springael D, Heuer H, van Elsas JD, Thomas C, Smalla K. Cultivation-independent screening revealed hot spots of IncP-1, IncP-7 and IncP-9 plasmid occurrence in different environmental habitats. Plos One. 2014;9 doi: 10.1371/journal.pone.0089922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shintani M, Takahashi Y, Yamane H, Nojiri H. The behavior and significance of degradative plasmids belonging to Inc groups in Pseudomonas within natural environments and microcosms. Microbes Environ. 2010;25:253–265. doi: 10.1264/jsme2.me10155. [DOI] [PubMed] [Google Scholar]
- 12.Dennis JJ. The evolution of IncP catabolic plasmids. Curr Opin Biotechnol. 2005;16:291–298. doi: 10.1016/j.copbio.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Bahl MI, Burmølle M, Meisner A, Hansen LH, Sørensen SJ. All IncP-1 plasmid subgroups, including the novel ε subgroup, are prevalent in the influent of a Danish wastewater treatment plant. Plasmid. 2009;62:134–139. doi: 10.1016/j.plasmid.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 14.Götz A, Pukall R, Smit E, Tietze E, Prager R, Tschäpe H, Van Elsas JD, Smalla K. Detection and characterization of broad-host-range plasmids in environmental bacteria by PCR. Appl Environ Microbiol. 1996;62:2621–2628. doi: 10.1128/aem.62.7.2621-2628.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dealtry S, Holmsgaard PN, Dunon V, Jechalke S, Ding GC, Krögerrecklenfort E, Heuer H, Hansen LH, Springael D, Zühlke S, Sørensen SJ, Smalla K. Shifts in abundance and diversity of mobile genetic elements after the introduction of diverse pesticides into an on-farm biopurification system over the course of a year. Appl Environ Microbiol. 2014;80:4012–4020. doi: 10.1128/AEM.04016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmsgaard PN, Sørensen SJ, Hansen LH. Simultaneous pyrosequencing of the 16S rRNA, IncP-1 trfA, and merA genes. J Microbiol Methods. 2013;95:280–284. doi: 10.1016/j.mimet.2013.09.016. [DOI] [PubMed] [Google Scholar]
- 17.Heuer H, Krögerrecklenfort E, Wellington EMH, Egan S, van Elsas JD, van Overbeek L, Collard JM, Guillaume G, Karagouni AD, Nikolakopoulou TL, Smalla K. Gentamicin resistance genes in environmental bacteria: prevalence and transfer. FEMS Microbiol Ecol. 2002;42:289–302. doi: 10.1111/j.1574-6941.2002.tb01019.x. [DOI] [PubMed] [Google Scholar]
- 18.Smalla K, Haines AS, Jones K, Krögerrecklenfort E, Heuer H, Schloter M, Thomas CM. Increased abundance of IncP-1 beta plasmids and mercury resistance genes in mercury-polluted river sediments: First discovery of IncP-1 beta plasmids with a complex mer transposon as the sole accessory element. Appl Environ Microbiol. 2006;72:7253–7259. doi: 10.1128/AEM.00922-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Binh CTT, Heuer H, Kaupenjohann M, Smalla K. Piggery manure used for soil fertilization is a reservoir for transferable antibiotic resistance plasmids. FEMS Microbiol Ecol. 2008;66:25–37. doi: 10.1111/j.1574-6941.2008.00526.x. [DOI] [PubMed] [Google Scholar]
- 20.Jutkina J, Heinaru E, Vedler E, Juhanson J, Heinaru A. Occurrence of plasmids in the aromatic degrading bacterioplankton of the Baltic Sea. Genes. 2011;2:853–868. doi: 10.3390/genes2040853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Y-G, Johnson TA, Su J-Q, Qiao M, Guo G-X, Stedtfeld RD, Hashsham SA, Tiedje JM. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1222743110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jechalke S, Schreiter S, Wolters B, Dealtry S, Heuer H, Smalla K. Widespread dissemination of class 1 integron components in soils and related ecosystems as revealed by cultivation-independent analysis. Front Microbiol. 2014;4 doi: 10.3389/fmicb.2013.