Abstract
Phosphatidylinositol 3-kinase α (PI3Kα) is a promising target for anticancer drug design. Oncogenic mutation H1047R in the catalytic domain is observed in many tumors and may enhance PI3Kα kinase activity by affecting loop confirmations as well as membrane binding. We applied docking methods to 33 PI3K inhibitors against the wild type (wt) PI3Kα, the H1047R mutant of PI3Kα and the γ isoform of PI3K (PI3Kγ). We also investigated the effect of protein flexibility on ligand binding by docking the same set of ligands to conformations of the wt and mutant PI3Kα generated by molecular dynamics simulations. Our data suggests that conformational differences in Gln859, Ser854, Tyr836, and Ser774 between the PI3Kα wt and H1047R mutant may be used to design ligands that are active against both the wt and H1047R mutant isoforms. Gln859, Ser854 and Ser774 may play critical roles in ligand binding to the α isoform H1047R mutant while formation of H-bonds with Ser806 of PI3Kγ may enhance γ-isoform-specific inhibition. In addition to H-bond interactions, structural and size differences in the activation and hydrophobic domains of PI3Kα, PI3Kγ, and the PI3Kα H1047R mutant could be exploited to direct the design of isoform- and/or mutant-specific PI3K inhibitors. Our data provide a reasonable explanation for the activity and selectivity of small molecular PI3K inhibitors and are in good agreement with available experimental and computational data.
1. INTRODUCTION
Phosphatidylinositol 3-kinases (PI3Ks) are lipid kinases that phosphorylate the 3-hydroxyl of phosphatidylinositol 4,5-bisphosphate (PIP2), generating phosphatidylinositol 3,4,5-trisphosphate (PIP3). The resulting PIP3 interacts with pleckstrin-homology- (PH-) domain-containing proteins, such as the Akt serine-threonine kinases, eliciting a series of signal transduction events that lead to DNA synthesis and cell proliferation via the activation of the MDM2 and mTOR (mammalian target of rapamycin) pathways.1 Under normal physiological conditions, PIP3 levels are tightly regulated by the phosphatase and tensin homologue protein (PTEN). The inactivation of PTEN by mutations in tumors leads to the accumulation of PIP3.2 One approach for antitumor drug design would be to design a small-molecule inhibitor of PI3K, thus lowering the PIP3 level.
Among four distinct PI3K subfamilies, only the class I PI3Ks are capable of catalyzing the conversion of PIP2 to PIP3. The class I PI3Ks can be further divided into IA and IB subclasses based on the mechanism of activation. The class IA PI3K proteins are activated by receptor tyrosine kinases (RTKs) through the regulatory unit (p85), whereas the class IB isoform (the PI3Kγ protein, encoded by PIK3CG) is activated by seven transmembrane G-protein-coupled receptors (GPCRs). The class IA subfamily contains three isoforms with different catalytic subunits: a p110α isoform (encoded by PIK3CA), a p110β isoform (encoded by PIK3CB), and a p110δ isoform (encoded by PIK3CD).3
Expression of PI3Kα and PI3Kβ is ubiquitous in mammalian tissues, whereas PI3Kδ and PI3Kγ show a more restricted distribution in leukocytes.4 PI3Kα is a heterodimeric protein consisting of a catalytic p110α subunit and a p85 regulatory subunit.5 The p110R subunit contains N-terminal adaptor-binding (ABD), Ras-binding, C2, helical, and catalytic kinase domains. The ABD domain was proposed to be responsible for p85α binding, and the C2 domain for cellular membrane binding. PI3Kα is activated by RTKs such as endothelial growth factor receptor (EGFR), human epidermal growth factor receptor 2 (HER2), and vascular endothelial growth factor receptor (VEGFR). The activated p110α catalytic subunit catalyzes the conversion of the PIP2 to PIP3.1
The implication of PI3Kα in cancer was confirmed by the observation that PI3Kα is frequently mutated in some human cancers. Perrone et al.6 observed four mutations in PIK3CA (E545K, E545A, E542K, and A1020V) in patients with advanced colorectal cancer who did not respond to the EGFR inhibitor cetuximab. Inhibition of the PI3K/AKT pathway restored the sensitivity of gefitinib (an EGFR inhibitor) in cell lines that were originally resistant to gefitinib treatment.7 Samuels et al.8 reported an observation of the incidence of tumors with PI3Kα mutations: colon, 32% (74/234); brain, 27% (4/15); and stomach, 25% (3/12). Recently, Liu et al.9 reported the incidence of tumors with PI3Kα mutations in a much larger population: breast, 27% (468/1766); endometrial, 24% (102/429); colon, 15% (448/3024); upper digestive tract, 11% (38/352); stomach, 8% (29/362); pancreas, 8% (29/362); and ovarian, 8% (61/787). These PI3Kα mutations exclusively cluster in two “hot-spot” regions in exons 9 and 20, corresponding to the helical (E545K) and kinase (H1047R) domains of p110α. E545K and H1047R account for ap- proximately 80% of PI3Kα mutants observed in diverse cancers (Catalog of Somatic Mutations in Cancer, www.sanger.ac.uk/genetics/CGP/cosmic). The PI3Kα mutant H1047R results in a 2-fold increase in lipid kinase activity, and its activity increases further upon phosphopeptide binding.10
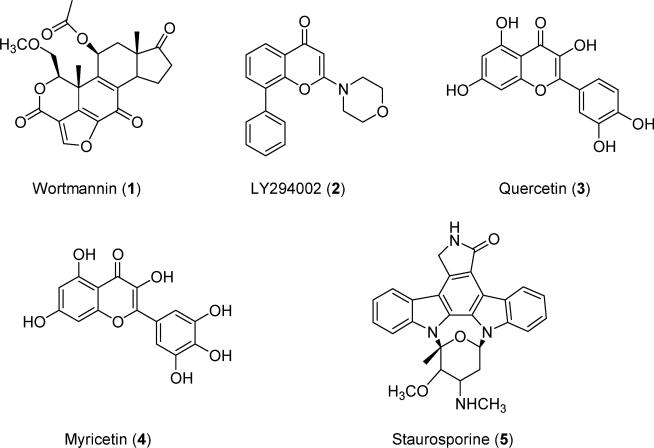
This gain-of-function character of PI3Kα mutants uniquely poses a potential to decrease the efficacy of many anticancer therapeutic drugs that target PI3Kα. On the other hand, these mutants make PI3Kα a unique drug target, in that other PI3K isoforms, such as PI3Kβ and PI3Kγ, do not exhibit the same types of mutations. Therefore, selective inhibition of the PI3Kα H1047R mutant might avoid the toxicity of pan-PI3K inhibitors such as wortmannin and LY294002 (Figure 1).
