Abstract
The human neocortex has numerous specialized functional areas whose formation is poorly understood. Here, we describe a 15–base pair deletion mutation in a regulatory element of GPR56 that selectively disrupts human cortex surrounding the Sylvian fissure bilaterally including “Broca’s area,” the primary language area, by disrupting regional GPR56 expression and blocking RFX transcription factor binding. GPR56 encodes a heterotrimeric guanine nucleotide–binding protein (G protein)–coupled receptor required for normal cortical development and is expressed in cortical progenitor cells. GPR56 expression levels regulate progenitor proliferation. GPR56 splice forms are highly variable between mice and humans, and the regulatory element of gyrencephalic mammals directs restricted lateral cortical expression. Our data reveal a mechanism by which control of GPR56 expression pattern by multiple alternative promoters can influence stem cell proliferation, gyral patterning, and, potentially, neocortex evolution.
Although most mammals have elaborate and species-specific patterns of folds (“gyri”) in the neocortex, the genetic and evolutionary mechanisms of cortical gyrification are poorly understood (1–3). Abnormal gyrification, such as polymicrogyria (too many small gyri), invariably signals abnormal cortical development, so regional disorders of gyrification are of particular interest, because they highlight mechanisms specific to cortical regions. The human cortex contains dozens of cortical regions specialized for distinct functions—such as language, hearing, and sensation—yet it is unsolved how these cortical regions form and how human cortical regions evolved from those of prehuman ancestors.
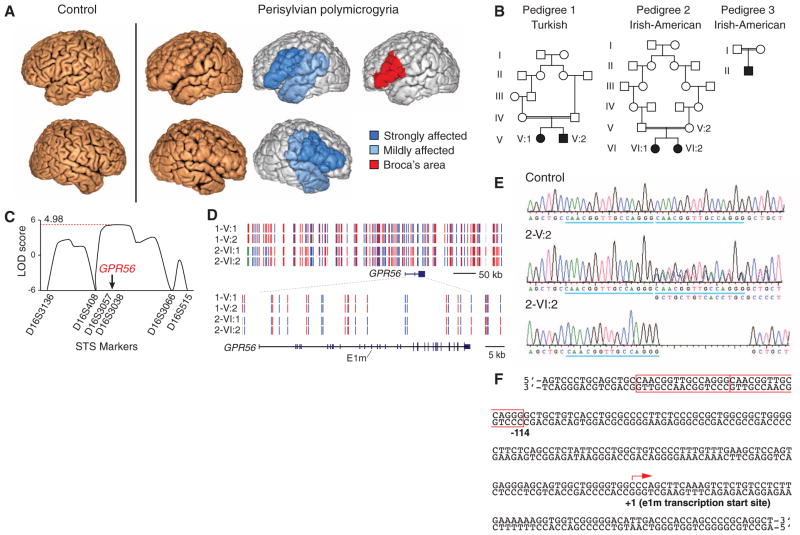
Examination of >1000 individuals with gyral abnormalities identified five individuals from three families (one Turkish and two Irish-American) with strikingly restricted polymicrogyria limited to the cortex surrounding the Sylvian fissure (Fig. 1, A and B; fig. S1; and movies S1 and S2), which suggests a rare, but genetically distinctive, condition. Affected individuals suffered intellectual and language difficulty, as well as refractory seizures (onset 7 months to 10 years), but had no motor disability (table S1). Magnetic resonance imaging (MRI) and quantitative gyral analysis showed abnormal inferior and middle gyri in pre-frontal and motor cortex, with mildly affected temporal lobes. Broca’s area—the “motor center for speech” (4)—in the left hemisphere and the corresponding areas of the right hemisphere were most severely affected. Affected neocortical surface showed abnormally numerous, small gyral-like folds that fused in coarse, irregular patterns, with abnormal and highly irregular white matter protrusions, consistent with polymicrogyria (5, 6), along with widening of the Sylvian fissure (Fig. 1A and fig. S1B).
Fig. 1. A noncoding mutation in the GPR56 gene disrupts perisylvian gyri.
(A) MRI shows polymicrogyria in the perisylvian area, in which abnormally thin cortex is folded in on itself, giving a paradoxical, but characteristic, thickened appearance (8). (B) Pedigrees of the three families with perisylvian polymicrogyria. (C) Linkage analysis isolates an interval containing GPR56. LOD, logarithm of the odds ratio for linkage. (D) The mutation arose independently in the Turkish and the Irish-American families. Haplotype mapping shows that pedigree 1 (1-V:1, 1-V:2) and pedigree 2 (2-VI:1, 2-VI:2) are unrelated. Homozygous single-nucleotide polymorphisms (SNPs) are shown in red or blue, heterozygous SNPs in green, and SNPs for which no genotype could be assigned in gray. (E and F) A homozygous deletion in one of two 15-bp tandem repeats (blue underscore and red box) upstream of GPR56 e1m causes perisylvian polymicrogyria. 2-V:2 stands for a heterozygous parent and 2-VI:2 for an affected individual from pedigree 2.
