Abstract
PolyPhen-2 (Polymorphism Phenotyping v2), available as software and via a Web server, predicts the possible impact of amino acid substitutions on the stability and function of human proteins using structural and comparative evolutionary considerations. It performs functional annotation of single-nucleotide polymorphisms (SNPs), maps coding SNPs to gene transcripts, extracts protein sequence annotations and structural attributes, and builds conservation profiles. It then estimates the probability of the missense mutation being damaging based on a combination of all these properties. PolyPhen-2 features include a high-quality multiple protein sequence alignment pipeline and a prediction method employing machine-learning classification. The software also integrates the UCSC Genome Browser’s human genome annotations and MultiZ multiple alignments of vertebrate genomes with the human genome. PolyPhen-2 is capable of analyzing large volumes of data produced by next-generation sequencing projects, thanks to built-in support for high-performance computing environments like Grid Engine and Platform LSF.
Keywords: human genetic variation, single-nucleotide polymorphism (SNP), mutation effect prediction, computational biology, PolyPhen-2
INTRODUCTION
Most human genetic variation is represented by single-nucleotide polymorphisms (SNPs), and many SNPs are believed to cause phenotypic differences between human individuals. We specifically focus on nonsynonymous SNPs (nsSNPs), i.e., SNPs located in coding regions and resulting in amino acid changes in protein products of genes. It has been shown in several studies that the impact of amino acid allelic variants on protein structure and function can be predicted via analysis of multiple sequence alignments and protein 3-D structures. As we demonstrated earlier (Sunyaev et al., 2001; Boyko et al., 2008), these predictions correlate with the effect of natural selection, demonstrated by an excess of rare alleles among alleles that are predicted to be functional. Therefore, predictions at the molecular level reveal SNPs affecting actual phenotypes.
PolyPhen-2 (Adzhubei et al., 2010) is an automatic tool for prediction of the possible impact of an amino acid substitution on the structure and function of a human protein. Automated predictions of this kind are essential for interpreting large datasets of rare genetic variants, which have many applications in modern human genetics research. Uses in recent research include identifying rare alleles that cause Mendelian disease (Bamshad et al., 2011), scanning for potentially medically actionable alleles in an individual’s genome (Ashley et al., 2010), and profiling the spectrum of rare variation uncovered by deep sequencing of large populations (Tennessen et al., 2012).
The prediction is based on a number of sequence, phylogenetic, and structural features characterizing the substitution. For a given amino acid substitution in a protein, PolyPhen-2 extracts various sequence and structure-based features of the substitution site and feeds them to a probabilistic classifier.
We describe here three basic protocols for accessing PolyPhen-2 through its Web interface: (a) predicting the effect of a single-residue substitution or reference SNP (Basic Protocol 1), (b) analyzing a large number of SNPs in a batch mode (Basic Protocol 2), and (c) searching in a database of precomputed predictions for the whole human exome sequence space, WHESS.db (Basic Protocol 3). Alternate protocols provide detailed instructions on how to install and use the stand-alone version of the software on a Linux computer. Support protocols explain how to check the status of a query using the Grid Gateway Interface, and how to update PolyPhen-2’s built-in protein annotation and sequence databases.
BASIC PROTOCOL 1: PREDICTING THE EFFECT OF A SINGLE-RESIDUE SUBSTITUTION ON PROTEIN STRUCTURE AND FUNCTION USING THE PolyPhen-2 WEB SERVER
The PolyPhen-2 Web interface can be reached at http://genetics.bwh.harvard.edu/pph2/. The input form at this URL allows querying for a single individual amino acid substitution or a coding, non-synonymous SNP annotated in the dbSNP database. After submitting a query, the user is transferred to the Grid Gateway Interface Web page (see Support Protocol 1), which is used to track the user’s query progress and retrieve results. Results of the analysis are linked to a separate Web page with more detailed output, formatted as text, graphics, and HTML cross-links to relevant sequence and structural database entries.
Materials
An up-to-date Web browser, such as Firefox, Internet Explorer, or Safari. JavaScript support and cookies should be enabled in the browser configuration; the Java browser plug-in is required for the protein 3-D structure viewer to function.
Example 1: Query variant in a known human protein
-
1Access the PolyPhen-2 Web interface at http://genetics.bwh.harvard.edu/pph2/. In the “Protein or SNP identifier” text box, enter a protein identifier, e.g., UniProtKB accession number or entry name. For this example, type:
P41567PolyPhen-2 uses the UniProtKB database as a reference source for all protein sequences and annotations. You can also enter human protein identifiers from other databases (e.g., RefSeq) or standard gene symbols; for the full list of supported databases, see Advanced Configuration Options, “Proteins not in UniProtKB.” However, UniProtKB identifiers are preferred as the most reliable and unambiguous. -
2
Leave the “Protein sequence in FASTA format” text box empty.
-
3Enter the position of the substitution in the protein sequence into the Position text box. For this example, type:
59
-
4Select the appropriate boxes for the wild-type (query sequence) amino acid residue AA1 and the substitution residue AA2. For this example, select:
L P
for AA1 and AA2, respectively.
-
5Enter an optional description into the “Query description” text box:
Sample query
A human-readable query description might help you more easily locate a particular query when you have a large number of pending, running, and completed queries in your PolyPhen-2 Web session. -
6
Click the Submit Query button; see Figure 7.20.1 for the filled-in query form example. You will be transferred to the Grid Gateway Interface Web page (see Support Protocol 1). Use the Refresh button to reload the page until your query job is listed under Completed.
-
7
Click the corresponding View link to browse the PolyPhen-2 prediction report for your query; see Figure 7.20.2.
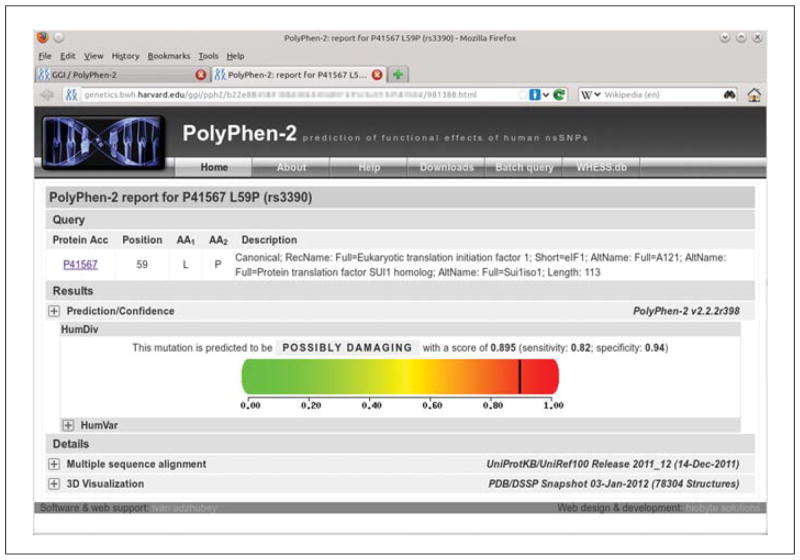
You will be presented with the Web page listing your Query (mapped to a reference UniPro-tKB protein if possible) and the Prediction/Confidence panel. Prediction outcome can be one of probably damaging, possibly damaging, or benign. “Score” is the probability of the substitution being damaging; “sensitivity” and “specificity” correspond to prediction confidence (see Background Information, Prediction Algorithm for explanation). The predicted damaging effect is also indicated by a vertical black marker inside a color gradient bar, where green is benign and red is damaging. -
8
By default, only the prediction based on the HumDiv model is shown; click on the [+] control box next to the HumVar label to display the HumVar-based prediction and scores.
For explanation of the differences between the HumDiv and HumVar classification models, see Background Information, Prediction Algorithm. -
9
Click on the [+] control box next to “Multiple sequence alignment” to display the multiple sequence alignment panel; see Figure 7.20.3.
This panel displays a multiple sequence alignment for 75 amino acid residues surrounding your variant’s position in the query sequence. Click the link at the bottom of the panel to open an interactive alignment viewer (Jalview, http://www.jalview.org/) (Waterhouse et al., 2009) to scroll through the complete alignment. -
10
Click on the [+] control box next to 3-D Visualization to display the interactive 3-D protein structure viewer applet (Jmol, http://jmol.sourceforge.net/); see Figure 7.20.3.
Most of the 3-D protein structure viewer controls are self-explanatory; visit the Jmol Web site for a complete user guide.
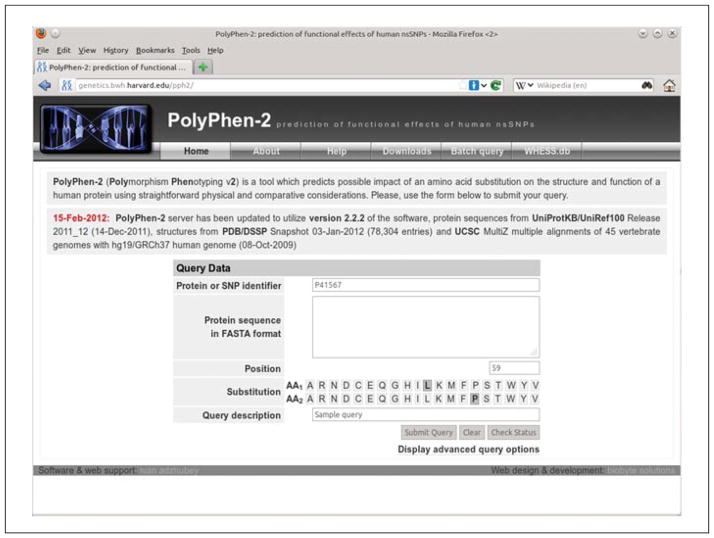
Figure 7.20.1.
PolyPhen-2 home Web page with the input form prepared to submit a single protein substitution query using Swiss-Prot accession as a protein identifier. Also supported are RefSeq and Ensembl protein identifiers; alternatively, a dbSNP reference SNP identifier can be entered, in which case no other input is required.
Figure 7.20.2.
Detailed results of the PolyPhen-2 analysis for single variant query. This format is used for all PolyPhen-2 query reports except the Batch Query. The top Query section includes UniProtKB/Swiss-Prot description of query protein, if it was recognized as a known database entry. The large “heatmap” color bar with the black indicator mark dominates the display, illustrating the strength of the putative damaging effect for the variant, assessed using the default HumDiv-trained predictor. Clicking on the [+] control boxes expands the Prediction/Confidence panel for the HumDiv-trained predictor, as well as additional panels with protein multiple sequence alignment and 3D-structure viewers. For the color version of this figure, go to http://www.currentprotocols.com/protocol/hg0720.
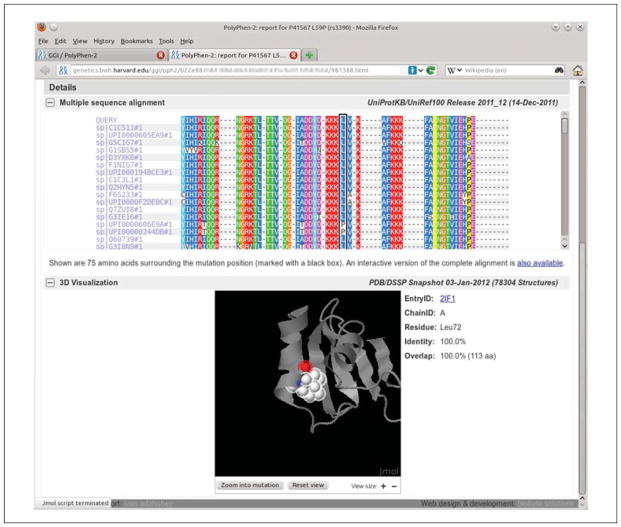
Figure 7.20.3.
Detailed results of the PolyPhen-2 analysis for a single variant query with the multiple sequence alignment and 3-D-structure protein viewer panels expanded the multiple sequence alignment panel displays a fixed 75-residue wide window surrounding the variant’s position (the column indicated by black frame), with the alignment colored using the ClustalX (Thompson et al., 1997) scheme for all columns above 50% conservation threshold. Clicking on the link at the bottom of the alignment panel opens the Jalview (Waterhouse et al., 2009) alignment viewer applet with the complete multiple alignment loaded. Displayed below is a 3-D-structure viewer applet (Jmol; http://www.jmol.org/) with the protein structure loaded and zoomed into the mutation residue using the Zoom into mutation button. The structure viewer window is fully interactive, and the protein structure can be rotated, moved, or zoomed in and out.