Figure 1.
Chemical structures of the pan-PI3K inhibitors.
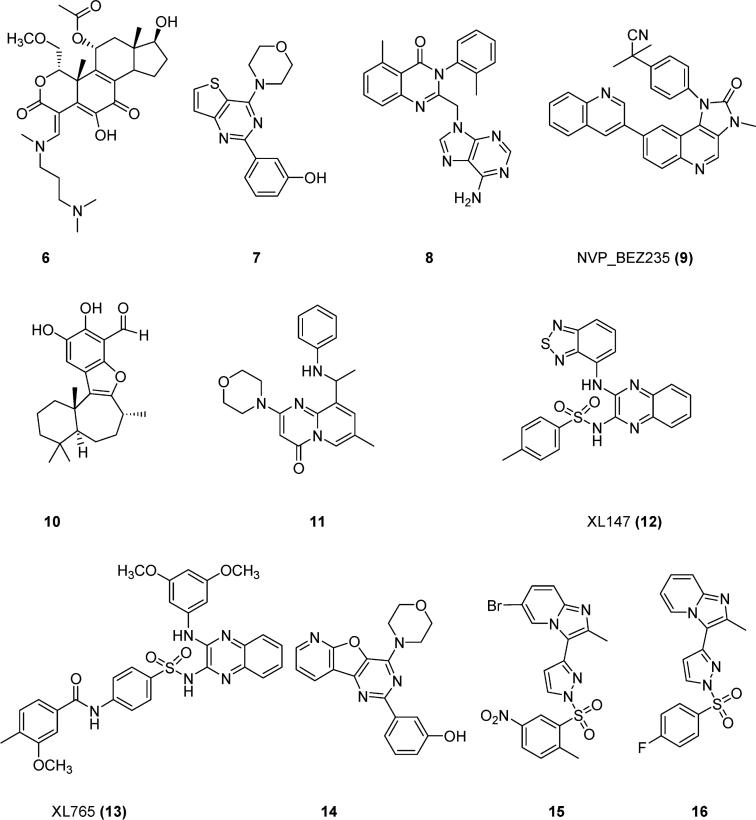
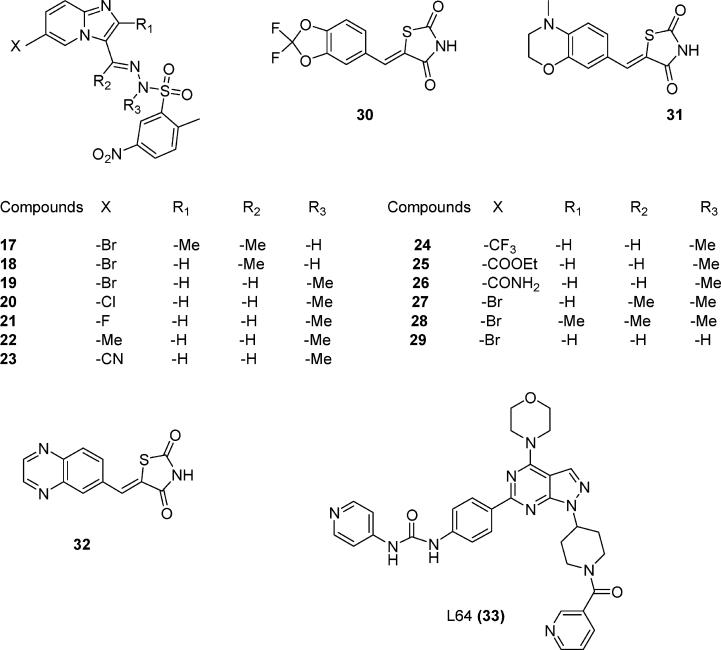
To discover molecules with anticancer potential, scientists have synthesized compounds targeting PI3Kα. Nonselective first-generation PI3K inhibitors such as wortmannin (1), LY294002 (2), quercetin (3), myricetin (4), and staurosporine (5) (Figure 1) inhibit both PI3Kα and PI3Kγ. Compound 6 (Figure 2), the ring-opened analogue of 1, inhibits both α and γ PI3Ks at nanomolar concentrations.11 Compound 7 (Figure 2) is a PI3Kα-selective inhibitor with a half-maximal inhibitory concentration (IC50) of 2.5 nM.12 Removal of a morpholine moiety in LY294002 generated 8 with much weaker PI3Kα activities.13 NVP_BEZ235 (9, Figure 2) is an imidazo[4,5-c]quinoline that inhibits both PI3Kα and PI3Kγ in the nanomolar range.14 Screening a library of marine invertebrate extracts in a fluorescent polarization assay identified liphagal (10, Figure 2) as a selective PI3Kα inhibitor with an IC50 value of 0.1 μM.15 Compound 11 has PI3Kγ selectivity,13 whereas XL147 (12) (Figure 2)16 and its structural homologue XL765 (13) (Figure 2)17 inhibit both PI3Kα and PI3Kγ at nanomolar concentrations. Oral administration of 12 resulted in significant inhibition of tumor growth in a mice model in which PI3K signaling was activated. In addition to inhibiting PI3K, 13 also inhibited the mammalian target of rapamycin (mTOR), another critical enzyme in the PI3K/AKT/mTOR pathway. IC50 values for 13 against PI3Kα, PI3Kγ, mTOR/Raptor (TORC1), and mTOR/Rictor (TORC2) were 0.039, 0.009, 0.190, and 0.908 μM respectively.36 Replacement of the thiophene in 7 with a pyridofuran yielded 14, which has a much better pharmacokinetic profile than 7.12 The imidazo[1,2-α]pyridine derivative (15, Figure 2) is a potent PI3Kα inhibitor, with an in vitro IC50 value of 3.1 nM.18 Structural modification of 15 generated compounds 16–29 (Figure 3).
Figure 2.
Chemical structures of reported PI3K inhibitors.
Figure 3.
Chemical structures of reported PI3Kα inhibitors.
Some docking studies investigating PI3Kα/ligand interactions have been published. However, these computational studies were based on either PI3Kγ19 or a PI3Kα homology model built on the PI3Kγ structure using homology modeling.13,20 A recent pharmacophore and docking study of PI3Kα inhibitors was reported by Li et al.21 However, no computational work on ligand interactions with the PI3Kα H1047R mutant has been reported. Given the prevalence of the H1047R mutant in cancer cells and the unique presence of H1047R in PI3Kα, targeting both the overexpressed wild-type (wt) PI3Kα and the PI3Kα mutant H1047R would have advantages over compounds that inhibit only the wt PI3Kα. Therefore, investigating factors dictating ligand interactions with the PI3Kα H1047R mutant might shed light on the development of isoform- and/or mutant-specific inhibitors with enhanced selectivity over other PI3K isoforms.