Genome-wide analysis identified a single linked locus on chromosome 16q12.2-21 (Fig. 1C) containing the GPR56 gene, which, when mutated in its coding region, leads to polymicrogyria of the entire neocortex, as well as cerebellar and white matter abnormalities (7–9). As we found no mutations in the exons of GPR56, we sequenced 38 conserved non-exonic elements (table S2), in one of which we identified a small deletion in all five individuals. The mutated element normally contains two copies of a 15–base pair (bp) tandem repeat, but all affected individuals have a homozygous deletion of one 15-bp repeat (Fig. 1, E and F). The deletion is heterozygous in parents of the affected individuals, who manifest no obvious clinical signs, and is absent from thousands of control chromosomes in the Single-Nucleotide Polymorphism Database and 1000 Genomes database. The two Irish-American families carry the mutation on the same chromosomal haplotype, which reflects a common founder. It is noteworthy that the Turkish family carries the same deletion on a distinct haplotype, which indicates that the mutation arose independently (Fig. 1D). The element is located <150 bp upstream of the transcriptional start site of noncoding exon 1m (e1m) of GPR56, which suggests that it may regulate e1m expression as a cis-regulatory element. GPR56 has at least 17 alternative transcription start sites, each beginning from a different non-coding first exon; all of the start sites are predicted to drive transcription of mRNAs whose coding sequence starts from exon 3 (Fig. 2A and fig. S2A) and all of which encode the same GPR56 protein (10, 11). The diverse noncoding first exons have distinct expression profiles, with e1m being the most robustly transcribed first exon in fetal human brain but with several other alternative transcripts also expressed in fetal and adult brain (Fig. 2A and fig. S2, B to D).
Fig. 2. The noncoding mutation ablates lateral gene expression.
(A) Schematic of the human GPR56 locus showing 17 alternative transcription start sites. E1m is highly expressed in the human fetal brain [mRNA-sequencing (mRNA-Seq) track, arrow]. The 15-bp deletion is upstream of e1m, located within a cis-regulatory element as one of two tandem 15-bp repeats. (B) A 23-kb upstream region of human GPR56 drives GFP expression throughout the transgenic mouse neocortex (E14.5), which mirrors endogenous GPR56 protein expression. The 15-bp deletion eliminates GFP expression from lateral cortex but preserves medial cortex expression, consistent with lesions observed by brain MRI (fig. S1) (n = 4 to 6 embryos with identical patterns per construct). Scale bar, 200 μm. (C) Y1H screening reveals Rfx transcription factor binding to the cis-regulatory element. See text for details. (D) The mutation decreases RFX binding to the cis-regulatory element in vitro. (E) RFX1 and GPR56 are colocalized in a human fetal brain 19 weeks after conception. Higher magnification of the outer subventricular zone is shown (right). v, ventricular zone; is, inner subventricular zone; os, outer subventricular zone; and i, intermediate zone. Scale bars, 100 μm (left) and 10 μm (right). (F) Dominant-negative RFX (white bars) abrogates normal e1m promoter activity. Black bars, GFP control. (G) Each RFX gene has distinct expression patterns in the fetal human brain. Each number means the corresponding RFX isoform. RFX3 and RFX7 are enriched in regions affected by perisylvian polymicrogyria (green boxes). pfc, prefrontal cortex; opfc, orbital pfc; dlpfc, dorsolateral pfc, mpfc, medial pfc; vlpfc, ventrolateral pfc; ms, motor-sensory cortex. *P < 0.001, t test.
To confirm that the 15-bp deletion disrupts perisylvian GPR56 expression, we generated transgenic mice with the 23-kb human GPR56 upstream region driving green fluorescent protein (GFP) expression. The 23-kb region encompasses 16 of the 17 transcription start sites containing e1m and ends before the translation start codon (Fig. 2A). This construct drives GFP expression in the entire central nervous system, including neocortex, and recapitulates the location and relative amount of expression of endogenous mouse GPR56 protein (Fig. 2B and fig. S3). In contrast, the 23-kb construct containing the 15-bp deletion drives expression in medial, but not lateral, cortex or lateral ganglionic eminence (Fig. 2B). These data suggest that the cis-regulatory element upstream of e1m drives GPR56 expression in the perisylvian and lateral cortex, whereas disruption of the element, with consequent impairment of e1m expression, causes the perisylvian malformation.