Example 2: Query a variant within user-submitted protein sequence
-
11
Leave the “Protein or SNP identifier” text box empty.
-
12In the “Protein sequence in FASTA format” text box, type or copy and paste your protein sequence in FASTA format, including the FASTA definition line with the protein identifiers:
>gi|5032133|ref|NP 005792.1| eukaryotic trans... MSAIQNLHSFDPFADASKGDDLLPAGTEDYIHIRIQQRNGRKTLTTVQGIA DDYDKKKLVKAFKKKFACNGTVIEHPEYGEVIQLQGDQRKNICQFLVEIGL AKDDQLKVHGF
It is important to include the FASTA definition line (as the first line starting with the “>” symbol); failure to do so will result in a fatal query processing error. However, you can use the simplest form of the definition line for your sequences:>NP 005792 MSAIQNLHSFDPFADASKGDDLLPAGTEDYIHIRIQQRNGRKTLTTVQ GIADDYDKKKLVKAFKKKFACNGTVIEHPEYGEVIQLQGDQRKNICQF LVEIGLAKDDQLKVHGF
Now proceed with steps 4 to 10 (see Example 1) to submit your query and obtain results.
Example 3: Query a dbSNP reference SNP
-
13Enter dbSNP reference SNP ID into the “Protein or SNP identifier” text box. For this example, type:
rs3390
Always include the rs prefix; enter only one identifier per query. -
14Enter an optional query description into the “Query description” text box:
Sample rsSNP query
Now proceed with steps 6 to 10 (see Example 1) to submit your query and obtain results.
The PolyPhen-2 SNP database only covers human reference SNPs annotated as part of the SwissVar project (http://swissvar.expasy.org/), which comprise a smaller, manually curated subset of all missense SNPs in the dbSNP database. If your reference SNP is not found by its reference ID, you will have to resubmit it using the full protein substitution variant specification, as described in Example 1.
BASIC PROTOCOL 2: ANALYZING A LARGE NUMBER OF SNPs IN A BATCH MODE USING THE PolyPhen-2 WEB SERVER
When analyzing large datasets of single nucleotide changes, it may be convenient to submit a large number of changes for analysis at once. For this purpose, the PolyPhen-2 Web interface includes a batch mode, which allows entry of multiple queries in a single form.
Materials
An up-to-date Web browser, such as Firefox, Internet Explorer, or Safari. Cookies should be enabled in the browser configuration.
-
1
Go to the PolyPhen-2 Batch Query page (http://genetics.bwh.harvard.edu/pph2/bgi.shtml).
-
2a
In the “Batch query” text box, enter one or more query lines, with the variants specified according to one of the following three supported formats:
-
Protein substitutions:
# Protein ID Position AA1 AA1 Q92889 706 I T Q92889 875 E G XRCC1 HUMAN 399 R Q NP 005792 59 L P
Protein identifiers in the first column can be from any of the supported databases; see Advanced Configuration Options/Proteins not in UniProtKB. - Reference SNP IDs:
# dbSNP rsID rs1799931 rs3390 rs1065757
-
Genomic variants:
# Chromosome:position Reference/Variant nucleotides chr1:1267483 G/A chr1:1158631 A/C,G,T chr2:167262274 C/T chr4:264904 G/A chr7:122261636 G/T chr16:53698869 T/C
Chromosome position coordinates are 1-based; reference/variant nucleotides should be specified on the plus strand of the assembly.
-
-
2bAlternatively, prepare a text file in the same format as described above and enter its full pathname into the “Upload batch file” text box, or click the Browse button to locate file on disk:
/home/username/my variants.txt
You can use tab or space character(s) as column delimiters in the batch file. -
3Enter an optional query description into the “Query description” text box:
Sample batch query
-
4Enter your e-mail address into the “E-mail address” text box to receive an automatic notification via e-mail when your Batch Query results will be ready (optional, recommended):
myself@example.com -
5Prepare a text file with all your protein sequences in FASTA format and enter its full pathname into the “Upload FASTA file” text box, or click the Browse button to select a file from disk (optional; only required when analyzing variants in nonstandard, novel, or otherwise unannotated protein sequences):
/home/username/my proteins.fas
-
6In the “File description” text box, enter a short description of your FASTA sequences (optional):
My protein sequences
User-supplied sequences are only supported when analyzing protein variants. Protein identifiers in the FASTA sequences file should match the corresponding protein identifiers in the first column of your Batch Query file. -
7
Under Advanced Options, select the Classifier model you want to use from the drop-down menu:
HumDiv
“HumDiv” is the default Classifier model used by probabilistic predictor; it is preferred for evaluating rare alleles, dense mapping of regions identified by genome-wide association studies, and analysis of natural selection. “HumVar” is better suited for diagnostics of Mendelian diseases, which requires distinguishing mutations with drastic effects from all the remaining human variation, including abundant mildly deleterious alleles. -
8Under Advanced Options, select the Genome assembly version matching the SNP coordinates in your input from the drop-down menu (genomic SNPs only):
GRCh37/hg19
-
9Under Advanced Options, select a set of gene Transcripts from the drop-down menu. These are used to map SNPs in the user input in order to obtain their functional class annotations (genomic SNPs only):
Canonical
The choices are All, which includes all UCSC known Gene transcripts (highly redundant set); Canonical (default), which includes the UCSC known Canonical subset only; and CCDS, which further restricts the known Canonical subset to those gene transcripts which are also annotated as part of NCBI CCDS database. See UCSC Genome Browser Web site for details (http://genome.ucsc.edu/). -
10Under Advanced Options, select Annotations from the drop-down menu to specify which SNP functional categories will be included in the output (genomic SNPs only):
Missense
The choices are All, which will output annotations for the following SNP categories coding-synon, intron, nonsense, missense, utr-3, and utr-5; Coding, which will output annotations for coding-synon, nonsense, and missense SNPs; and Missense (default), which outputs only missense SNP annotations. Note that PolyPhen-2 predictions are always produced for missense mutations only. -
11
Click the Submit Query button; see Figure 7.20.4 for the filled-in Batch Query form example.
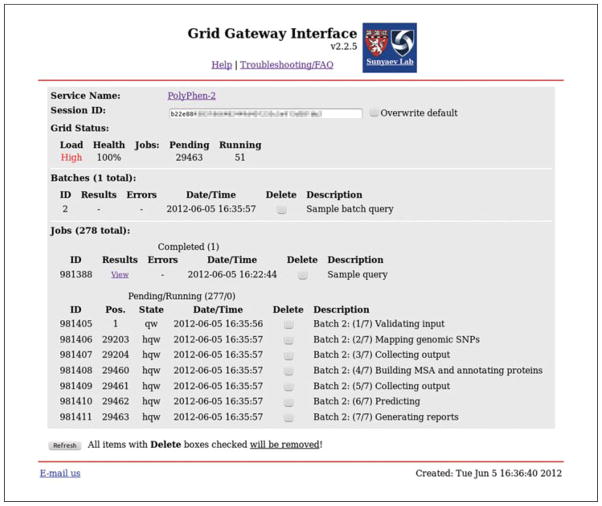
You will be transferred to the Grid Gateway Interface Web page; see Figure 7.20.5. Use the Refresh button to reload the page until your query batch is listed under Batches; see Figure 7.20.6. See Support Protocol 1 for detailed description of the Grid Gateway Interface. -
12
Under Batches/Results, click the “SNPs” link to view SNP functional annotations, or right-click on the link to download the file; see Table 7.20.1 for the file format description.
-
13
Under Batches/Results, click the Short link to view PolyPhen-2 predictions and scores, or right-click on the link to download the file; see Table 7.20.2 for the file format description.
-
14
Under Batches/Results, click the Full link to view the complete set of features and parameters used for calculating PolyPhen-2 predictions and scores, or right-click on the link to download the file; see Table 7.20.2 for the file format description.
-
15
Under Batches/Results, click the Logs link to view errors/warnings generated by the analysis pipeline, or right-click on the link to download the file.
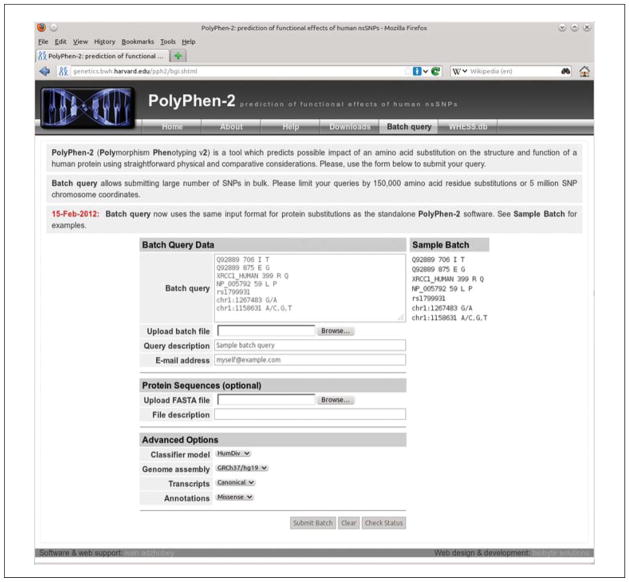
Figure 7.20.4.
The PolyPhen-2 Batch Query Web page allows submitting large number of variants for analysis in a single operation. Type or paste your variants into the Batch Query text input area (one variant per line) or upload a text file listing variants using Upload batch file text box (locate the file using the Browse button). If you enter your e-mail address into the corresponding text box, you will be notified via e-mail when your query completes. To analyze protein variants in nonstandard or unannotated proteins, you can upload your own protein sequences in FASTA format using the Upload FASTA file text box. Genomic variants are also supported; see the Sample Batch panel for the various input format examples. Do not forget to select the genome assembly version matching your genomic SNP data under Advanced Options; default assembly version used is GRCh37/UCSC hg19.
Figure 7.20.5.
Grid Gateway Interface (GGI) Web page showing a PolyPhen-2 user session with one single-variant query completed and a Batch Query pending execution. Click on the View link to access results of a single-variant query (no errors were reported). This Batch Query was queued as a 7-stage pipeline; the status of each pipeline stage is tracked and displayed separately, with short stage explanations printed in the Description column. The batch will be completed when the last stage finishes. Grid Status shows high Grid Load and large number of other Pending jobs; batch completion waiting time is likely to be substantial. Click on the Refresh button periodically to update session status. You can also close your browser and check your session at a later time—go to the PolyPhen-2 home page, click on the Check Status button, and you will be transferred to your session automatically.
Figure 7.20.6.
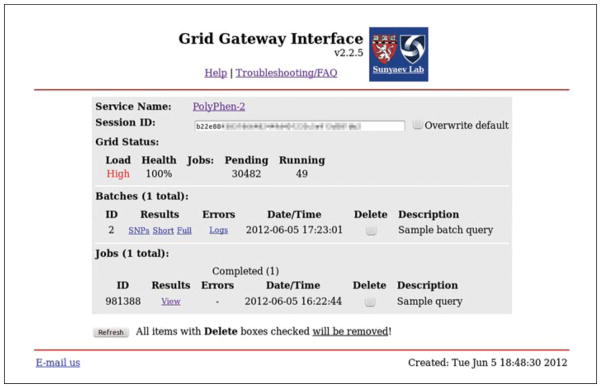
Grid Gateway Interface (GGI) Web page showing a completed PolyPhen-2 Batch Query. Right-click on one of the SNPs, Short, or Full links in the Batches/Results column to download results to your computer; see Basic Protocol 2 for description of the three various types of report files. Click on the Logs link under Batches/Errors to view all error and warning messages generated by the pipeline. Note that most of the warnings are for your information only and do not indicate failure of the analysis. After downloading results, all batch data can be deleted by checking corresponding Batches/Delete checkbox and clicking on the Refresh button. Be warned that the delete operation is irreversible and deleted data cannot be restored.
Table 7.20.1.