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jechalke S, Dealtry S, Smalla K, Heuer H. Quantification of IncP-1 plasmid prevalence in environmental samples. Appl Environ Microbiol. 2013;79:1410–1413. doi: 10.1128/AEM.03728-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jechalke S, Heuer H, Siemens J, Amelung W, Smalla K. Fate and effects of veterinary antibiotics in soil. Trends Microbiol. 2014;22:536–545. doi: 10.1016/j.tim.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 25.Rosche TM, Siddique A, Larsen MH, Figurski DH. Incompatibility protein IncC and global regulator KorB interact in active partition of promiscuous plasmid RK2. J Bacteriol. 2000;182:6014–6026. doi: 10.1128/jb.182.21.6014-6026.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herman D, Thomas CM, Stekel DJ. Global transcription regulation of RK2 plasmids: a case study in the combined use of dynamical mathematical models and statistical inference for integration of experimental data and hypothesis exploration. Bmc Systems Biology. 2011;5 doi: 10.1186/1752-0509-5-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jechalke S, Focks A, Rosendahl I, Groeneweg J, Siemens J, Heuer H, Smalla K. Structural and functional response of the soil bacterial community to application of manure from difloxacin-treated pigs. FEMS Microbiol Ecol. 2013;87:78–88. doi: 10.1111/1574-6941.12191. [DOI] [PubMed] [Google Scholar]
- 28.Kopmann C, Jechalke S, Rosendahl I, Groeneweg J, Krögerrecklenfort E, Zimmerling U, Weichelt V, Siemens J, Amelung W, Heuer H, Smalla K. Abundance and transferability of antibiotic resistance as related to the fate of sulfadiazine in maize rhizosphere and bulk soil. FEMS Microbiol Ecol. 2013;83:125–134. doi: 10.1111/j.1574-6941.2012.01458.x. [DOI] [PubMed] [Google Scholar]
- 29.Schreiter S, Ding G-c, Heuer H, Neumann G, Sandmann M, Grosch R, Kropf S, Smalla K. Effect of the soil type on the microbiome in the rhizosphere of field-grown lettuce. Front Microbiol. 2014;5:144. doi: 10.3389/fmicb.2014.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neumann G, Bott S, Ohler MA, Mock HP, Lippmann R, Grosch R, Smalla K. Root exudation and root development of lettuce (Lactuca sativa L. cv. Tizian) as affected by different soils. Front Microbiol. 2014;5 doi: 10.3389/fmicb.2014.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown CJ, Sen DY, Yano H, Bauer ML, Rogers LM, Van der Auwera GA, Top EM. Diverse broad-host-range plasmids from freshwater carry few accessory genes. Appl Environ Microbiol. 2013;79:7684–7695. doi: 10.1128/AEM.02252-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Król JE, Penrod JT, McCaslin H, Rogers LM, Yano H, Stancik AD, Dejonghe W, Brown CJ, Parales RE, Wuertz S, Top EM. Role of IncP-1 beta plasmids pWDL7::rfp and pNB8c in chloroaniline catabolism as determined by genomic and functional analyses. Appl Environ Microbiol. 2012;78:828–838. doi: 10.1128/AEM.07480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tauch A, Schneiker S, Selbitschka W, Puhler A, van Overbeek LS, Smalla K, Thomas CM, Bailey MJ, Forney LJ, Weightman A, Ceglowski P, Pembroke T, Tietze E, Schroder G, Lanka E, van Elsas JD. The complete nucleotide sequence and environmental distribution of the cryptic, conjugative, broad-host-range plasmid pIPO2 isolated from bacteria of the wheat rhizosphere. Microbiology-(UK) 2002;148:1637–1653. doi: 10.1099/00221287-148-6-1637. [DOI] [PubMed] [Google Scholar]
- 34.Gstalder ME, Faelen M, Mine N, Top EM, Mergeay M, Couturier M. Replication functions of new broad host range plasmids isolated from polluted soils. Res Microbiol. 2003;154:499–509. doi: 10.1016/S0923-2508(03)00143-8. [DOI] [PubMed] [Google Scholar]