To elucidate how the 15-bp deletion in the cis-regulatory element disrupts e1m expression, we performed yeast one-hybrid (Y1H) screening of a mouse forebrain cDNA library with the human cis-regulatory element as bait and obtained multiple yeast colonies encoding members of the regulatory factor X (Rfx) transcription factor family (Fig. 2C) (12). RFX1 and RFX3 bind the normal element in vitro, with binding decreased 60 to 70% by the 15-bp deletion (Fig. 2D). Chromatin immunoprecipitation sequencing confirmed RFX3 binding to the element (fig. S4) (13). RFX1 and GPR56 colocalize in germinal zones of fetal human brain (Fig. 2E). Dominant-negative RFX abrogates normal, but not mutant, e1m promoter activity on embryonic day 13.5 (E13.5) in mouse cortical cultures (Fig. 2F). Furthermore, genetic ablation of Rfx4 decreases Gpr56 expression in developing mouse brain (14). RFX and GPR56 expression patterns are correlated (fig. S5, A and B) (15), with RFX3 and RFX7 most prominent in human ventrolateral prefrontal cortex, the region affected in perisylvian polymicrogyria (Fig. 2G), which suggests that multiple RFX proteins regulate the element.
GPR56 encodes an adhesion heterotrimeric guanine nucleotide–binding protein (G protein)–coupled receptor that is highly expressed in cortical progenitors (7, 16) and binds extracellular matrix proteins (17). Loss of GPR56 disrupts radial glia and causes breaches in the pial basement membrane, through which some neurons over-migrate (9, 16). However, even where the pia is intact, we found that neocortical thickness and organization are irregular, with occasional thin regions in Gpr56 knockout mice (Fig. 3A). Postmortem analysis of a human with biallelic GPR56 coding mutations showed a very thin cortex, which suggested potential roles of GPR56 in neurogenesis as well (9). GPR56 protein is most highly expressed in progenitors in the ventricular and subventricular zones during neurogenesis in mice (16, 18). GPR56 expression in developing human and marmoset neocortex is highest in the ventricular zone, as well as in the outer subventricular zone, which is expanded in mammals with larger brains (2) (Fig. 3B and fig. S5, C and D).
Fig. 3. GPR56 regulates neuroprogenitor proliferation.
(A) In Gpr56 knockout mice, neurons overmigrate through breached pial basement membrane (arrowheads) or undermigrate (arrows) forming irregular cortical layers, as shown by immunostaining of Cux1, an upper layer (II to IV) marker (p9). Thin cortex is occasionally observed (asterisks). (B) GPR56 is highly expressed in human ventricular zone and outer subventricular zone at 12 to 17 weeks of gestation (GW), which suggests roles in neuroprogenitors. v, ventricular zone; s, subventricular zone; is, inner subventricular zone; os, outer subventricular zone; i, intermediate zone; c, cortical plate; and m, marginal zone. (C to D) Human GPR56 transgenic (Tg) mice have significantly more mitotic (PH3+) neuroprogenitor cells and intermediate progenitor (TBR2+) cells than wild-type (WT). In contrast, Gpr56 knockout (KO) mice have significantly fewer mitotic cells and intermediate progenitors than WT (E13.5 to E14.5). (n = 7 mice per genotype; *P < 0.005; **P < 0.001; paired t test). (E) The cells that are in utero electroporated (from E13.5 to E15.5) with human GPR56-IRES-GFP [either side of the internal ribosome entry site (IRES), GFP expressing] persist in the germinal zones longer that the GFP control cells. Red, TBR2; blue, Hoechst. (n = 11 mouse embryos per construct; *P < 0.0001; chi-squared test). Scale bars, 500 μm (A) and 100 μm (B) to (E).
Impairment and overexpression of GPR56 show that its expression regulates proliferation. Gpr56 knockout mice show fewer phosphohistone H3 (PH3)–positive mitotic progenitor cells and TBR2-positive intermediate progenitors than wild-type mice in the neocortex at E14.5. Conversely, mice carrying a transgene that directs overexpression of human GPR56 show increased mitotic progenitor cells and intermediate progenitor cells (Fig. 3, C and D). In utero electroporation (at E13.5 with analysis 48 hours later at E15.5) of a plasmid encoding GPR56 (as well as GFP, to mark the cells) caused cells to persist in proliferative zones compared with cells expressing GFP alone (Fig. 3E). Changes in the number of intermediate progenitors in transgenic and knockout mice may be secondary to changes in the radial progenitor cells that generate them or might reflect a direct role of GPR56 in intermediate progenitors but is consistent with a report that loss of TBR2 (EOMES) also causes polymicrogyria in humans (19).