MapSNPs Annotation Summary Reporta
| Column no. | Column name | Description |
|---|---|---|
| 1 | snp_pos | input SNP chromosome:position (chromosome coordinates are 1-based) |
| 2 | str | transcript strand (+ or −) |
| 3 | gene | gene symbol |
| 4 | transcript | UCSC transcript name |
| 5 | ccid | UCSC canonical cluster ID (number) |
| 6 | ccds | NCBI CCDS cluster ID |
| 7 | cciden | NCBI CCDS CDS similarity level by genomic overlap with the corresponding UCSC known gene transcript |
| 8 | refa | reference allele/variant allele (+ strand) |
| 9 | type | SNP functional category (coding-synon, intron, nonsense, missense, utr-3, utr-5) |
| 10 | ntpos | mutation position in the full transcript nucleotide sequence (in the direction of transcription) |
| 11 | nt1 | reference nucleotide (transcript strand) |
| 12 | nt2 | variant nucleotide (transcript strand) |
| 13 | flanks | nucleotides flanking mutation position in the transcript sequence, enumerated in the direction of transcription (5′3′) |
| 14 | trv | transversion mutation (0, transition; 1, transversion) |
| 15 | cpg | CpG context: 0, non-CpG context retained; 1, mutation removes CpG site; 2, mutation creates new CpG site; 3, CpG context retained: C(C/G)G |
| 16 | jxdon | distance from mutation position to the nearest donor exon/intron junction (“−” for upstream, “+” for downstream) |
| 17 | jxacc | distance from mutation position to the nearest acceptor intron/exon junction (−for upstream, “+” for downstream) |
| 18 | exon | mutation in exon #/of total exons (exons are enumerated in the direction of transcription) |
| 19 | cexon | same as above but for coding exons only |
| 20 | jxc | mutation in a codon that is split across two exons (?, no; 1, yes) |
| 21 | dgn | degeneracy index for mutated codon position (Nei and Kumar, 2000:, p. 64): 0, non-degenerate; 2, simple 2-fold degenerate; 3, complex 2-fold degenerate; 4, 4-fold degenerate |
| 22 | cdnpos | number of the mutated codon within transcript’s CDS (1-base) |
| 23 | frame | mutation position offset within the codon (0..2) |
| 24 | cdn1 | reference codon nucleotides |
| 25 | cdn2 | mutated codon nucleotides |
| 26 | aa1 | wild type (reference) amino acid residue |
| 27 | aa2 | mutant (substitution) amino acid residue |
| 28 | aapos | position of amino acid substitution in protein sequence (1-base) |
| 29 | spmap | CDS protein sequence similarity to known UniProtKB protein (?, no match) |
| 30 | spacc | UniProtKB protein accession |
| 31 | spname | UniProtKB protein entry name |
| 32 | refs_acc | RefSeq protein accession |
| 33 | dbrsid | dbSNP SNP rsID |
| 34 | dbobsrvd | dbSNP observed alleles (transcript strand) |
| 35 | dbavHet | dbSNP average heterozygosity from all observations |
| 36 | dbavHetSE | dbSNP standard error for the average heterozygosity |
| 37 | dbRmPaPt | dbSNP reference orthologous alleles in macaque (Rm), orangutan (Pa), and chimp (Pt) |
| 38 | Comments | optional user comments, copied from input |
The MapSNPs genomic SNP annotation tool is part of the PolyPhen-2 Batch Query Web service. Whenever you submit genomic SNPs in the form of chromosome coordinates/alleles, a report formatted as described below will appear under the “SNPs” link on the GGI Web page. It is a plain text tab-separated file with each line annotating a corresponding protein sequence variant (amino acid residue substitution) for each missense allelic variant found in user input. Columns 18 to 32 will contain ?placeholders for SNPs annotated as non-coding; columns 33 to 37 will have values only for reference SNPs annotated in dbSNP build 135. MapSNPs filters SNP annotations in the output depending on the user selection of SNP functional categories via the Advanced Options/Annotations drop-down menu on the Batch Query Web page. Selecting “All” disables filtering and results in annotations for all SNP categories reported. However, PolyPhen-2 predictions are produced for missense SNPs only, regardless of the Advanced Options/Annotations menu selection.
Table 7.20.2.
PolyPhen-2 Annotation Summary Reporta
| Column no. | Column name | Description |
|---|---|---|
| Original query (copied from user input): | ||
| 1b | o_acc | original protein identifier |
| 2 | o_pos | original substitution position in the protein sequence |
| 3 | o_aa1 | original wild type (reference) amino acid residue |
| 4 | o_aa2 | original mutant (substitution) amino acid residue |
| Annotated query: | ||
| 5b | rsid | dbSNP reference SNP identifier (rsID) if available |
| 6b | acc | UniProtKB accession if known protein, otherwise same as o_acc |
| 7b | pos | substitution position in UniProtKB protein sequence, otherwise same as o_pos |
| 8b | aa1 | wild-type amino acid residue in relation to UniProtKB sequence |
| 9b | aa2 | mutant amino acid residue in relation to UniProtKB sequence |
| 10 | nt1 | wild-type (reference) allele nucleotide |
| 11 | nt2 | mutant allele nucleotide |
| PolyPhen-2 prediction outcome: | ||
| 12b | prediction | qualitative ternary classification appraised at 5%/10% (HumDiv) or 10%/20% (HumVar) FPR thresholds (benign, possibly damaging, probably damaging) |
| PolyPhen-1 prediction description (obsolete, please ignore): | ||
| 13 | based_on | prediction basis |
| 14 | effect | predicted substitution effect on the protein structure or function |
| PolyPhen-2 classifier outcome and scores: | ||
| 15 | pph2_class | probabilistic binary classifier outcome (damagingor neutral) |
| 16b | pph2_prob | classifier probability of the variation being damaging |
| 17b | pph2_FPR | classifier model False Positive Rate (1- specificity) at the above probability |
| 18b | pph2_TPR | classifier model True Positive Rate (sensitivity) at the above probability |
| 19 | pph2_FDR | classifier model False Discovery Rate at the above probability |
| UniProtKB/Swiss-Prot derived protein sequence annotations: | ||
| 20 | site | substitution SITE annotation |
| 21 | region | substitution REGION annotation |
| 22 | PHAT | PHAT matrix element for substitutions in the TRANSMEM region |
| Multiple sequence alignment scores: | ||
| 23 | dScore | difference of PSIC scores for two amino acid residue variants (Score1-Score2) |
| 24 | Score1 | PSIC score for wild type amino acid residue (aa1) |
| 25 | Score2 | PSIC score for mutant amino acid residue (aa2) |
| 26 | MSAv | version of the multiple sequence alignment used in conservation scores calculations: 1, pairwise BLAST HSP (obsolete); 2, MAFFT-Leon-Cluspack (default); 3, MultiZ CDS |
| 27 | Nobs | number of residues observed at the substitution position in multiple alignment (without gaps) |
| Protein 3D-structure features: | ||
| 28 | Nstruct | initial number of BLAST hits to similar proteins with 3-D structures in PDB |
| 29 | Nfilt | number of 3-D BLAST hits after identity threshold filtering |
| 30 | PDB_id | PDB entry identifier |
| 31 | PDB_pos | position of substitution in PDB protein sequence |
| 32 | PDB_ch | PDB polypeptide chain identifier |
| 33 | ident | sequence identity between query sequence and aligned PDB sequence |
| 34 | length | PDB sequence alignment length |
| 35 | NormAcc | normalized accessible surface area |
| 36 | SecStr | DSSP secondary structure assignment |
| 37 | MapReg | region of the phi-psi map (Ramachandran map) derived from the residue dihedral angles |
| 38 | dVol | change in residue side chain volume |
| 39 | dProp | change in solvent accessible surface propensity resulting from the substitution |
| 40 | B-fact | normalized B-factor (temperature factor) for the residue |
| 41 | H-bonds | number of hydrogen sidechain-sidechain and sidechain-mainchain bonds formed by the residue |
| 42 | AveNHet | number of residue contacts with heteroatoms, average per homologous PDB chain |
| 43 | MinDHet | closest residue contact with a heteroatom, Å |
| 44 | AveNInt | number of residue contacts with other chains, average per homologous PDB chain |
| 45 | MinDInt | closest residue contact with other chain, Å |
| 46 | AveNSit | number of residue contacts with critical sites, average per homologous PDB chain |
| 47 | MinDSit | closest residue contact with a critical site, Å |
| Nucleotide sequence context features: | ||
| 48 | Transv | whether substitution is a transversion |
| 49 | CodPos | position of the substitution within a codon |
| 50 | CpG | whether substitution changes CpG context: 0, non-CpG context retained; 1, removes CpG site; 2, creates new CpG site; 3, CpG context retained |
| 51 | MinDJnc | substitution distance from closest exon/intron junction |
| Pfam protein family: | ||
| 52 | PfamHit | Pfam identifier of the query protein |
| Substitution scores: | ||
| 53 | IdPmax | maximum congruency of the mutant amino acid residue to all sequences in multiple alignment |
| 54 | IdPSNP | maximum congruency of the mutant amino acid residue to the sequences in multiple alignment with the mutant residue |
| 55 | IdQmin | query sequence identity with the closest homologue deviating from the wild type amino acid residue |
| Comments: | ||
| 56b | Comments | optional user comments, copied from input |
Reports in this format are produced by both the PolyPhen-2 Batch Query Web service as well as by standalone PolyPhen-2 software. It is a plain text, tab-separated file with each line annotating a single protein variant (amino acid residue substitution).
The eleven columns (1, 5 to 9, 12, 16 to 18, 56) included in the short version of the report available via the Short link on the GGI Web page. These are sufficient if you are only interested in the PolyPhen-2 prediction outcome and prediction confidence scores. The rest of the columns in the full report version (available via the Full link) are mostly useful only if you want to investigate all features supporting the prediction in detail.
BASIC PROTOCOL 3: QUICK SEARCH IN A DATABASE OF PRECOMPUTED PREDICTIONS
Quick access to a precomputed set of the PolyPhen-2 predictions for the whole human exome sequence space is provided by WHESS.db. It contains annotations for all single-nucleotide non-synonymous (missense) codon changes enumerated at each CDS codon position in the exons of 43,043 UCSC knownGene transcripts (hg19/GRCh37) with maximum sequence overlap and identity to known UniProtKB proteins.
Materials
An up-to-date Web browser, such as Firefox, Internet Explorer, or Safari. JavaScript support should be enabled in the browser configuration.
Go to the WHESS.db Web page (http://genetics.bwh.harvard.edu/pph2/dbsearch.shtml).
-
In the “Enter search” box, enter a search term (single variant specification) in one of the supported formats. For this example, type:
P06241 445 I F
WHESS.db quick search supports the same variant specifications as the PolyPhen-2 Batch Query, except that only the hg19/GRCh37 assembly version is supported for genomic SNPs. Click the Search button.
-
On the WHESS.db search results page, click the View links to open the detailed PolyPhen-2 prediction report.
WHESS.db search results are presented in a tabular format similar to the “Short” Batch Query output format; both HumDiv- and HumVar-based predictions and scores are included in the same table. The detailed WHESS.db search report format is identical to the PolyPhen-2 single variant query report (Basic Protocol 1).
SUPPORT PROTOCOL 1: CHECKING THE STATUS OF YOUR QUERY WITH THE GRID GATEWAY INTERFACE
Grid Gateway Interface (GGI) is a simple Web-based interface to the Grid Engine distributed computing management system. GGI is used to submit computationally heavy tasks directly from the Web browser query input form into our high-performance computing cluster. Queries are queued and then run as resources permit. Users are able to track the progress of their queries and retrieve results at a later time. It is also possible to submit large number of queries at once without having to wait for each one to complete (Batch Query).
Terminology
Query, Job, Task
These terms are mostly used interchangeably throughout this protocol to describe individual computational tasks processed by a grid, in the form of user-defined set of parameters (a query) submitted to an underlying algorithm that carries out the analysis (e.g., PolyPhen-2).
Cluster, Grid, Node
Refers to a number of networked computers capable of running noninteractive computational jobs submitted and managed remotely via a centralized queuing and scheduling system. Clusters are often built of identical compute nodes attached to a high-speed dedicated network; Grid is a more generic term for network-connected computers and/or clusters cooperating in distributed job execution, either across LAN or WAN (e.g., the Internet).
Materials
An up-to-date Web browser, such as Firefox, Internet Explorer, or Safari. Cookies should be enabled in the browser configuration.
-
1
Click the Submit Query button on the PolyPhen-2 home page, or on the Batch Query page, to access the GGI Web page. You can also get to this page without submitting a new query by clicking the Check Status button on the PolyPhen-2 home page or the Batch Query page.