- 35.Frost LS, Thomas CM. Naming and annotation of plasmids. Springer Science + Business Media; New York: 2014. [DOI] [Google Scholar]
- 36.Goris J, Dejonghe W, Falsen E, De Clerck E, Geeraerts B, Willems A, Top EM, Vandamme P, De Vos P. Diversity of transconjugants that acquired plasmid pJP4 or pEMT1 after inoculation of a donor strain in the A- and B-horizon of an agricultural soil and description of Burkholderia hospita sp nov and Burkholderia terricola sp nov. Syst Appl Microbiol. 2002;25:340–352. doi: 10.1078/0723-2020-00134. [DOI] [PubMed] [Google Scholar]
- 37.Sentchilo V, Mayer AP, Guy L, Miyazaki R, Tringe SG, Barry K, Malfatti S, Goessmann A, Robinson-Rechavi M, Van der Meer JR. Community-wide plasmid gene mobilization and selection. Isme J. 2013;7:1173–1186. doi: 10.1038/ismej.2013.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norman A, Riber L, Luo WT, Li LL, Hansen LH, Sørensen SJ. An improved method for including upper size range plasmids in metamobilomes. Plos One. 2014;9 doi: 10.1371/journal.pone.0104405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li LL, Norman A, Hansen LH, Sørensen SJ. Metamobilomics — expanding our knowledge on the pool of plasmid encoded traits in natural environments using high-throughput sequencing. Clin Microbiol Infect. 2012;18:5–7. doi: 10.1111/j.1469-0691.2012.03862.x. [DOI] [PubMed] [Google Scholar]
- 40.Umbarger MA, Toro E, Wright MA, Porreca GJ, Bau D, Hong SH, Fero MJ, Zhu LJ, Marti-Renom MA, McAdams HH, Shapiro L, Dekker J, Church GM. The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation. Mol Cell. 2011;44:252–264. doi: 10.1016/j.molcel.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burton JN, Liachko I, Dunham MJ, Shendure J. Species-level deconvolution of metagenome assemblies with Hi-C-based contact probability maps. G3-Genes Genomes Genetics. 2014;4:1339–1346. doi: 10.1534/g3.114.011825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beitel CW, Froenicke L, Lang JM, Korf IF, Michelmore RW, Eisen JA, Darling AE. Strain- and plasmid-level deconvolution of a synthetic metagenome by sequencing proximity ligation products. PeerJ. 2014;2:e415. doi: 10.7717/peerj.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones BV, Marchesi JR. Transposon-aided capture (TRACA) of plasmids resident in the human gut mobile metagenome. Nature Methods. 2007;4:55–61. doi: 10.1038/nmeth964. [DOI] [PubMed] [Google Scholar]
- 44.Warburton PJ, Allan E, Hunter S, Ward J, Booth V, Wade WG, Mullany P. Isolation of bacterial extrachromosomal DNA from human dental plaque associated with periodontal disease, using transposonaided capture (TRACA) FEMS Microbiol Ecol. 2011;523:349–354. doi: 10.1111/j.1574-6941.2011.01166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang T, Zhang X-X, Ye L. Plasmid metagenome reveals high levels of antibiotic resistance genes and mobile genetic elements in activated sludge. PLoS ONE. 2011;6:e26041. doi: 10.1371/journal.pone.0026041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Low HH, Gubellini F, Rivera-Calzada A, Braun N, Connery S, Dujeancourt A, Lu F, Redzej A, Fronzes R, Orlova EV, Waksman G. Structure of a type IV secretion system. Nature. 2014;508:550–553. doi: 10.1038/nature13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Elsas JD, Gardener BBM, Wolters AC, Smit E. Isolation, characterization, and transfer of cryptic gene-mobilizing plasmids in the wheat rhizosphere. Appl Environ Microbiol. 1998;64:880–889. doi: 10.1128/aem.64.3.880-889.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneiker S, Keller M, Droge M, Lanka E, Puhler A, Selbitschka W. The genetic organization and evolution of the broad host range mercury resistance plasmid pSB102 isolated from a microbial population residing in the rhizosphere of alfalfa. Nucleic Acids Res. 2001;29:5169–5181. doi: 10.1093/nar/29.24.5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heuer H, Kopmann C, Binh CTT, Top EM, Smalla K. Spreading antibiotic resistance through spread manure: characteristics of a novel plasmid type with low %G plus C content. Environ Microbiol. 2009;11:937–949. doi: 10.1111/j.1462-2920.2008.01819.x. [DOI] [PubMed] [Google Scholar]