To identify the structural basis for PI3K isoform- or mutant-specific binding, we carried out a series of docking studies of reported PI3K inhibitors (Figures 1-3) with the native structures of PI3Kα, PI3Kα H1047R, and PI3Kγ. To study the effect of protein flexibility on ligand docking, we carried out molecular dynamics (MD) simulations on the wild-type and mutant PI3Kα and docked the same set of ligands to the protein conformations at the well-equilibrated 2000th-ps snapshot. Because of sequence and/or structural differences among these three proteins, we suggest that docking studies based on the native and H1047R mutant PI3Kα structures might provide useful insights for inhibitor design.
2. COMPUTATIONAL METHODS
2.1. Preparation of Protein Structures
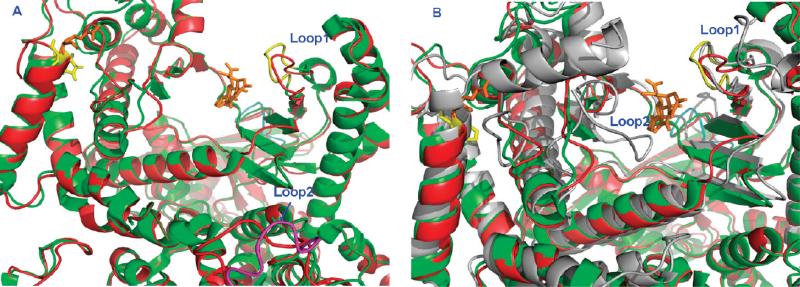
The X-ray crystal structures of apo PI3Kα (PDB id: 2RD0),5 PI3Kγ complexed with the ligand L64 (PDB id: 3IBE),22 and the PI3Kα H1047R mutant/wortmannin complex (PDB id: 3HHM)23 were retrieved from the RCSB Protein Data Bank. Structural alignment between these three proteins using the DaliLite program24 showed that these proteins are homologous to each other (Figure 4). Structural alignment of 3IBE and 1E8W,25 a PI3Kγ/quercetin complex, showed that L64 and quercetin share the same binding pocket. The coordinates of wortmannin in 3HHM were adopted to 2RD0 and were used as the ligand to generate the grid file for the docking study. There are four sections of missing residues in the crystal structure of 2RD0: section 1 (Tyr307–Thr324), section 2 (Ala415–Ala423), section 3 (Phe506–Asp527), and section 4 (Lys941–950). The structures of the missing residues were built using the homology modeling module in MOE.26 The template for section 1 was adopted from the chain B of 1A9X (1A9X.B);27 section 2, 2JRJ.A;28 section 3, 1JD5.A;29 and section 4, 1HUL.A.30 Each insertion of the missing section and its surrounding region (within 4.5 Å of the modeled section) was subjected to energy minimization to reduce steric clash. The overall 2RD0 was subsequently prepared using the Protein Preparation Wizard in the Schrödinger software suite.31 During the Protein Preparation Wizard procedure, the side-chain structures of Gln and Asn were allowed to flip to maximize H-bond interactions. The H1047R mutant of PI3Kα model was built based on the crystal 3HHM. The same missing residues in 2RD0 were also identified in 3HHM. Therefore, 3HHM was structurally aligned to 2RD0 using the DaliLite program, and the missing residues were adopted from the modified 2RD0 model. For docking studies, we adopted the kinase domain (residues between Gly725 and Ile1094) for the protein PI3Kγ. There were two sections of missing residues in the kinase domain of 3IBE (PI3Kγ). The first section of missing residues (Thr895–Asn898) was adopted from the protein 1E8W.25 The second section of missing residues (Ile968–Asn979) was modeled from the template structure 1RYK.A32 based on the homology modeling protocol as described in MOE. The modified regions were subjected to the same treatment as 2RD0 for the minimization and protein preparation procedures. All three proteins, PI3Kα wt (2RD0), PI3Kα H1047R (3HHM), and PI3Kγ (3IBE) were then subjected to 500 iterations of energy minimization with backbone atoms being restrained using the OPLS force field in the MacroModel module in the Schrödinger software suite.