-
2
Find the line marked Service Name at the top of the page, below the Grid Gateway Interface header with the logo and documentation links. This line should read “PolyPhen-2”; if it does not, return to the PolyPhen-2 home page or Batch Query page and try to access the GGI page again.
This line identifies and links back to a home page for the Web service you are using. There are several Web services installed on our server which utilize the same GGI system; by checking the Service Name, you should always know which one you are currently using. -
3
Check the status of your pending queries under the Batches and/or Jobs sections. Queries are listed by an internally generated numerical ID, as well as a description if one was entered when the query was submitted. Jobs are listed either under Pending or Running. A more detailed status code is also listed in the State column. Usually, this is qw for queued, awaiting execution; t for being transferred to one of the Grid’s nodes for execution; or r for running jobs. There are other status codes, but you should not encounter them in normal execution. For instance, when the letter E is included in the status code (e.g., Eqw), it means that the job caused an internal Grid Engine error and cannot be executed.
-
4a
For pending jobs, check how long it is likely to be before the job starts running:
The position of the job in the queue is indicated under the Pos column. Smaller numbers indicate higher positions in the queue, with a value of 1 indicating the job is to be dispatched for execution immediately, as soon as the next free CPU slot is available.
The time of the last status change of the job is listed under the Date column. For pending jobs, this is usually the time the job was first added to the queue.
The overall load and health of the cluster in the Cluster Status section near the top of the page. This information will give you some idea of how long it might take for your job to be executed.
-
4b
For running jobs, check how long the job has been running:
The time the job started running is listed under the Date column.
For batch queries, the number of jobs from the batch running at the same time is shown in parentheses after the status (e.g., r(10)).
-
5
Check whether the job is complete by looking at the Results and Errors columns. For jobs that have completed successfully, the Results column will contain links to view the results. For jobs that have completed with errors, the Errors column will contain links to documents describing the errors.
For a successfully completed job, click on the View link under the Results column to display the results in the browser, or right-click on the link to save the file to your computer. (Batch jobs have several different kinds of results linked from the Results section, these being SNPs, Short, and Full; see Basic Protocol 2). Contents of the Results column may also indicate errors, often caused by incorrect values in the user input. You should be able to fix such errors yourself by returning to the original query input page and re-entering corrected values in the query form, then resubmitting the query.
For jobs with errors, click on the View or Logs links under the Errors column to display the error messages in the browser, or right-click on the links to save the files to your computer. The View link only appears if there were exceptions caught during the execution of the analysis pipeline. This should rarely happen, and might indicate a bug in the software or a problem with the analysis pipeline. If you get the same error message repeatedly, please report it to us so we can get it fixed.
-
6
To load the latest results, press the Refresh Status button. The page does not refresh automatically, so pressing this button is the only way to keep the status current. Please do not abuse the server by refreshing excessively. The time when the last report was generated is listed at the bottom of the page, in the server’s local time (US/Eastern).
-
7
To remove unwanted jobs from the list, check the Delete box next to the job you want to delete and press the Refresh Status button. Running jobs will be terminated, pending jobs will be removed from the queue, and results of completed jobs will be removed from the server.
-
8
If you want to access these results later from a different computer, write down the 40-character Session ID listed near the top of the page. When you load the page on a different computer, type this value into the Session ID box and press Refresh Status to load your previous results. The new session ID will last until you close the browser window, at which point it will revert to its default. To overwrite the default session ID, check the “Overwrite default” box to the right of the Session ID text box before pressing Refresh Status. It is typically not necessary to save the session ID to view your results later on the same computer, since the Session ID is stored in the browser’s cookies for 3 months.
ALTERNATE PROTOCOL 1: AUTOMATED BATCH SUBMISSION
If you hate clicking through the browser’s query input page, you can automate batch submission to the PolyPhen-2 Batch Query Web interface easily, since it is a simple REST-compliant service. Use the curl command-line utility (http://curl.haxx.se/) to script your batch submission with shell scripting. This protocol contains a sample script that can do this for you on a Linux machine, and describes its use. This protocol may require modification to work on other systems.
Materials
A text editor and curlcommand-line utility (http://curl.haxx.se/)
Prepare your batch query and save it in a text file on your computer. Allowed file formats are as described in step 2 of Basic Protocol 2, above.
- Open a text editor and paste the following text into it:
#!/bin/sh curl -F_ggi_project=PPHWeb2 -F_ggi_origin=query -F _ggi_target_pipeline=1 -F MODELNAME=HumDiv -F UCSCDB=hg19 -F SNPFUNC=m -F NOTIFYME= myemail@myisp.com -F_ggi_batch_file=@example_batch.txt -D -http://genetics.bwh.harvard.edu/cgi-bin/ggi/ggi2.cgi -
Replace example_batch.txt with the name of the file containing your batch query. Keep the @symbol in front of the filename. Replace myemail@myisp.com with your e-mail address, or remove that line entirely to disable e-mail notifications.
Save the file as batch_submission.sh(or another filename of your choice).
The other parameters correspond to the options set in batch mode on the Web interface, described in Basic Protocol 2:MODELNAME corresponds to the “Classifier model” option, and can be HumDiv or HumVar.UCSCDB corresponds to the “Genome assembly” option, and can be hg18 or hg19.SNPFUNC corresponds to the “Annotations” option, and can be m for missense, c for coding, or empty (SNPFUNC=) for allOther possible options that are not included in this example script include:SNPFILTER, which corresponds to the “Transcripts” option, and can be 0 for all, 1 for canonical (the default), or 3 for CCDSuploaded_sequences_1, which corresponds to the “upload FASTA file option,” and should be the filename of a sequence file in FASTA format on your computer, preceded by the @ symbol -
On the command line, set the executable bit of the script you just created and run it. In a bash-like shell (the default in most Linux systems), use the following commands:
$ chmod +x batch_submission.sh $./batch_submission.sh
The $ represents the prompt, and should be omitted when actually entering the commands. On Windows systems or other shells, these commands may need to be modified.
-
Watch the terminal for the result of your submission. There are several useful pieces of information to be extracted:
-
Find your session ID by looking for a line like this:
Set-Cookie: polyphenweb2=98ba900751d509ce6dc262c078f37c023395782b;
This 40-character hash (not including the polyphenweb2=and the semicolon) is your session ID, as described in Support Protocol 1. It can be entered into the Grid Gateway Interface Web page in your browser to track your batch query progress and access its results later.
-
Find the job ID of the last job in your batch by looking for a line like this:
name=“lastJobSubmitted” value=“42145”
The number after value=is the job ID number of the last job in the batch. The grid system will identify that job by this number. The other jobs in the batch will usually have a contiguous block of ID numbers, with the last ID number being the highest.
-
Find the batch ID number by looking for a line like this:
Batch 1: (1/7) Validating input
The number before the colon is your batch ID number. The grid system will identify that batch by the combination of your session ID and the batch ID.
Your batch ID number is always 1 for newly-created sessions unless you have reused an existing session during submission. The latter can be achieved by adding the following parameter to the curl command line:-F sid=98ba900751d509ce6dc262c078f37c023395782b
-
-
To track your query progress, poll the server for the semaphore files located inside your session/batch directory. Use the following URL scheme to access the files:
http://genetics.bwh.harvard.edu/ggi/pph2/ <sessionid>/<batch number>/<filename>Replace <session id> with your session ID, <batch number> with your batch number, and <filename> with one of the following:
started.txt: created when the batch is dispatched for execution on the compute gridcompleted.txt: created when the batch has been fully processedBoth files contain server-generated timestamps in human-readable format.
Automatically polling the server is left as an exercise for the user. Be warned however, that polling the server too frequently from the same client may result in blocking of its IP ddress. Automatic polling should not exceed a rate of once every 60 sec. -
To access the result of a job once it is completed, download the result files in the same directory:
pph2-short.txt pph2-full.txt pph2-snps.txt pph2-log.txt
These four files correspond to the Short, Full, SNPs, and Log files available through the Grid Gateway Interface Web page.
ALTERNATE PROTOCOL 2: INSTALLING PolyPhen-2 STANDALONE SOFTWARE
These installation instructions are for the Linux operating system but should also work on Mac OS X and other Unix- or BSD-based systems with minor modifications. Some familiarity with basics of the bashshell and Linux in general are assumed. The Windows operating system is currently not supported.
In the examples below, all commands start with a $ symbol, which indicates a bash shell command-line prompt; the $symbol should be omitted when entering commands into your shell.
System Requirements
The following software needs to be present in the system before attempting to install PolyPhen-2.
Perl
Perl is required to run PolyPhen-2. Minimal recommended Perl version is 5.8.0; version 5.14.3 was the latest successfully tested. To check the version of Perl interpreter on your system, execute:
$ perl -v
The following extra Perl modules should also be present in the system:
XML::Simple LWP::Simple DBD::SQLite
If you do not have the modules installed already, you can do this by using standard software management tools for your system. On Ubuntu Linux, for example, run the following command to install these modules:
$ sudo apt-get install libxml-simple-perl
libwww-perl libdbd-sqlite3-perl
Build Tools
Build tools (C/C++ compiler, make, etc) are required during installation in order to compile several helper programs from their sources. Compilation has been tested with GCC 4.1.2 (minimal) / GCC 4.5.1. To install build tools in Ubuntu Linux, execute:
$ sudo apt-get install build-essential
Java
You will also need Java 6 installed. The latest version tested is: Oracle (Sun) Java Runtime Environment 1.6.0.26.
Several Linux systems, including Ubuntu, recommend using OpenJDK instead. On Ubuntu, install OpenJDK with this command:
$ sudo apt-get install openjdk-6-jre
OpenJDK Java 6 builds were not extensively tested, but should work. Java 7 was not tested (but may work).
Disk Space and Internet Connection
Download and installed size estimates for the database components of PolyPhen-2 are listed in Table 7.20.3. These estimates are for the versions from December, 2011. Since databases tend to grow in size constantly, your mileage may vary slightly. Be prepared to have at least 60 GB of free disk space available to accommodate a full PolyPhen-2 install.
Table 7.20.3.
Downloaded and Installed Sizes for Database Components
| Database | Download size (GB) | Installed size (GB) |
|---|---|---|
| Bundled Databases | 3.7 | 9.9 |
| MLC Alignments | 2.4 | 19.0 |
| MultiZ Alignments | 0.9 | 5.8 |
| UniRef100 Non-Redundant | 3.1 | 8.1 |
| Sequence Database | ||
| PDB | 12.0 | 12.0 |
| DSSP | 6.0 | 6.0 |
You will also need a fast and reliable Internet connection in order to download all of the components and databases required. While it is possible to install and use PolyPhen-2 on a computer without an Internet connection, such an installation would require substantial extra effort and is not discussed in detail herein.
Installation Steps
Download the latest PolyPhen-2 source code from: http://genetics.bwh.harvard.edu/pph2/dokuwiki/downloads.
-
Extract the source tarball:
$ tar vxzf polyphen-2.2.2r402.tar.gz
This will create a PolyPhen-2 installation tree in the current directory, which will be called something like polyphen-2.2.2 (this will be different for different versions of the software).
-
Download the database tarballs from the same site.
The bundled database tarball is required. The two precomputed alignment tarballs are recommended, but not required. If you choose not to install the MLC alignments, PolyPhen-2 will attempt to build MLC alignments for your proteins automatically on its first invocation and subsequently use them for all further runs. This is a highly computationally intensive task and may take very long time if you are going to analyze variants in more than just a handful of unique protein sequences. The MultiZ alignments cannot be recreated by PolyPhen-2, and if they are not installed, only MLC alignments are used for conservation inference, significantly reducing the coverage of PolyPhen-2 predictions for difficult-to-align sequences. -
Extract the tarballs you just downloaded, by entering commands similar to the following:
$ tar vxjf polyphen-2.2.2-databases-2011_12.tar.bz2 $ tar vxjf polyphen-2.2.2-alignments-mlc-2011_12.tar.bz2 $ tar vxjf polyphen-2.2.2-alignments-multiz-2009_10.tar.bz2
All contents will be extracted into the same installation directory created in step 2. -
If you want to rename your PolyPhen-2 installation directory or move it to another location, you should do so at this point:
$ mv polyphen-2.2.2 pph2
Replace polyphen-2.2.2 in the example above with the name of the directory as unpacked from the tarballs, and pph2 with the path to the desired directory.