- 50.Jechalke S, Kopmann C, Rosendahl I, Groeneweg J, Weichelt V, Krögerrecklenfort E, Brandes N, Nordwig M, Ding G-C, Siemens J, Heuer H, Smalla K. Increased abundance and transferability of resistance genes after field application of manure from sulfadiazine-treated pigs. Appl Environ Microbiol. 2013;79:1704–1711. doi: 10.1128/AEM.03172-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heuer H, Smalla K. Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months. Environ Microbiol. 2007;9:657–666. doi: 10.1111/j.1462-2920.2006.01185.x. [DOI] [PubMed] [Google Scholar]
- 52.Heuer H, Binh CTT, Jechalke S, Kopmann C, Zimmerling U, Krögerrecklenfort E, Ledger T, González B, Top EM, Smalla K. IncP-1ε plasmids are important vectors of antibiotic resistance genes in agricultural systems: diversification driven by class 1 integron gene cassettes. Front Microbiol. 2012;3 doi: 10.3389/fmicb.2012.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smalla K, Heuer H, Götz A, Niemeyer D, Krögerrecklenfort E, Tietze E. Exogenous isolation of antibiotic resistance plasmids from piggery manure slurries reveals a high prevalence and diversity of IncQ-like plasmids. Appl Environ Microbiol. 2000;66:4854–4862. doi: 10.1128/aem.66.11.4854-4862.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dronen AK, Torsvik V, Top EM. Comparison of the plasmid types obtained by two distantly related recipients in biparental exogenous plasmid isolations from soil. FEMS Microbiol Lett. 1999;176:105–110. [Google Scholar]
- 55.Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother. 2009;53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alvarado A, Garcillan-Barcia MP, de la Cruz F. A degenerate primer MOB typing (DPMT) method to classify gamma-Proteobacterial plasmids in clinical and environmental settings. Plos One. 2012;7 doi: 10.1371/journal.pone.0040438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Toro M, Garcillán-Barcia MP, de la Cruz F. Plasmid diversity and adaptation analyzed by massive sequencing of Escherichia coli plasmids. Microbiol Spectrum. 2014;2 doi: 10.1128/microbiolspec.PLAS-0031-2014. [DOI] [PubMed] [Google Scholar]
- 58.Lanza VF, de Toro M, Garcillán-Barcia MP, Mora A, Blanco J, Coque TM, De la Cruz F. Plasmid flux in Escherichia coli ST131 sublineages, analyzed by plasmid constellation network (PLACNET), a new method for plasmid reconstruction from whole genome sequences. doi: 10.1371/journal.pgen.1004766. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amos GCA, Hawkey PM, Gaze WH, Wellington EM. Waste water effluent contributes to the dissemination of CTX-M-15 in the natural environment. J Antimicrob Chemother. 2014;69:1785–1791. doi: 10.1093/jac/dku079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eltlbany N, Prokscha ZZ, Castaneda-Ojeda MP, Krogerrecklenfort E, Heuer H, Wohanka W, Ramos C, Smalla K. A new bacterial disease on Mandevilla sanderi, caused by Pseudomonas savastanoi: lessons learned for bacterial diversity studies. Appl Environ Microbiol. 2012;78:8492–8497. doi: 10.1128/AEM.02049-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klümper U, Riber L, Dechesne A, Sannazzarro A, Hansen LH, Sørensen SJ, Smets BF. Broad host range plasmids can invade an unexpectedly diverse fraction of a soil bacterial community. ISME J. 2014 doi: 10.1038/ismej.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pukall R, Tschäpe H, Smalla K. Monitoring the spread of broad host and narrow host range plasmids in soil microcosms. FEMS Microbiol Ecol. 1996;20:53–66. [Google Scholar]