Figure 4.
Kinase domains of PI3Ks and their interactions with ligands. (A) Active site of PI3Kα and the H1047R mutant. Color code: native, red cartoon; H1047R mutant (3HHM), green; activation loop (Loop1, Met772-Arg777), yellow (H1047R); C2 domain loop (Loop2, Val346-Lys353), pink (H1047R); Ligand, orange; His1047 (α native), yellow; Arg1047 (α mutant), orange. (B) Active site of PI3Kα, H1047R mutant (3HHM), and PI3Kγ proteins (3IBE). Color code: wt PI3Kα, red cartoon; H1047R mutant (3HHM), green; PI3Kγ, gray; activation loop (Loop1, Met772-Arg777), yellow, H1047R; red, wt PI3Kα; and gray, wt PI3Kγ. Ligand: yellow, His1047; orange, PI3Kα Arg1047 mutant; gray, Arg1076 of PI3Kγ; activation loop 2 (Loop2, His962-His967) of PI3Kγ, gray.
2.2 Preparation of Ligand Structures
Thirty-three PI3K inhibitors collected from various sources were built based on the template structure quercetin in 1E8W.25 Ligands 1–5 in Figure 1 are pan-PI3K inhibitors. Molecules 6–13 and 30–32 are inhibitors of both PI3Kα and PI3Kγ. Ligands 14–29 were designed to be PI3Kα-selective inhibitors.18 All ligands were built using the Maestro Build panel and minimized by the MacroModel program using the OPLS2005 force field. A database of 260,071 molecules was downloaded from the National Cancer Institute33 and was filtered according to the Lipinski's rule of five,34 yielding 33,778 druglike molecules. One hundred druglike molecules were randomly selected from this database and were then subjected to energy minimization using the MacroModel program.
2.3 Glide Docking Procedures
Three grid files for 2RD0, 3HHM, and 3IBE were generated using the Glide Grid Generation protocol with the bound ligands as centroids. All 33 PI3K inhibitors were docked to each of the three grid files, and the 100 druglike molecules were docked to the native 2RD0 grid file. During the docking process, the scaling factor for receptor van der Waals for the nonpolar atoms was set to 0.8 to allow for some flexibility of the receptor. All other parameters were used as defaults. The binding affinity of the PI3K/ligand complexes was expressed in terms of docking scores. The more negative the docking score, the more favorable the interaction of the complex. Unless stated otherwise, the figures of protein/ligand interactions were generated with the Maestro program in the Schrödinger software suite. In these figures, residues important for interactions are shown as stick models and labeled, all ligands are shown as ball-and-stick models, and H-atoms are hidden for clarity purposes.
2.4 Molecular Dynamics (MD) Simulations
Established procedures35 for MD simulations were adopted for this experiment. Briefly, each system (3HHM and 2RD0) was neutralized with sodium ions and soaked in a rectangular box of TIP3P water molecules36 extended 10 Å in all directions from any protein atom. Each system underwent 1000 steps of minimization using the steepest-descent algorithm to reduce the steric clash, followed by 30 ps of ramping the temperature from 10 to 300 K and 100 ps of equilibration at 300 K. Then, 2000 snapshots were collected during the 2000-ps production run using the NPT ensemble with a time step of 2 fs. The long-range electrostatic interactions were calculated with the particle–mesh Ewald (PME) method,37 and the nonbonded cutoff for the van der Waals interaction was set to 10 Å. All energy minimizations and MD simulations were carried out using the AMBER 10 package38 with the AMBER 99SB force field.39 The root-mean-square deviation (rmsd) of each collected snapshot in comparison to the crystal structure was obtained using the PTRAJ module in the AMBER 10 package and is included in Figure S1 in the Supporting Information. The 2000th-ps snapshot was used for studying the effect of protein flexibility on docking by docking the same set of ligands to these MD-derived structures.