Renaming or moving the top-level PolyPhen-2 directory after the installation steps below have been completed will render your PolyPhen-2 installation unusable, as will altering the internal subdirectory structure of the PolyPhen-2 installation. -
Set up the shell environment for your PolyPhen-2 installation by typing the following commands (if you are using Linux and the bash shell; different commands may be required for different systems).
$ cat ≫ ~/.bashrc export PPH=/home/login/pph2 export PATH=“$PATH:$PPH/bin” <Ctrl-D> $ source ~/.bashrc
Replace /home/login/pph2 in the example above with the correct path to your PolyPhen-2 installation directory.
Throughout this protocol, $PPH will be used to denote the path to your PolyPhen-2 installation directory. With the shell environment set up according to these instructions, command examples below should work by copying and pasting or typing them at your bash shell prompt. -
Download the NCBI BLAST+ tools from: ftp://ftp.ncbi.nih.gov/blast/executables/LATEST/.
The recommended version of BLAST+ is 2.2.26. Avoid BLAST+ v2.2.24 at all costs. Due to a nasty bug in this version, the makeblastdb command will take an excessively long time to format a database (up to several days). This issue has been fixed in BLAST+ v2.2.25. - Install BLAST+ binaries by typing commands like the following (for a 64-bit Linux system):
$ wget ftp://ftp.ncbi.nih.gov/blast/executables/LATEST/ncbi-blast-2.2.26+-x64-linux.tar.gz $ tar vxzf ncbi-blast-2.2.26+-x64-linux.tar.gz $ mv ncbi-blast-2.2.26+/* $PPH/blast/ -
Optionally, download and install Blat.
Download Blat binaries or sources according to instructions here: http://genome.ucsc.edu/FAQ/FAQblat.html#blat3.
If you need to build Blat from source, follow the instructions on the site above.
- If you chose to download Blat, copy the files required by PolyPhen-2 to the PolyPhen-2 installation directory.
$ cp blat twoBitToFa $PPH/bin/
-
Ensure that the executable bit is set for all downloaded binaries:
$ chmod +x $PPH/bin/* $ chmod +x $PPH/blast/bin/*
Without Blat tools, PolyPhen-2 is limited to gene annotations from the UCSC hg19 database and protein sequences and annotations fromUniProtKB. Blat tools are necessary in order to analyze variants in novel, unannotated, or otherwise nonstandard genes and proteins, including RefSeq and Ensembl genes.
-
Download and install the UniRef100 nonredundant protein sequence database:
$ cd $PPH/nrdb $ wget ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/uniref/uniref100/uniref100.fasta.gz $ gunzip uniref100.fasta.gz $ $PPH/update/format_defline.pl uniref100.fasta >uniref100-formatted.fasta $ $PPH/blast/bin/makeblastdb -in uniref100-formatted.fasta -dbtype prot -out uniref100 -parse seqids $ rm -f uniref100.fasta uniref100-formatted.fasta
Note that on a 32-bit Linux system, you may encounter the following error on the second-to-last step: Unable to open input uniref100- formatted.fasta as either FASTA file or BLAST db. It is recommended that you prepare your database on another, 64-bit computer using a 64-bit version of BLAST+, and then copy uniref100*.p?? files to the $PPH/nrdb/ folder on your 32-bit system. Alternatively, a simple workaround is to pipe the contents of the FASTA files into the standard input of makeblastdb, using the cat command. Replace the second-to-last command above with the following:$ cat uniref100-formatted.fasta | $PPH/blast/bin/makeblastdb -dbtype prot -title “UniRef100” -out uniref100 -parse seqidsIt is possible to use a sequence database other than UniRef100, but it will require modifying the format_defline.pl script. Consult the following link for descriptions of FASTA definition line formats supported by NCBI BLAST+: ftp://ftp.ncbi.nih.gov/blast/documents/formatdb.html. -
Download a copy of the PDB database:
$ rsync -rltv --delete-after --port=33444 rsync.wwpdb.org::ftp/data/structures/divided/pdb/$PPH/wwpdb/divided/pdb/ $ rsync -rltv --delete-after --port=33444 rsync.wwpdb.org::ftp/data/structures/all/pdb/$PPH/wwpdb/all/pdb/RCSB may occasionally change the layout of the PDB directories on their FTP site. If you encounter errors while mirroring PDB contents, please consult the instructions on the RCSB Web site: http://www.rcsb.org/pdb/static.do?p=download/ftp/index.html. -
Download a copy of the DSSP database:
$ rsync -rltvz --delete-after rsync://rsync.cmbi.ru.nl/dssp/ $PPH/dssp/Quote from the DSSP Web site: Please do these rsync jobs between midnight and 8:00am Dutch time! -
Download all remaining packages with the automated download procedure:
$ cd $PPH/src $ make download
If automatic downloading fails for whatever reason, you will need to manually (using your Web browser or wget command) download missing packages via the URLs below and save them to your $PPH/src directory:
After the packages are downloaded, change into $PPH/src directory and repeat the make download command.
- Build and install these remaining programs:
$ cd $PPH/src $ make clean $ make $ make install
-
Run the configure script to configure your installation:
$ cd $PPH $ ./configure
This will launch a series of interactive prompts to configure PolyPhen-2. At every prompt, you can safely stick to defaults by pressing the Enter key. The purpose of the script is to create a reasonable default configuration, which you can fine-tune later.
The resulting configuration will be stored in $PPH/config/*.cnf files. You can edit these files later, or remove them all and repeat these steps to revert back to distribution defaults. See Advanced Configuration Options under Critical Parameters for more details on these files. -
Optionally, test your PolyPhen-2 installation by running the PolyPhen-2 pipeline with the test set of protein variants and compare the results to the reference output files in the $PPH/sets folder:
$ cd $PPH $ bin/run_pph.pl sets/test.input 1>test.pph.output 2>test.pph.log $ bin/run_weka.pl test.pph.output >test.humdiv.output $ bin/run_weka.pl -l models/HumVar.UniRef100.NBd.f11.model test.pph.output >test.humvar.output $ diff test.humdiv.output sets/test.humdiv.output $ diff test.humvar.output sets/test.humvar.outputThe last two commands (the diff commands) should produce no output, indicating that the output from your PolyPhen-2 installation and the reference output is the same. If you update PolyPhen-2 built-in databases manually, then some differences (e.g., in the PSIC scores) are expected and normal since the exact values depend on the particular versions of databases installed; see Troubleshooting for details.
ALTERNATE PROTOCOL 3: USING PolyPhen-2 STANDALONE SOFTWARE
The PolyPhen-2 analysis pipeline consists of three separate components, each one executed by a dedicated Perl program:
MapSNPs (mapsnps.pl) Genomic SNP annotation tool PolyPhen-2 (run_pph.pl) Protein variant annotation tool PolyPhen-2 (run_weka.pl) Probabilistic variant classifier
The complete PolyPhen-2 analysis pipeline involves optionally running mapsnps.pl to translate genomic SNPs to protein substitutions, and then running the other two scripts in order:
[ (SNPs) → mapsnps.pl] → (Substitutions) → run_pph.pl → run_weka.pl →(Predictions)
Materials
A Linux computer with the PolyPhen-2 standalone software installed as described in Alternate Protocol 2
Prepare the input file. The file may be in any of the three batch formats described under Basic Protocol 2, above (protein substitutions, reference SNP IDs, or genomic variants).
-
If the input file is in the genomic variants format, map the variants into protein substitutions using the MapSNPs software. A typical command line is as follows:
$ $PPH/bin/mapsnps.pl -g hg19 -m -U -y subs.pph.input snps.list 1>snps.features 2>mapsnps.log &where:
snps.list is an input text file with chromosome coordinates and allele nucleotides prepared by a user.
subs.pph.input is an output file with the protein substitution specifications in run_pph.pl input format.
snps.features output file with functional annotations of the SNPs in user input (see Table 7.20.1).
-
mapsnps.log is a log file with the program’s errors and warnings.
If the input file is already in the form of protein substitutions or reference SNP IDs, skip this step; it is only necessary for genomic variants.
-
Run the PolyPhen-2 protein annotation tool. A typical command line is as follows:
$ $PPH/bin/run_pph.pl subs.pph.input 1>pph.features 2>run_pph.log &where:
subs.pph.input is an input text file with the protein substitution specifications, either prepared by a user or generated by the mapsnps.pl program.
pph.features is an output file with detailed functional annotations of the SNPs in user input.
run_pph.log is a log file with program’s errors and warnings.
-
Run the PolyPhen-2 probabilistic classification tool. A typical command line is as follows:
$ $PPH/bin/run_weka.pl pph.features 1>pph.predictions
where:
pph.features is an input file with SNPs functional annotations produced by the run_pph.pl program.
-
pph.predictions output file with predictions and scores (see Table 7.20.2).
This uses the default HumDiv-based model for predictions (see Background Information). To use the HumVar-based model instead, add the following option:-l $PPH/models/HumVar.UniRef100.NBd.f11.model
SUPPORT PROTOCOL 2: UPDATING BUILT-IN DATABASES
Updates for the PolyPhen-2 built-in protein annotation and sequence databases may occasionally be provided via the PolyPhen-2 downloads page (http://genetics.bwh.harvard.edu/pph2/dokuwiki/downloads). These updates are checked for errors and guaranteed to match the most current version of the software. The recommended way of updating your installation is to download and install these packages from this page, using the same procedure as in steps 3 and 4 of Alternate Protocol 2. After downloading and installing a new database bundle, you should then update your sequence and structural databases by repeating steps 10 to 12 from Alternate Protocol 2.
The following protocol describes the process of updating local copies of the built-in databases manually, instead of downloading the updated bundle from the Web site. Normally, this is not recommended, but is possible to do with the help of the scripts in the $PPH/update/ directory. It is important to update all of the databases at the same time to ensure annotations and sequences always stay in sync. A few of the update steps will require substantial computer resources to complete in reasonable time. A powerful multi-CPU workstation or a Linux cluster may be required to run them.
Materials
A Linux computer with the PolyPhen-2 standalone software installed as described in Alternate Protocol 2 and a sufficiently fast Internet connection. Steps 5 and 6 require Blat tools installed (see Alternate Protocol 2, step 9) and involve substantial amounts of calculation. In order for these steps to complete within a reasonable time, it is recommended to use a powerful multi-CPU workstation or a Linux cluster.
Update your sequence and structural databases, following instructions in steps 10 to 12 of Alternate Protocol 2.
- Create a temporary directory and switch into it:
$ mkdir dbupdates $ cd dbupdates
- Create and update the combined UniProtKB/Pfam sequence/annotation databases:
$ $PPH/update/uniprot.pl $ $PPH/update/unipfam.pl $ mv -f * $PPH/uniprot/
-
Create and update the PDB sequences database:
$ $PPH/update/pdb2fasta.pl $PPH/wwpdb>pdb2fasta.log 2>&1 & $ mv -f * $PPH/pdb2fasta/Substitute your local PDB mirror directory path for $PPH/wwpdb if different. Note that the pdb2fasta.pl script may take several hours to complete. -
MapUniProtKB protein sequences to translated CDS sequences for all UCSC (hg19) knownGene transcripts (requires blat):
$ $PPH/update/seqmap.pl $PPH/uniprot/human.seq $PPH/ucsc/hg19/genes/knownGeneAA.seq 1>up2kg.tab 2>up2kg.log & $ $PPH/update/seqmap.pl $PPH/ucsc/hg19/genes/ knownGeneAA.seq $PPH/uniprot/human.seq 1>kg2up.tab 2>kg2up.log &Each command will take up to several days to complete if run on a single CPU. Consider using techniques described in the “Parallel execution support” section under Critical parameters (the seqmap.pl script supports both the -r N/M option and cluster array mode). - Create and update the SQLite database with sequence maps:
$ $PPH/update/map2sqlite.pl up2kg.tab upToKg.sqlite $ $PPH/update/map2sqlite.pl kg2up.tab kgToUp.sqlite $ mv -f upToKg.sqlite kgToUp.sqlite $PPH/ucsc/hg19/genes/ Repeat steps 5 and 6 for the hg18 assembly version if necessary.