- 63.Heuer H, Ebers J, Weinert N, Smalla K. Variation in permissiveness for broad-host-range plasmids among genetically indistinguishable isolates of Dickeya sp. from a small field plot. FEMS Microbiol Ecol. 2010;73:190–196. doi: 10.1111/j.1574-6941.2010.00880.x. [DOI] [PubMed] [Google Scholar]
- 64.De Gelder L, Vandecasteele FPJ, Brown CJ, Forney LJ, Top EM. Plasmid donor affects host range of promiscuous IncP-1 beta plasmid pB10 in an activated-sludge microbial community. Appl Environ Microbiol. 2005;71:5309–5317. doi: 10.1128/AEM.71.9.5309-5317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bellanger X, Guilloteau H, Bonot S, Merlin C. Demonstrating plasmid-based horizontal gene transfer in complex environmental matrices: A practical approach for a critical review. Sci Total Environ. 2014;493:872–882. doi: 10.1016/j.scitotenv.2014.06.070. [DOI] [PubMed] [Google Scholar]
- 66.Musovic S, Oregaard G, Kroer N, Sorensen SJ. Cultivation-independent examination of horizontal transfer and host range of an IncP-1 plasmid among Gram-positive and Gram-negative bacteria indigenous to the barley rhizosphere. Appl Environ Microbiol. 2006;72:6687–6692. doi: 10.1128/AEM.00013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Musovic S, Dechesne A, Sørensen J, Smets BF. Novel assay to assess permissiveness of a soil microbial community toward receipt of mobile genetic elements. Appl Environ Microbiol. 2010;76:4813–4818. doi: 10.1128/AEM.02713-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shintani M, Matsui K, Inoue J, Hosoyama A, Ohji S, Yamazoe A, Nojiri H, Kimbara K, Ohkuma M. Single-cell analyses revealed transfer ranges of IncP-1, IncP-7, and IncP-9 plasmids in a soil bacterial community. Appl Environ Microbiol. 2014;80:138–145. doi: 10.1128/AEM.02571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thomas CM, Smith CA. Incompatibility group-P plasmids - genetics, evolution, and use in genetic manipulation. Annu Rev Microbiol. 1987;41:77–101. doi: 10.1146/annurev.mi.41.100187.000453. [DOI] [PubMed] [Google Scholar]
- 70.Musovic S, Klumper U, Dechesne A, Magid J, Smets BF. Long- term manure exposure increases soil bacterial community potential for plasmid uptake. Environ Microbiol Rep. 2014;6:125–130. doi: 10.1111/1758-2229.12138. [DOI] [PubMed] [Google Scholar]
- 71.Takahashi Y, Shintani M, Takase N, Kazo Y, Kawamura F, Hara H, Nishida H, Okada K, Yamane H, Nojiri H. Modulation of primary cell function of host Pseudomonas bacteria by the conjugative plasmid pCAR1. Environ Microbiol. 2014 doi: 10.1111/1462-2920.12515. [DOI] [PubMed] [Google Scholar]
- 72.Jechalke S, Kopmann C, Richter M, Moenickes S, Heuer H, Smalla K. Plasmid-mediated fitness advantage of Acinetobacter baylyi in sulfadiazine-polluted soil. FEMS Microbiol Lett. 2013;348:127–132. doi: 10.1111/1574-6968.12284. [DOI] [PubMed] [Google Scholar]
- 73.Gullberg E, Albrecht LM, Karlsson C, Sandegren L, Andersson DI. Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. mBio. 2014;5 doi: 10.1128/mBio.01918-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Suzuki H, Sota M, Brown CJ, Top EM. Using Mahalanobis distance to compare genomic signatures between bacterial plasmids and chromosomes. Nucleic Acids Res. 2008;36:e147. doi: 10.1093/nar/gkn753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suzuki H, Yano H, Brown CJ, Top EM. Predicting plasmid promiscuity based on genomic signature. J Bacteriol. 2010;192:6045–6055. doi: 10.1128/JB.00277-10. [DOI] [PMC free article] [PubMed] [Google Scholar]