3. RESULTS AND DISCUSSION
The sequence alignment between 3IBE and 2RD0 revealed that the percentage identity between these two sequences was only 35.5% based on 915 aligned residues (Table S2, Supporting Information). The structural alignment, however, showed that they are structurally homologous with a root-mean-square deviation (rmsd) of 2.5 Å between the backbones of these two structures. The alignment of the PI3Kα 3HHM mutant with 3IBE shows that both α and γ subtypes share the same binding pocket and that wortmannin (3HHM), quercetin (1E8W), and L64 (3IBE) occupy the same binding site (Figure 4B). The active site of PI3Kα H1047R (3HHM), defined as within 4.5 Å of the ligand wortmannin consists of Met772, Ser774, Lys776, Pro778, Trp780, Ile800, Tyr836, Glu849, Val850, Val851, Gln859, Thr856, Ser919, Met922, Ile932, and Asp933. The active site of the PI3Kγ (1E8W) is enclosed by the residues Typ812, Lys833, Asp836, Leu838, Asp841, Tyr867, Glu880, Ile881, Val882, Ala885, Thr886, Thr887, Lys890, Met953, Phe961, Ile963, Asp964, Phe965, and Gly966.
3.1 Validation of the Glide Docking Method
We carried out docking studies of 33 PI3K inhibitors against wt and mutant PI3Kα and PI3Kγ. Inhibitor docking scores against the three models are listed in Table 1, and the docking scores of the 100 druglike molecules against the wt 2RD0 model are listed in Table S1 (Supporting Information). To validate whether the docking results were sufficiently accurate for studying protein–ligand interactions in the PI3K system, we calculated the enrichment factor (ER) by docking a library of 33 PI3K inhibitors and 100 druglike molecules against the native PI3Kα model. The ER is defined as the concentration of active inhibitors in a subset divided by the concentration of active inhibitors in the database and is a general measurement of the efficiency of a docking program: the higher the ER, the more accurate the docking program. Docking of 133 molecules against the wt PI3Kα (2RD0) model produced an ER score of 3.30 (Table 2), indicating an approximately 3-fold increase in the probability of finding an active molecule in the top 10% of hits compared to an otherwise random selection. In the ideal case, the enrichment factor for our data set would be 4.29. The ER score of 3.30 for the Glide dock indicates that active ligands can be identified using this software.
Table 1.
Docking Scores of 33 PI3K Inhibitors against the Native PI3Kα (2RD0), PI3Kα H1047R Mutant (3HHM), PI3Kγ (3IBE), and MD-Generated Models of 2RD0 and 3HHM at the 2000th-ps Snapshot Conformations
| compound | IC50 PI3Kα (μM) |
IC50 PI3Kγ (μM) |
selectivity index |
2RD0 (PI3Kα native) |
3HHM (PI3Kα H1047R) |
3IBE (PI3Kγ) |
2RD0 (MD model) |
3HHM (MD model) |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.012a | 0.0042b | 0.35 | –9.91 | –12.04 | –5.73 | –11.61 | –12.90 |
| 2 | 0.550a | 1.4b | 2.55 | –9.79 | –10.51 | –9.34 | –11.82 | –11.51 |
| 3 | 3.8b | –9.08 | –11.17 | –10.89 | –12.83 | –11.11 | ||
| 4 | 1.8b | –9.24 | –10.71 | –11.18 | –12.22 | –9.92 | ||
| 5 | 9.0b | –10.67 | –11.23 | –9.30 | –11.04 | –10.74 | ||
| 6 | 0.0064c | 0.008c | 1.25 | –9.39 | –11.21 | –6.95 | –7.78 | –10.38 |
| 7 | 0.0025f | 0.66f | 264.00 | –8.44 | –9.05 | –11.15 | –11.27 | –10.55 |
| 8 | >5i | >5i | –10.52 | –9.56 | –10.92 | –8.75 | –7.57 | |
| 9 | 0.004j | 0.005j | 1.25 | –13.48 | –12.99 | –11.52 | –7.11 | –13.67 |
| 10 | 0.100a | –9.64 | –11.56 | –7.09 | –10.58 | –10.52 | ||
| 11 | >100i | 29i | –9.20 | –10.97 | –10.43 | –10.07 | –11.46 | |
| 12 | 0.039d | 0.023d | 0.59 | –8.61 | –9.22 | –6.52 | –12.89 | –12.27 |
| 13 | 0.039e | 0.009e | 0.23 | –10.87 | –8.47 | –9.52 | –14.74 | –9.88 |
| 14 | 0.0036f | 0.25f | 69.44 | –8.62 | –10.55 | –8.78 | –9.02 | –10.73 |
| 15 | 0.0031g | –10.22 | –10.49 | –9.95 | –10.19 | –11.31 | ||
| 16 | 0.67g | –9.82 | –9.20 | –11.08 | –8.72 | –10.22 | ||
| 17 | 0.4g | –8.64 | –8.96 | –8.24 | –7.74 | –9.06 | ||
| 18 | 0.017g | –8.00 | –9.44 | –7.11 | –6.62 | –9.80 | ||
| 19 | 0.0003g | 0.040g | 133.33 | –8.47 | –8.67 | –7.69 | –8.04 | –9.82 |
| 20 | 0.00077g | –9.70 | –9.28 | –9.03 | –8.18 | –9.76 | ||
| 21 | 0.0053g | –8.56 | –8.70 | –7.33 | –6.71 | –7.94 | ||
| 22 | 0.0060g | –8.90 | –8.46 | –8.21 | –8.08 | –8.97 | ||
| 23 | 0.00026g | –9.26 | –9.09 | –8.82 | –8.26 | –7.79 | ||
| 24 | 0.014g | –8.11 | –8.73 | –8.47 | –7.75 | –7.95 | ||
| 25 | 0.34g | –9.33 | –9.25 | –8.70 | –8.91 | –10.30 | ||
| 26 | 0.78g | –8.70 | –9.05 | –7.97 | –8.27 | –9.03 | ||
| 27 | 0.081g | –8.09 | –7.40 | –7.11 | –7.34 | –9.15 | ||
| 28 | 0.17g | –7.77 | –8.12 | –7.46 | –7.23 | –8.29 | ||
| 29 | 0.021g | –8.21 | –8.72 | –6.88 | –7.99 | –6.46 | ||
| 30 | 4.5i | 0.25i | –8.90 | –10.18 | –8.01 | –9.49 | –12.91 | |
| 31 | >10i | >10i | –9.26 | –9.73 | –13.05 | –9.16 | –9.32 | |
| 32 | 0.06i | 0.008i | 0.13 | –11.24 | –11.33 | –10.37 | –9.19 | –10.97 |
| 33 | 0.548h | –9.08 | –6.42 | –9.26 | –10.80 | –6.51 |
The listed IC50 values were used to define active ligands that were used to derive the enrichment factor. Compounds with IC50 > 10.0 μM were considered inactive.