COMMENTARY
Background Information
Predictive features
PolyPhen-2 predicts the functional effect of a single-nucleotide change based on a variety of features derived from sequence annotations, multiple sequence alignments, and, where available, 3-D structures. These parameters include sequence annotations downloaded from the UniProtKB database and sequence features calculated by PolyPhen-2 and other programs. For sites thatmap to 3-D structures, they include annotations downloaded from the DSSP database and structural features calculated by PolyPhen-2. All these features are listed in Table 7.20.4.
Table 7.20.4.
Predictive Features Used by PolyPhen-2
| Feature | Source | Notes |
|---|---|---|
| Bond annotation | UniProtKB/Swiss-Prot annotation | Includes annotation codes DISULFID(disulfide bond), CROSSLNK(covalent link between proteins) |
| Functional site annotation | UniProtKB/Swiss-Prot annotation | Includes annotation codes BINDING(binding site for any chemical group), ACT_SITE(enzyme active site), METAL (binding site for a metal ion), LIPID(lipidated residue), CARBOHYD(glycosylated residue), MOD_RES(other covalent modification), NON_STD(non-standard amino acid), SITE (other interesting site) |
| Region annotation | UniProtKB/Swiss-Prot annotation | Includes annotation codes TRANSMEM(membrane-crossing region), INTRAMEM(membrane-contained region with no crossing), COMPBIAS(region with compositional bias), REPEAT(repetitive sequence motif or domain), COILED (coiled coil region), SIGNAL(endoplasmic reticulum targeting sequence), PROPEP(sequence cleaved during maturation) |
| PHAT score | PHAT trans-membrane specific matrix (Ng et al., 2000) | Measures effect of substitutions in trans-membrane regions; only used for positions annotated as trans-membrane. |
| PSIC score | PSIC software (Sunyaev et al., 1999) | See text for details. |
| Secondary structure annotation | DSSP database (Joosten et al., 2011) | Only used for sites that map to a 3-D structure. |
| Solvent-accessible surface area | DSSP database (Joosten et al., 2011) | Value in Å2. Only used for site that map to a 3-D structure. |
| Phi-psi dihedral angles | DSSP database (Joosten et al., 2011) | Only used for sites that map to a 3-D structure. |
| Normalized accessible surface area | Calculated by PolyPhen-2 | Calculated by dividing the value retrieved from DSSP by the maximal possible surface area. The maximal possible surface area is defined by the 99th percentile of the surface area distribution for this particular amino acid type in PDB. Only used for sites that map to a 3-D structure. |
| Change in accessible surface propensity | Calculated by PolyPhen-2 | Accessible surface propensities (knowledge-based hydrophobic “potentials”) are logarithmic ratios of the likelihood of a given amino acid occurring at a site with a particular accessibility to the likelihood of this amino acid occurring at any site (background frequency). Only used for sites that map to a 3-D structure. |
| Change in residue side chain volume | Calculated by PolyPhen-2 | Value in Å3. Only used for sites that map to a 3-D structure. |
| Region of the phi-psi map (Ramachandran map) | Calculated by PolyPhen-2 | Calculated from the dihedral angles retrieved from DSSP. Only used for sites that map to a 3-D structure. |
| Normalized B-factor | Calculated by PolyPhen-2 | B-factor is used in crystallographic studies of macromolecules to characterize the “mobility” of an atom. It is believed that the values of B-factor of a residue may be correlated with its tolerance to amino acid substitutions (Chasman and Adams, 2001). Only used for sites that map to a 3-D structure. |
| Ligand contacts | Calculated by PolyPhen-2 | Contacts of the query residue with heteroatoms, excluding water and “non-biological” crystallographic ligands that are believed to be related to the structure determination procedure rather than to the biological function of the protein. Only used for sites that map to a 3-D structure. |
| Interchain contacts | Calculated by PolyPhen-2 | Contacts of the query residue with residues from other polypeptide chains present in the PDB file. Only used for sites that map to a 3-D structure. |
| Functional site contacts | Calculated by PolyPhen-2 | Contacts of the query residue with sites annotated as BINDING, ACT_SITE, LIPID, or METALin the site annotation retrieved from UniProtKB. Only used for sites that map to a 3-D structure. |
Sequence annotations
A substitution may occur at a specific site, e.g., active or binding, or in a non-globular, e.g., transmembrane, region. Given a query protein, PolyPhen-2 tries to locate the corresponding entry in the human proteins subset of the UniProtKB/Swiss-Prot database (The UniProt Consortium, 2011). It then uses the feature table (FT) section of the corresponding entry to check if the amino acid replacement occurs at a site which is annotated as participating in a covalent bond, as a site of interest, or as part of a region of interest. The list of specific annotations that are retrieved can be found in Table 7.20.4.
Based on these annotations, it is determined whether the substitution is in an annotated or predicted transmembrane region. For substitutions in transmembrane regions, PolyPhen- 2 uses the PHAT trans-membrane specific matrix score (Ng et al., 2000) to evaluate possible functional effect.
Multiple sequence alignment and PSIC score
The amino acid replacement may be incompatible with the spectrum of substitutions observed at the position in the family of homologous proteins. PolyPhen-2 identifies homologs of the input sequences via BLAST search in the UniRef100 database. The set of BLAST hits is filtered to retain hits that have:
sequence identity to the input sequence in the range 30% to 94%, inclusively, and alignment with the query sequence not smaller than 75 residues in length.
Sequence identity is defined as the number ofmatches divided by the complete alignment length.
The resulting multiple alignment is used by the PSIC software (Position-Specific Independent Counts) to calculate the so-called profile matrix (Sunyaev et al., 1999). Elements of the matrix (profile scores) are logarithmic ratios of the likelihood of a given amino acid occurring at a particular position to the likelihood of this amino acid occurring at any position (background frequency). PolyPhen-2 computes the difference between the profile scores of the two allelic variants in the polymorphic position. Large positive values of this difference may indicate that the studied substitution is rarely or never observed in the protein family. PolyPhen-2 also shows the number of aligned sequences at the query position. This number may be used to assess the reliability of profile score calculations.
Mapping of the substitution site to known protein 3-D structures
Mapping of an amino acid replacement to the known 3-D structure reveals whether the replacement is likely to destroy the hydrophobic core of a protein, electrostatic interactions, interactions with ligands, or other important features of a protein. If the spatial structure of the query protein itself is unknown, one can use homologous proteins with known structure.
PolyPhen-2 BLASTs the query sequence against the protein structure database (PDB; http://www.pdb.org/) and retains all hits that meet the given search criteria. By default, these criteria are:
sequence identity threshold is set to 50%, since this value guarantees the conservation of basic structural characteristics
minimal hit length is set to 100
maximal number of gaps is set to 20.
By default, a hit is rejected if its amino acid at the corresponding position differs from the amino acid in the input sequence. The position of the substitution is then mapped onto the corresponding positions in all retained hits. Hits are sorted according to the sequence identity or E-value of the sequence alignment with the query protein.
Structural parameters
Further analysis performed by PolyPhen-2 is based on the use of several structural parameters. Some of these features are retrieved from the DSSP (Dictionary of Secondary Structure in Proteins) database (Joosten et al., 2011), while others are calculated by PolyPhen-2. These features are listed in Table 7.20.4. Importantly, although all parameters are reported in the output, only some of them are used in the final decision rules.
Contacts
The presence of specific spatial contacts of a residue may reveal its role for the protein function. The suggested default threshold for all contacts to be displayed in the output is 6 Å. However, the internal threshold for contacts to be included in the prediction is 3Å. For evaluation of a contact between two atom sets PolyPhen-2 finds the minimal distance between the two sets.
By default, contacts are calculated for all found hits with known structure. This is essential for the cases when several PDB entries correspond to one protein, but carry different information about complexes with other macromolecules and ligands.
Specific types of contacts are listed in Table 7.20.4.
Prediction algorithm
PolyPhen-2 predicts the functional significance of an allele replacement from its individual features by a Naïve Bayes classifier trained using supervised machine learning.
Two pairs of datasets were used to train and test PolyPhen-2 prediction models. The first pair, HumDiv, was compiled from all damaging alleles with known effects on the molecular function causing human Mendelian diseases, present in the UniProtKB database, together with differences between human proteins and their closely related mammalian homologs, assumed to be non-damaging. The second pair, HumVar, consisted of all human disease-causing mutations from UniProtKB, together with common human nsSNPs (MAF>1%) without annotated involvement in disease, which were treated as non-damaging.
The user can choose between HumDiv- and HumVar-trained PolyPhen-2 models. Diagnostics ofMendelian diseases requires distinguishing mutations with drastic effects from all the remaining human variation, including abundant mildly deleterious alleles. Thus, the HumVar-trained model should be used for this task. In contrast, the HumDiv-trained model should be used for evaluating rare alleles at loci potentially involved in complex phenotypes, dense mapping of regions identified by genome-wide association studies, and analysis of natural selection from sequence data, where even mildly deleterious alleles must be treated as damaging.
For a mutation, PolyPhen-2 calculates the Naïve Bayes posterior probability that this mutation is damaging and reports estimates of the prediction sensitivity (True Positive Rate, the chance that the mutation is classified as damaging when it is indeed damaging) and specificity (1 – False Positive Rate, the chance that the benign mutation is correctly classified as benign). A mutation is also appraised qualitatively as benign, possibly damaging, or probably damaging based on pairs of False Positive Rate (FPR) thresholds, optimized separately for each of the two models (HumDiv and HumVar).
Currently, the thresholds for this ternary classification are 5%/10% FPR for the HumDiv model and 10%/20% FPR for the HumVar model. Mutations whose posterior probability scores correspond to estimated false positive rates at or below the first (lower) FPR value are predicted to be probably damaging (more confident prediction). Mutations whose posterior probabilities correspond to false positive rates at or below the second (higher) FPR value are predicted to be possibly damaging (less confident prediction). Mutations with estimated false positive rates above the second (higher) FPR value are classified as benign. If no prediction can be made due to a lack of data then the outcome is reported as unknown.
Critical Parameters
Documentation of command-line tools
As described in Alternate Protocol 3 above, the PolyPhen-2 pipeline is composed of three command-line tools: mapsnps.pl, run_pph.pl, and run_weka.pl.
To get help with each program’s options and arguments, execute the script without arguments; to get extended help, as well as the input format description, use the -h option:
$ $PPH/bin/mapsnps.pl -h $ $PPH/bin/run_pph.pl -h $ $PPH/bin/run_weka.pl -h
MapSNPs (mapsnps.pl) is an optional tool which can be used when you have a list of chromosome positions and allele nucleotides as input. The main PolyPhen-2 module (run_pph.pl) requires protein substitutions as its input, so MapSNPs will perform the conversion.
MapSNPs maps genomic SNPs to human genes using the UCSC human genome assembly and knownGene set of transcripts, reports all missense variants found, fetches a UniProtKB/Swiss-Prot protein entry with a sequence matching that of the transcript’s CDS, and outputs a list of corresponding amino acid residue substitutions in the UniProtKB protein in a format accepted by the next pipeline step (run_pph.pl).
MapSNPs has some limitations. It only works with single-nucleotide variants; neither insertions/deletions nor other multi-nucleotide sequence changes are supported. It can only annotate biallelic variants—if you specify several alternative nucleotides as alleles, each one in turn will be paired with reference nucleotide and analyzed as a separate biallelic variant. Finally, MapSNPs only works with the human genome assembly (but both hg19 and hg18 are supported).
See Table 7.20.1 for complete description of the MapSNPs functional annotations.
PolyPhen-2 protein annotation tool (run_pph.pl) extracts protein annotations from various sequence and structural databases and calculates several evolutionary conservation scores from multiple sequence alignments (building an MSA in the process if it is not already present). The output of run_pph.pl is only an intermediate step in the analysis pipeline and is not intended to be directly interpreted by a user. The reason for this separate step is that it wraps up the most computationally-intensive operations in a single program and allows to run it on a distributed parallel high-performance computing system. See below for a description of built-in support for Grid Engine and Platform LSF parallel execution environments.
PolyPhen-2 probabilistic classification tool (run_weka.pl) takes annotations, conservation scores, and other features generated by run_pph.pl and produces, for each variant input, a qualitative prediction outcome (benign, possibly damaging, probably damaging) and a probability score for the variant to have a damaging effect on the protein function. Several prediction confidence scores are also included in the output, these being model sensitivity, specificity, and false discovery rate at the particular probability threshold level.