The cis-regulatory element upstream of GPR56 e1m is found in genomes of all placental mammals, but not monotremes, marsupials, or non-mammals, which suggests that it emerged after placental and nonplacental mammals diverged 85 to 100 million years ago (fig. S7B). The cis-regulatory element sequence is only found at the e1m locus in GPR56 but not elsewhere in the genome. E1m itself shows homology at its 3′ end to a long interspersed nuclear element (LINE)–4/RTE, a family of retrotransposons active in early mammals after divergence from marsupials (20). Another noncoding GPR56 exon (exon 2), present only in primates, derives from a primate-specific Alu insertion (fig. S7B). In contrast to the >17 alternative first exons in humans, we found only five noncoding first exons in the mouse Gpr56 gene (Fig. 4A and fig. S7A) (10, 11). Thus, GPR56 acquired many noncoding upstream exons and generated alternative splice forms with distinct expression patterns (fig. S2, B and D), in the lineage leading to humans. Transposable element insertion played a role in generating this diversity.
Fig. 4. GPR56 gene evolution.
(A) The mouse Gpr56 locus has only 5 transcription start sites compared with 17 in human. (B) A 300-bp human GPR56 e1m promoter sequence containing the cis-regulatory element (Fig. 2A) directs β-gal expression to lateral cortex in mice (arrow), whereas the orthologous mouse e1c(m) promoter directs more widespread expression. Orthologous promoters from other mammals with larger cortical sizes, abundant gyri, or both (marmoset, dolphin, and cat) drive relatively limited expression patterns generally similar to human (n = 3 to 10 embryos with identical patterns per promoter). Scale bar, 2 mm. (C) Human, marmoset, dolphin, and cat brains are gyrencephalic or near-gyrencephalic with a Sylvian fissure (arrowhead). Mouse brain lacks both gyri and Sylvian fissure. Scale bar, 1 cm. Images from the University of Wisconsin and Michigan State Comparative Mammalian Brain Collections and/or the National Museum of Health and Medicine are reproduced with permission from Brainmuseum.org.
To test directly whether evolutionary changes in GPR56’s alternative splice forms have functional effects, we generated transgenic mice in which the β-galactosidase (β-gal) gene is driven by a minimal 300-bp e1m promoter from human, mouse, marmoset, dolphin, and cat (Fig. 2A and fig. S6A). The mouse e1m promoter drives β-gal expression broadly in the nervous system in diverse cell types, which suggests that this simple 300-bp e1m promoter is sufficient to recapitulate major features of the endogenous mouse GPR56 expression (16–18) (Fig. 4B and fig. S6B). In contrast, the corresponding human e1m promoter has a variety of deletions and single-nucleotide variants, relative to the mouse sequence (fig. S6A), and drives much more limited expression in rostral-lateral cortex (Fig. 4B and fig. S6B). Weak lateral cortical expression is visible in embryos carrying the mouse e1m promoter:β-gal transgene, which suggests that, in humans, additional elements besides the 300-bp e1m promoter region are required to drive the full complement of GPR56 expression. E1m promoters from marmoset, dolphin, and cat drive expression patterns generally similar to human. The shared expression patterns in the four mammals that have a Sylvian fissure (Fig. 4C) suggest that elaboration of GPR56 noncoding regulation is consistent with the larger number of noncoding first exons in gyrencephalic mammals and humans. Elaboration of additional alternative splice forms provides a mechanism for potentially independent evolution of these multiple forms.
Our studies show that levels of GPR56 control proliferation of progenitors in the neocortex. Loss of GPR56 expression impairs neurogenesis, and overexpression enhances proliferation and progenitor number. Selective GPR56 loss causes strikingly regional defects of cortical development. GPR56 likely influences progenitor proliferation by stabilizing the basal process of radial neuroepithelial progenitors, because (i) GPR56 protein localizes to the basal processes of radial neuroepithelial cells (16); (ii) GPR56 binds extracellular matrix proteins in the pial basement membrane, such as collagen type III (17) and tetraspanins, which bind integrins as well (21); (iii) GPR56 is required for normal attachment of basal processes to the pial basement membrane in mice (16); and (iv) basal processes regulate progenitor proliferation via integrin signaling (22, 23), and GPR56 interacts genetically with α3β1 integrin (24). The elaboration of the GPR56 locus in gyrencephalic mammals, and especially humans, to produce many alternative splice forms with diverse expression patterns presents GPR56 as a key target that could influence the dramatic changes in shape and folding that characterize the forebrain of many mammals. Elaboration and specialization of alternative transcripts with distinct transcription start sites is an evolutionary mechanism that has been difficult to study because of the lack of comprehensive catalogs of RNA splice forms, but continued RNA sequencing studies may soon provide the opportunity to assess its importance systematically.