Table 2.
Enrichment Factor of the Glide Docking against PI3Kα (2RD0)
| number of active PI3Kα inhibitors | 31 |
| number of total molecules in the database | 133 |
| number of PI3Kα inhibitors in the top 10% of poses (the top 13) | 10 |
| enrichment factor (ERa) | 3.30 |
ER = (10/13)/(31/133) = 3.30. The ER for ideal case = (13/13)/(31/133) = 4.29.
The performance of the Glide dock program was also evaluated by comparing the docked poses to native conformations in the crystal structures. The rmsd values for heavy atoms between the Glide-generated docked poses and the native poses for quercetin, myricetin, LY294002, wortmannin, and L64 are listed in Table 3. All docked poses have rmsd values lower than 2.0 Å. Figure S2 (Supporting Information) shows the superposition of the Glide-generated LY294002 and the native conformation in 1E7V, indicating that the Glide program is able to successfully reproduce the native conformation. The rmsd between these two poses is 0.41 Å. Other than the very flexible L64, the rmsd values of all other ligand docked poses are either lower than or very close to 1.0 Å (Table 3). These low rmsd values indicate that Glide dock is capable of identifying the native poses in crystal structures and can be reliably used to predict the binding conformations of other ligands.
Table 3.
Ligand rmsd Values (Heavy Atoms, Å) between the Docked Posed from Glide and the Crystal-Structure Native Conformations
| Glide | |
|---|---|
| 1E8W (quercetin/γ) | 0.84 |
| 1E90 (myricetin/γ) | 1.09 |
| 1E7V (LY294002/γ) | 0.41 |
| 3IBE (L64/γ) | 1.60 |
| 1E7U (wortmannin/γ) | 0.93 |
| 3HHM (wortmannin/α) | 0.88 |
3.2 Binding Mode of pan-PI3K Inhibitors
Among 33 ligands, wortmannin (1, Figure 1) is a special case in that it forms a covalent bond with the Lys833 nitrogen atom of PI3Kγ through the furan ring,25 an important determinant of the low nanomolar binding affinity of this natural product. Structural analogs of 1 without the furan heterocyclic ring do not form a covalent bond with Lys833 and have much weaker or no binding affinity to PI3Kγ.40 In our docking study, 1 was docked to PI3Kγ in a noncovalent manner; not surprisingly, this resulted in a weak docking score. This result is in good agreement with the necessity of covalent bond formation for wortmannin binding to PI3Kγ. The crystal structure of 1E7U (PI3Kγ/wortmannin) shows no H-bond between the protein and the ligand. Our docking study generated one H-bond interaction between them via Lys890.
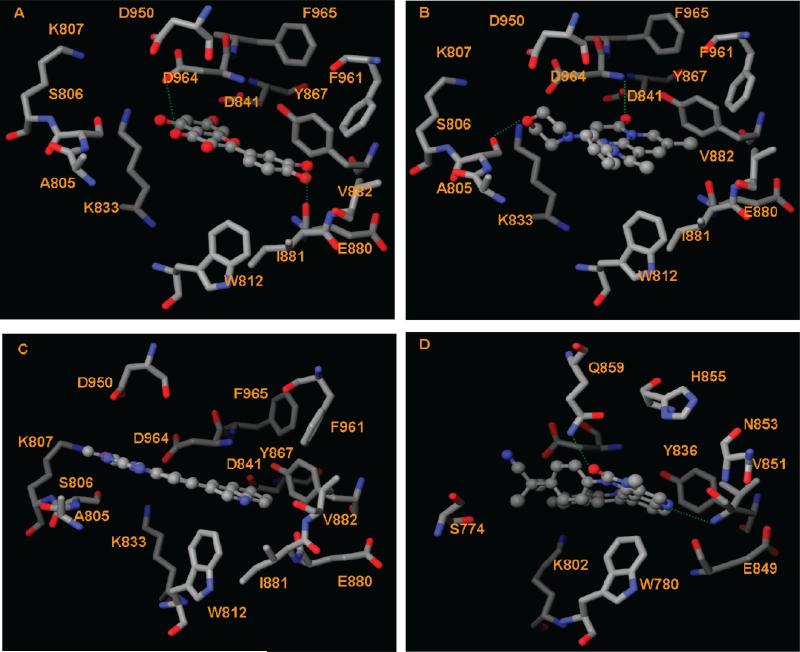
The crystal structure of the PI3Kα H1047R/wortmannin complex shows that residues Val851 and Asp933 of PI3Kα provide two H-bonds to wortmannin. The corresponding residues in the PI3Kγ isoform, Val882 and Asp964 (Table S2, Supporting Information), form H-bonds to quercetin (3) in the crystal structure 1E8W. The H-bond interactions between 33 PI3K inhibitors and the four PI3K models (Table 4) show that Val851 provides H-bonds with the majority of inhibitors for both wt and mutant H1047R PI3Kα. The corresponding residue Val882 of PI3Kγ forms H-bonds with more than 25% of the 33 PI3K inhibitors. For example, these docking studies show that both pan-PI3K inhibitors quercetin (3, Figure 1) and myricetin (4, Figure 1) form H-bonds to 2RD0 via Val851 and Asp933 and to 3IBE (PI3Kγ) via Asp964 and Glu880 (Figure 5A). Therefore, ligands capable of forming H-bonds with Val851 and/or Asp933 in PI3Kα and