Note that run_weka.pl is fairly fast and should not produce any warnings, so there is normally no need to run it in the background.
See Table 7.20.2 for complete description of the run_weka.pl output. Most likely, you will be interested in the contents of the prediction and pph2_prob columns; the rest of the annotations are only useful if you want to investigate some of the supporting features in detail.
In addition to these tools, there is also a pph wrapper script available, which combines run_pph.pl and run_weka.pl program calls in a single command and can be used for analyzing small batches of protein variants:
$ $PPH/bin/pph subs.pph.input >pph.predictions
The pph script utilizes the default HumDiv classifier and provides almost no diagnostics in case of errors. Its use is discouraged due to limited functionality.
Advanced configuration options for standalone software
Storing large databases outside main installation tree
It is often convenient to keep the databases and large datasets PolyPhen-2 uses in a separate location (or locations) outside the rest of the PolyPhen-2 files, e.g., on a dedicated file server. This can be achieved by moving selected subdirectories from the installation tree and editing the corresponding lines in the $PPH/config/databases.cnf configuration file to point to the new locations. See the comments inside the databases.cnf file for exact format of each database path configuration option.
Below is the full list of variables that can be reconfigured (original PolyPhen-2 installation subdirectories for the corresponding databases are given in brackets).
NRDB_BLAST (nrdb)
Nonredundant protein sequence database (UniRef100).
UNIPROT (uniprot)
UniProtKB protein sequences and annotations.
PDB (wwpdb)
RCSB PDB structures.
DSSP (dssp)
DSSP structural database.
GOLDENPATH (ucsc)
UCSC human genome annotations and sequences.
You can also move the precomputed protein alignments to another location in a similar way. Edit $PPH/config/paths.cnf to reflect the change:
PRECOMPATH (precomputed)
UniRef100-based multiple sequence alignments.
Be careful to move or copy each subdirectory as a whole, preserving its internal tree. PolyPhen-2 relies on a predefined internal structure for each of its data directories.
Shared installation
If you want several users on a multiuser system to be able to share the same PolyPhen-2 installation, you might need to make a few changes to the default configuration.
First of all, make sure file/directory permissions are set properly, in order to allow all users read access to the PolyPhen-2 installation directory and all its subdirectories.
Edit $PPH/config/paths.cnf and change the following line:
From (default):
SCRATCH = $CONFIG{PPH}/scratch
to:
SCRATCH = $ENV{HOME}/scratch
Alternatively, users can specify a path to their personal scratch directory via the -d dumpdir option each time they execute run_pph.pl:
$ $PPH/bin/run_pph.pl -d ~/tmp/scratch
Personal and project configuration files
It is possible to create and use several different PolyPhen-2 configurations with the same installation. PolyPhen-2 looks for its configuration directories in several locations in the following order of preference:
./.pph/(in the current directory) ~/.pph/(in the user’s home directory) $PPH/config/(in the PolyPhen-2 installation directory).
Whichever configuration directory is found first will be used. All other configuration sources elsewhere in the system will be ignored, so ensure that all configuration files are present in each alternative configuration directory.
To create your own user-specific configuration, make a full copy of the system-wide defaults first:
$ mkdir ~/.pph $ cp $PPH/config/*.cnf ~/.pph/
Now, edit the configuration files inside ~/.pph/to match your preferences. Remember that you always need to copy a complete set of configuration files to your alternative configuration directory, even if you only want to change a single configuration option.
Proteins not in UniProtKB
If you know the UniProtKB accession or entry name for your protein, then this is all you need to submit to run_pph.pl, together with the variant’s sequence position and a pair of amino acid residue codes. If, however, your variants are in some other known protein (e.g., from the RefSeq or Ensembl databases) or you have a novel/unannotated sequence to analyze, you can still submit it to run_pph.pl, with a little extra effort.
If your protein is from one of the supported “alien” databases, you can simply use an “alien” database accession in place of a UniProtKB one. PolyPhen-2 knows about and automatically recognizes protein accessions from the following databases:
GeneID RefSeq GI PDB GO IPI UniParc PIR UniGene
EMBL EMBL-CDS Ensembl Ensembl TRS Ensembl PRO
Note, however, that even when your query is defined by one of these “alien” databases, PolyPhen-2 always uses UniProtKB accessions internally. The software uses cross-reference data from UniProtKB to translate all non-UniProt identifiers to UniProtKB accessions. These cross-references are ambiguous in many cases: while the entries in different databases may be related, their sequences may not be necessarily identical. This may cause PolyPhen-2 to report sequence errors, because it may not be able to locate the correct residues at the sequence positions specified.
To avoid this problem, or if you want to analyze new or unannotated proteins, you may prefer to submit full protein sequences to run_pph.pl, in addition to the input file with the variants specifications.
Prepare a text file with all your protein sequences in standard FASTA format. The following is an excerpt from the NCBI BLAST User Guide (http://www.ncbi.nlm.nih.gov/BLAST/blastcgi-help.shtml):
A sequence in FASTA format begins with a single-line description, followed by lines of sequence data. The description line (defline) is distinguished from the sequence data by a greater-than (>) symbol at the beginning. It is recommended that all lines of text be shorter than 80 characters in length. An example sequence in FASTA format is:
>gi|129295|sp|P01013|OVAX_CHICK GENE X PROTEIN QIKDLLVSSSTDLDTTLVLVNAIYFKGMWKTAFNAEDTREMPFHVTKQESKPVQMMCMNNSFNVATLPAEKMKILELPFASGDLSMLVLLPDEVSDLERIEKTINFEKLTEWTNPNTMEKRRVKVYLPQMKIEEKYNLTSVLMALGMTDLFIPSANLTGISSAESLKISQAVHGAFMELSEDGIEMAGSTGVIEDIKHSPESEQFRADHPFLFLIKHNPTNTIVYFGRYWSP >gi|6754226|ref|NP_034580.1|homeobox protein Hox-A11 MMDFDERGPCSSNMYLPSCTYYVSGPDFSSLPSFLPQTPSSRPMTYSYSSNLPQVQPVREVTFREYAIEPATKWHPRGNLAHCYSAEELVHRDCLQAPSAAGVPGDVLAKSSANVYHHPTPAVSSNFYSTVGRNGVLPQAFDQFFETAYGTPENLASSDYPGDKNAEKGPQAAAATSAAAVAAAATGAPATSSSDGGGGGGCQEAAAEEKERRRRPESSSSPESSSGHTEDKAGGSGGQRTRKKRCPYTKYQIRELEREFFFSVYINKEKRLQLSRMLNLTDRQVKIWFQNRRMKEKKINRDRLQYYSANPLL
Blank lines are not allowed in the middle of FASTA input.
Sequences are expected to be represented in the standard IUB/IUPAC amino acid and nucleic acid codes, with these exceptions: lower-case letters are accepted and are mapped into upper-case; a single hyphen or dash can be used to represent a gap of indeterminate length; and in amino acid sequences, U and *are acceptable characters.
If you are preparing the FASTA file manually, you can use simplified FASTA identifiers:
>P01013 QIKDLLVSSSTDLDTTLVLVNAIYFKGMWKTAFNAEDTREMPFHVTKQESKPVQMMCMNNSFNVATLPAEKMKILELPFASGDLSMLVLLPDEVSDLERIEKTINFEKLTEWTNPNTMEKRRVKVYLPQMKIEEKYNLTS >NP_034580 MMDFDERGPCSSNMYLPSCTYYVSGPDFSSLPSFLPQTPSSRPMTYSYSSNLPQVQPVREVTFREYAIEPATKWHPRGNLAHCYSAEELVHRDCLQAPSAAGVPGDVLAKSSANVYHHPTPAVSSNFYSTVGRNGVLPQA
The only mandatory rule is that identifiers entered after the >symbol should be unique strings without embedded blanks in them.
Take care never to use standard UniProtKB accessions or entry names as identifiers for non-UniProtKB sequences in your FASTA files. UniProtKB identifiers are used internally by PolyPhen-2 to access canonical UniProtKB sequences in its built-in database. If you edit a UniProtKB sequence, remember always to change the accession in its definition line, e.g., by appending a unique version string to the identifier:
>P01013.X10
Prepare your input file for run_pph.pl using protein identifiers that match the primary accessions from your FASTA file. Note that PolyPhen-2 always ignores “gi” accession numbers present in GenBank FASTA deflines, so the primary accessions for the two proteins listed above would be P01013 and NP_034580.1, and the input file would look like this:
P01013 5 L A
P01013 10 S F
NP 034580.1 15 Y P
NP 034580.1 25 G R
...
Now, submit both files to the run_pph.pl program using the -s seqfile.fa command-line option:
$ $PPH/bin/run_pph.pl -s myproteins.fa myvariants.input
where myproteins.fa is the file with your sequences in FASTA format and myvariants.input is the input file with the variants specified using unique protein identifiers matching the ones in myproteins.fa.
Note that analyzing novel proteins may involve building multiple sequence alignments. This may take plenty of computational resources and CPU time if the number of proteins is large. Consider running run_pph.pl in parallel mode on a multi-CPU server or cluster; see “Parallel execution support,” below.
Parallel execution support
Both the mapsnps.pl and run_pph.pl programs have some level of support for parallel execution built in. This makes them capable of efficiently utilizing multi-CPU and/or multi-core computer hardware for processing very large datasets.
If your computer has a multi-core CPU, you can start multiple instances of run_pph.pl via this simple bash (v3) wrapper script (shown here for 4 cores, but you can change the number of instances according to the number of cores your system has):
---cut here---
#!/bin/bash
M=4 # number of program instances to run
for ((N=1; N<=$M; N++)); do
$PPH/bin/run_pph.pl -r $N/$M “$@” 1>pph$N.features
2>pph$N.log &
done
wait
rm -f pph.features pph.log
for ((N=1; N<=$M; N++)); do
cat pph$N.features ≫pph.features
cat pph$N.log ≫pph.log
rm -f pph$N.features pph$N.log
done
---cut here---
Copy and save the above code to a file (e.g., run_pph4.sh), set the executable bit on it, and run it in the background:
$ chmod +x run_pph4.sh
$ ./run_pph4.sh -d ~/tmp/scratch $PPH/sets/test.input
>run_pph4.log 2>&1 &
This will create a pph.features output file identical to $PPH/sets/test.pph.output, except that it will take approximately one-fourth of the time that the single process would take on the same quad-core computer.
If you have PolyPhen-2 installed on a Linux cluster with either the Grid Engine or Platform LSF distributed execution environment management systems, you can submit multiple instances of run_pph.pl or mapsnps.pl without any additional scripts, simply as standard array jobs. Both programs will recognize they are running on a cluster in array mode and split the input file automatically, processing it in parallel.
Submitting a Grid Engine array job:
$ qsub -cwd -b y -N pph -t 1-16 $PPH/bin/run_pph.pl
pph.input
Submitting a Platform LSF array job:
$ bsub -cwd ‘pwd’ -J ’pph[1-16]’ -o pph.o%J.%I -e
pph.e%J.%I $PPH/bin/run_pph.pl pph.input
Both examples will submit 16 instances of run_pph.pl to the default cluster queue. Results will be written to files named by appending the job ID and sequential task number to the job name (pph in these examples). You will then have to collect the results manually from these files:
$ cat pph.o*.{1..16} >pph.features
$ cat pph.e*.{1..16} >pph.log
You may need to enable the PATH environment variable adjustment configuration option for cluster jobs by uncommenting the following line in your $PPH/config/paths.cnf file:
GRID_PATH = /bin:/usr/bin:/usr/java/latest/bin:/usr/local/bin
Edit the PATH directories search list to match your cluster’s configuration.
Analyzing reciprocal substitutions
The conservation scores PolyPhen-2 computes are directional, i.e., they depend on the direction of substitution. PolyPhen-2 will check amino acid residues in user input (AA1 and AA2) against the reference residue found in the query protein sequence at the substitution position to make sure AA1 matches the reference AA, so that the observed change follows the rule:
If PolyPhen-2 finds instead that AA2 in the user input matches the reference AA, it will swap the two user-submitted residues before calculating scores, so that AA1 always matches the query sequence. A warning message will be posted in such cases, e.g.:
WARNING: Swapped input residues AA1 (V) and AA2 (M) for P12259 query sequence at position (1764)
This is done in an attempt to automatically correct putatively polymorphic sites in user input, under the assumption that the query sequence always has a correct reference/ancestral state. This is mostly true for Swiss-Prot “canonical” entry sequences, but may not be the case for other proteins, especially user-submitted sequences.