Supplementary Material
Acknowledgments
Research performed on samples of human origin was conducted according to protocols approved by participating institutions, including Boston Children’s Hospital and Beth Israel Deaconess Medical Center. The human embryonic and fetal material was provided by the Joint Medical Research Council (grant no. G0700089)–Wellcome Trust (grant no. GR082557) Human Developmental Biology Resource (www.hdbr.org) and the National Institute of Child Health and Human Development, NIH, Brain and Tissue Bank at the University of Maryland (contract no. HHSN275200900011C, reference no. NO1-HD-9-0011). Gpr56 knockout mice are from Genentech. This work was supported by the Strategic Research Program for Brain Sciences and from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) Japan (H.O.); Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST Program) (H.O.); U01MH081896 from National Institute of Mental Health, NIH (N.S.); 2R01NS035129 from National Institute of Neurological Disorders and Stroke, NIH (C.A.W.); and The Paul G. Allen Family Foundation (C.A.W.). Additional funding support listed in supplementary materials. C.A.W. is an investigator of the Howard Hughes Medical Institute. Gpr56 knockout mice are available from Genentech subject to a Material Transfer Agreement.
Footnotes
References and Notes
- 1.Rakic P. Nat Rev Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lui JH, Hansen DV, Kriegstein AR. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zilles K, Palomero-Gallagher N, Amunts K. Trends Neurosci. 2013;36:275–284. doi: 10.1016/j.tins.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Amunts K, et al. PLOS Biol. 2010;8:e1000489. doi: 10.1371/journal.pbio.1000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Golden JA, Harding BN. Nat Rev Neurol. 2010;6:471–472. doi: 10.1038/nrneurol.2010.118. [DOI] [PubMed] [Google Scholar]
- 6.Barkovich AJ. Neuroradiology. 2010;52:479–487. doi: 10.1007/s00234-009-0644-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piao X, et al. Science. 2004;303:2033–2036. doi: 10.1126/science.1092780. [DOI] [PubMed] [Google Scholar]
- 8.Piao X, et al. Ann Neurol. 2005;58:680–687. doi: 10.1002/ana.20616. [DOI] [PubMed] [Google Scholar]
- 9.Bahi-Buisson N, et al. Brain. 2010;133:3194–3209. doi: 10.1093/brain/awq259. [DOI] [PubMed] [Google Scholar]
- 10.Thierry-Mieg D, Thierry-Mieg J. Genome Biol. 2006;7(suppl 1):S12–S14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki Y, Yamashita R, Nakai K, Sugano S. Nucleic Acids Res. 2002;30:328–331. doi: 10.1093/nar/30.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ansley SJ, et al. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- 13.Jolma A, et al. Genome Res. 2010;20:861–873. doi: 10.1101/gr.100552.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang D, et al. J Neurochem. 2006;98:860–875. doi: 10.1111/j.1471-4159.2006.03930.x. [DOI] [PubMed] [Google Scholar]
- 15.Kang HJ, et al. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li S, et al. J Neurosci. 2008;28:5817–5826. doi: 10.1523/JNEUROSCI.0853-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo R, et al. Proc Natl Acad Sci USA. 2011;108:12925–12930. doi: 10.1073/pnas.1104821108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeong SJ, Luo R, Li S, Strokes N, Piao X. J Comp Neurol. 2012;520:2930–2940. doi: 10.1002/cne.23076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baala L, et al. Nat Genet. 2007;39:454–456. doi: 10.1038/ng1993. [DOI] [PubMed] [Google Scholar]
- 20.Mikkelsen TS, et al. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- 21.Xu L, Hynes RO. Cell Cycle. 2007;6:160–165. doi: 10.4161/cc.6.2.3760. [DOI] [PubMed] [Google Scholar]
- 22.Radakovits R, Barros CS, Belvindrah R, Patton B, Müller U. J Neurosci. 2009;29:7694–7705. doi: 10.1523/JNEUROSCI.5537-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fietz SA, et al. Nat Neurosci. 2010;13:690–699. doi: 10.1038/nn.2553. [DOI] [PubMed] [Google Scholar]
- 24.Jeong SJ, et al. PLOS ONE. 2013;8:e68781. doi: 10.1371/journal.pone.0068781. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.