the corresponding Val882 and/or Asp964 in PI3Kγ should inhibit both isoforms. This set of H-bond interactions provides a reasonable explanation for the observation that many PI3K inhibitors are effective against both α and γ isoforms. In addition, Ser806 (PI3Kγ)/Ser774 (PI3Kα) appears to be important for ligand binding, more so for the γ isoform in that this residue provides H-bonds with 10 of the 33 ligands for PI3Kγ but less than 4 of the 33 ligands for the three PI3Kα models (Table 4). The selectivity of 11 toward the γ isoform (Table 1) might be attributed to its ability to form H-bonds with Ser806 and Asp964. The docked pose of 11 in the γ protein pocket shows that the morpholino group of 11 lies in the polar region of PI3Kγ, forming favorable H-bond interactions with Ser806; in addition, the carbonyl of 11 forms a H-bond with the backbone amide of Asp964 (Figure 5B). The PI3Kγ-selective 12 and 13 also form H-bonds with Ser806.
Table 4.
Hydrogen-Bond Interactions between 33 PI3K Inhibitors and the Four PI3K Models
| compound | 2RD0 (PI3Kα native) | 3HHM (PI3Kα H1047R) | 3IBE (PI3Kγ) | 2RD0 (MD model) | 3HHM (MD model) |
|---|---|---|---|---|---|
| 1 | Val851 | Val851, Gln859, Ser774 | Lys890 | Lys776 | Ser774, Val851 |
| 2 | Val851, Tyr836, Asp933 | Gln859 | Ser806 | Val851 | Gln859, Val851 |
| 3 | Val851, Asp933 | Val851, Asp933, Gln859 | Glu880, Asp964 | Asp933, Lys802 | Glu849, Val851 |
| 4 | Val851, Lys802, Asp933 | Glu849, Gln859 | Glu880, Asp964 | Asp810, Lys802, Asp933 | Met922, Val851, Glu849, Ser774 |
| 5 | Val851 | Val851 | NAa | Val851 | Val851 |
| 6 | Val851 | Val851 | Lys890 | NAa | Ser774 |
| 7 | Val851, Ser774 | Val851 | Asp841, Val882 | NAa | Glu849 |
| 8 | Ser774 | Ser774 | Val882 | NAa | Gln859, Glu849 |
| 9 | Val851, Gln859 | Val851 | NAa | Lys776 | Gln859 |
| 10 | Tyr836, Lys802, Asp933 | NAa | NAa | Glu849, Val851 | |
| 11 | Val851, Gln859 | Val851, Gln859 | Ser806, Asp964 | NAa | Gln859, Val851 |
| 12 | Val851 | Val851 | Ser806, Val882 | Val851 | Ser854, Val851 |
| 13 | Tyr836 | Asp933, Asn920 | Ser806, Ala805 | Val851 | Asp933, His917, Asn920 |
| 14 | Val851, Ser774, Tyr836 | Val851, Glu849, Gln859 | Asp841, Val882 | Glu849, Gln859 | Glu849, Val851 |
| 15 | Val851 | Val851, Ser919 | Val882 | Val851 | Val851 |
| 16 | Val851, Gln859 | Gln859 | Val882 | Val851 | Val851 |
| 17 | Val851 | Val851, Arg770, Gln859 | NAa | NAa | |
| 18 | Val851 | Val851, Ser854 | Ser806, Asp964 | NAa | Val851 |
| 19 | Asp933 | Val851 | Ser806 | Val851 | Val851 |
| 20 | Val851, Tyr836 | Val851 | Lys807, Asp964 | NAa | NAa |
| 21 | Val851, Gln859, His855 | Val851, Asp933 | Ser806 | NAa | NAa |
| 22 | Val851 | NAa | NAa | NAa | Ser774 |
| 23 | Val851, Asn853 | Ser854 | Val882 | Val851, Ser854 | Ser774, Asn920, His917 |
| 24 | Val851 | Ser854 | Lys807, Asn951 | Val851 | His917 |
| 25 | Val851, His855 | Val851 | Val882 | Val851 | Ser854 |
| 26 | Val851, His855 | Ser774 | Ser806, Val882 | Val851, Ser854 | Gln859, Ser854 |
| 27 | Val851 | Ser774 | S806, Asp964 | NAa | NAa |
| 28 | NAa | Val851 | NAa | NAa | NAa |
| 29 | Val851 | Val851 | Ser806 | Val851 | Ser774 |
| 30 | Val851 | Val851 | Asp964 | Val851 | Ser774, Val851 |
| 31 | Val851, Asn853 | NAa | Val882 | Val851 | Val851 |
| 32 | Val851, Glu849, Tyr836 | NAa | Val882, Lys890 | Lys802, Asp810 | Ser774 |
| 33 | Lys802, Val851 | Gln859 | Lys833, Asp841 | Asp933 | His917 |
NA: not available, i.e., no H-bond interactions were observed.