This behavior can be disabled by editing $PPH/options.cnf and disabling the SWAPRESIDUES option by changing it from 1(default) to 0:
SWAPRESIDUES = 0
When SWAPRESIDUES option is disabled, all variants where AA1 does not match the reference (query) sequence will be reported as fatal input errors and their processing will be skipped.
However, in some specific cases you may want to analyze variants assuming they have actually occurred in the reciprocal direction:
One such example would be analyzing substitutions that occurred in the human lineage since the split from a close sister species (e.g., chimp). Here, AA1 would be a reconstructed common ancestral state residue and AA2 would be a variant that has become fixed in humans. To enable the reciprocal mode, you will need to change the REVERSEDIRECTION option in $PPH/options.cnf:
REVERSEDIRECTION = 1
Note that while in reciprocal mode, PolyPhen-2 will still use multiple sequence alignments created with human protein sequences as queries for all its inference of conservation scores. It will not attempt to reconstruct a proper common ancestor sequence for homology searches and MSA building. In practice, however, the difference in alignments for close enough sister sequences should be insignificant.
Analyzing variants in other species
While PolyPhen-2 evolutionary models were trained on human proteins, it is quite plausible to assume they should work for other mammals as well, and perhaps even across other classes, at least to some extent. The HumDiv model in particular does not incorporate any population-specific information, and can be applied to classify variants in proteins from other species besides humans.
Note, however, that all prediction confidence scores (FPR, TPR, FDR) are calculated for models trained on human-derived data and will no longer be reliable when human models are used with mutations in other species.
PolyPhen-2 has some experimental support for analyzing variants in other species built-in. This is the list of supported species common names (besides human):
orangutan mouse rat dog zebrafish fruitfly
Some broader taxonomic groups of species are also supported, based on UniProtKB set of taxonomic divisions, e.g., bacteria. For a full list, please see: ftp://ftp.uniprot.org/pub/data-bases/uniprot/current_release/knowledgebase/taxonomicdivisions/.
To set up this feature, you need to perform a fresh PolyPhen-2 install, according to the instructions in Alternate Protocol 2, above. Skip downloading and installing the MLC and MultiZ multiple alignments. If you already have MLC alignments installed into $PPH/precomputed directory, you will need to delete or rename the directory and replace it with an empty stub tree instead:
$ cd $PPH $ mv precomputed/precomputed.human.bak $ mkdir precomputed $ cd precomputed $ mkdir alignments blastfiles lock profiles structures $ cd ..
In this case, make sure also to remove all previously generated files stored under the $PPH/scratch/tree:
$ find $PPH/scratch -depth -type f -delete
Then, you must download and prepare the extra annotation and sequence databases for your species of interest:
$ cd $PPH/uniprot $ $PPH/update/uniprot.pl -n mouse $ $PPH/update/unipfam.pl -n mouse
Finally, edit $PPH/options.cnf and enter correct common and scientific species names, e.g.:
REFORGANISM = Mus musculus REFORGCOMMON = mouse
You can leave the scientific name empty if not applicable:
REFORGANISM =
Also in the $PPH/options.cnf file, disable the MAPGENE option:
MAPGENE = 0
Note that MapSNPs supports human genome assemblies only. Genomic variants from other species must first be mapped to proteins/AA substitutions by some other tool, such as snpEff (http://snpeff.source-forge.net/) or Variant Tools (http://variant-tools.sourceforge.net/).
After you convert missense mutations to protein substitutions, format them according to the PolyPhen-2 input format specification and submit to run_pph.pl. A new set of multiple sequence alignments will be built automatically. Note that building MSA for your species from scratch may require plenty of computational resources if you want to analyze variants in a large number of proteins. Consider running run_pph.pl in parallel mode on a multi-CPU server or cluster (see “Parallel execution support,” above).
After obtaining run_pph.pl annotations for the variants, you can run the PolyPhen-2 classifier with the default HumDiv model in the usual way:
$ $PPH/run_weka.pl mouse.pph.features >mouse.predictions
Troubleshooting
Inconsistent predictions
You might notice a substantial degree of variance in the predictions and scores output by PolyPhen-2 Web server as compared to your local PolyPhen-2 installation. The most common source of discrepancies in PolyPhen-2 output is different versions of the nonredundant protein sequence database (UniRef100) utilized for constructing multiple sequence alignments (MSA).
PolyPhen-2 relies heavily on sequence conservation estimates derived from MSA, and both alignment coverage and quality depend on the set of homologous sequences sourced from the UniRef100 database. The UniProt Consortium normally updates its databases on a monthly basis, while the PolyPhen-2 Web service is updated quarterly. If you have updated PolyPhen-2 built-in databases for your installation manually, following the instructions given in Support Protocol 2, chances are that your local copy of the UniRef100 database (as well as of other databases) is more recent than the one utilized by the PolyPhen-2 Web server.
PolyPhen-2 is not the only method to suffer from this issue. In fact, a recent publication (Hicks et al., 2011) claims PolyPhen-2 was the least affected among the four different SNP prediction tools tested.
There are other factors that can affect prediction outcome, among them changes in structural databases (PDB and DSSP), in the classifier models, and in the analysis pipeline itself, but these are less likely to introduce noticeable inconsistencies.
Versions of all databases utilized for analysis are displayed on the PolyPhen-2 Web report page and included in all Batch Query report files.
UNKNOWN prediction
Sometimes PolyPhen-2 reports “This mutation is predicted to be UNKNOWN (score is not available)” as a prediction outcome. What PolyPhen-2 is trying to say is that predicting the substitution’s effect was not possible in this particular case due to lack of multiple sequence alignment. The issue was largely addressed in PolyPhen-2 v2.2.2, thanks to integration of MultiZ genomic multiple alignments. This allowed for expanding prediction coverage significantly, especially in non-globular domains. Overall, close to 95% of all sequence positions in known UniProtKB proteins can be now successfully classified. However, you may still encounter UNKNOWN predictions in rare cases.
The most likely reason for such reports is if much of the sequence of your protein of interest is non-alignable due to the presence of large stretches of repeats and/or high compositional biases. For known proteins, you can easily check this by browsing the UniProt sequence annotations for the protein: http://www.uniprot.org/uniprot/ <UniProtKB-accession>.
Such sequence features often make it impossible to search for homologous sequences, build reliable multiple alignments, and ultimately infer conservation scores. The issue affects many non-globular proteins, including collagens, matrix proteins, DNA/RNA-binding proteins, muscle proteins, and more.
You can see this for yourself in the PolyPhen-2 Web interface by clicking on the [+] icon next to the “Multiple sequence alignment” label to inspect the MSA for your protein. You can also start an interactive MSA browser via the link at the bottom of the in-line alignment viewer section (requires Java browser plug-in).
Anticipated Results
The output of the PolyPhen-2 prediction pipeline is a prediction of probably damaging, possibly damaging, or benign, along with a numerical score ranging from 0.0 (benign) to 1.0 (damaging). (In the long report format detailed in Table 7.20.2, these are found in the prediction and pph2_prob columns.) A prediction of probably damaging means that the query substitution is predicted to be damaging with high confidence, while a prediction of benign means that the query substitution is predicted to be benign with high confidence. A prediction of possibly damaging means that the query substitution is predicted to be damaging, but with low confidence. Though the possibly damaging score is often interpreted as an indication of a mild effect or low penetrance, in general, it is intended as a measure of prediction confidence rather than effect size.
It is difficult to estimate the accuracy of PolyPhen-2’s predictions in a systematic way, but in general we expect them to be reasonably accurate. Our own published estimate (for version 2.0.0) is that, for a false positive rate of 20%, PolyPhen-2 achieves true positive prediction rates of 92% on the HumDiv dataset and 73% on the HumVar dataset (Adzhubei et al. 2010), and our unpublished estimates for newer versions show slightly better performance. Estimates from other sources arrive at similar numbers (for example, Hicks et al. 2011).
Time Considerations
Running PolyPhen-2 analysis for a known protein entry in the UniProtKB database is usually very fast. On the Web server, such queries typically take less than 30 sec to complete once started. The time it takes to run batches increases with the number of queries in the batch, but is still fast: hundreds or thousands of queries can often be completed in minutes, with typical average server performance reaching 1000 queries per minute. While execution time is fast, the time it takes for a job to start running once it is submitted can be highly variable. If the cluster is healthy and not under an unusually heavy load, it should take only a few minutes for your job to reach the top of the queue. When many large jobs are running on the cluster at once, or when the server is undergoing maintenance, it can sometimes take as long as several days for a job to reach the top of the queue. If you have many large jobs to run, or need your jobs to finish quickly, it is recommended that you install the standalone software, as described in Alternate Protocol 2.
How quickly the standalone software runs depends a great deal on your hardware. On a modern computer or cluster, it should remain very fast, especially with parallel processing enabled (as described in Advanced Configuration Options above). One factor that will impact the speed a great deal is the presence of precomputed multiple sequence alignments.
The MLC set of precomputed alignments covers more than 130,000 sequences of human proteins from the UniProtKB database, which includes all known isoforms. The same set is also utilized by the PolyPhen-2 Web server. When a user-provided sequence in FASTA format is submitted as part of the Web query (or via the corresponding option of the standalone software), the software will first compare it to all sequences in the built-in UniProtKB database. If an exact match is found, precomputed alignments for the matching UniProtKB sequence will be utilized, ensuring fastest possible performance. If, however, the sequence appears not to be in the database, new alignments have to be constructed for each unique non-UniProtKB sequence encountered, resulting in a much slower execution.
Literature Cited
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley EA, Butte AJ, Wheeler MT, Chen R, Klein TE, Dewey FE, Dudley JT, Ormond KE, Pavlovic A, Morgan AA, Pushkarev D, Neff NF, Hudgins L, Gong L, Hodges LM, Berlin DS, Thorn CF, Sangkuhl K, Hebert JM, Woon M, Sagreiya H, Whaley R, Knowles JW, Chou MF, Thakuria JV, Rosenbaum AM, Zaranek AW, Church GM, Greely HT, Quake SR, Altman RB. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- Boyko AR, Williamson SH, Indap AR, Degenhardt JD, Hernandez RD, Lohmueller KE, Adams MD, Schmidt S, Sninsky JJ, Sunyaev SR, White TJ, Nielsen R, Clark AG, Bustamante CD. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 2008;4:e1000083. doi: 10.1371/journal.pgen.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasman D, Adams RM. Predicting the functional consequences of non-synonymous single nucleotide polymorphisms: Structure-based assessment of amino acid variation. J Mol Biol. 2001;307:683–706. doi: 10.1006/jmbi.2001.4510. [DOI] [PubMed] [Google Scholar]
- Hicks S, Wheeler DA, Plon SE, Kimmel M. Prediction of missense mutation functionality depends on both the algorithm and sequence alignment employed. Hum Mutat. 2011;32:661–668. doi: 10.1002/humu.21490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten RP, te Beek TAH, Krieger E, Hekkelman ML, Hooft RWW, Schneider R, Sander C, Vriend G. A series of PDB related databases for everyday needs. Nucleic Acids Res. 2011;39:D411–D419. doi: 10.1093/nar/gkq1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular Evolution and Phylogenetics. Oxford University Press; New York: 2000. [Google Scholar]
- Ng PC, Henikoff JG, Henikoff S. PHAT: A transmembrane-specific substitution matrix. Predicted hydrophobic and transmembrane. Bioinformatics (Oxford) 2000;16:760–766. doi: 10.1093/bioinformatics/16.9.760. [DOI] [PubMed] [Google Scholar]
- Sunyaev SR, Eisenhaber F, Rodchenkov IV, Eisenhaber B, Tumanyan VG, Kuznetsov EN. PSIC: Profile extraction from sequence alignments with position-specific counts of independent observations. Protein Eng. 1999;12:387–394. doi: 10.1093/protein/12.5.387. [DOI] [PubMed] [Google Scholar]
- Sunyaev S, Ramensky V, Koch I, Lathe W, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM, Broad GO, Seattle GO on behalf of the NHLBI Exome Sequencing Project. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2011;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics (Oxford) 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]