Figure 5.
Interactions between PI3K inhibitors and the α/γ isoform. The H-bond interactions are depicted as green dotted lines. (A) Interactions between PI3Kγ and quercetin (3). (B) Interactions between PI3Kγ and compound 11. (C) Interactions between PI3Kγ and NVP_BEZ235 (9). (D) Interactions between PI3Kα and NVP_BEZ235 (9).
However, formation of H-bonds with Val882, Ser806, and/or Asp964 might not be a prerequisite for strong PI3Kγ binding. For instance, 9 is a strong PI3Kγ inhibitor with an IC50 value of 5.0 nM14 yet has no H-bond interactions with PI3Kγ. The docking score of the PI3Kγ/9 complex is −11.52 kcal/mol, placing it as one of the top hits for PI3Kγ and consistent with experimental observation that 9 is a potent inhibitor against PI3Kγ.14 The loss of H-bond interactions in 9 appears to be compensated by favorable van der Waals contacts with the hydrophobic pocket that is enclosed by residues Trp812, Tyr867, Phe961, Ile881, and Val882, as well as stabilizing electrostatic interactions between residues Asp950, Asp964, Lys833, Lys807, and Ser806 and the polar functional groups of 9 (Figure 5C). In addition, the T-shaped π–π stacking interactions between the aromatic rings of 9 and Trp812 of PI3Kγ (Figure 5C) might help stabilize the interactions between PI3Kγ and 9.
The antitumor inhibitory activity of 9 can be attributed to its strong interaction with PI3Kα: the IC50 value of 9 against PI3Kα is 4.0 nM. The nitrogen atom of the imidazo[4,5-c]quinoline core structure of 9 provides a key H-bond interaction with the backbone of the conserved PI3Kα Val851, and the carbonyl oxygen atom of the imidazo[4,5-c]quinoline of 9 forms a H-bond with Gln859 (Figure 5D). Formation of H-bonds with Gln859 of PI3Kα is also observed for other inhibitors. The corresponding residue in the γ isoform is Lys890. Differences between Gln859 of PI3Kα and Lys890 of PI3Kγ might be used to develop isoform-specific inhibitors.
3.3 Binding Mode of the PI3Kα-Selective Inhibitors
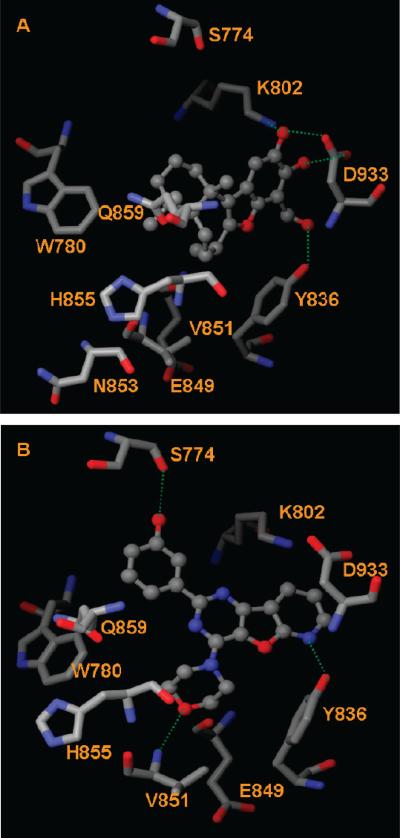
The high correlation between PI3Kα expression and tumor progression has prompted researchers to develop α-isoformspecific PI3K inhibitors. Anticancer compound 10 is the first example of a PI3Kα inhibitor with a terpenoid carbon skeleton and is approximately 10-fold more potent against PI3Kα than PI3Kγ. Our docking studies indicate that 10 interacts with PI3Kα through H-bonds with residues Tyr836, Lys802, and Asp933 (Figure 6A), whereas no H-bond interactions were observed between 10 and PI3Kγ. These three H-bonds were also observed in the crystal structure PI3Kα H1047R mutant/wortmannin.23
Figure 6.
Interactions between PI3Kα and (A) 10 and (B) 14. The H-bond interactions are depicted as green dotted lines.
A thieno[3,2-d]pyrimidine derivative (7, Figure 2) is a highly potent and selective PI3Kα inhibitor. However, the poor pharmacokinetic profile of 7 rendered it inactive in vivo.12 Replacement of the thieno[3,2-d]pyrimidine in 7 with a pyrido[3′,2′:4,5]furo[3,2-d]pyrimidine produced a compound (14, Figure 2) that was both potent and selective against PI3Kα. The selectivity index (SI) of 14, measured by the ratio IC50(PI3Kγ)/IC50(PI3Kα), was 69. Although the selectivity of 14 was lower than that of 7 (SI of 264), 14 exhibited a much better pharmacokinetic profile than 7.41 Inspection of PI3K/ligand interactions shows that a H-bond is formed between Val851 and the morpholino oxygen atoms of 7 and 14. The key 3-phenol functional group of both 7 and 14 makes a H-bond with Ser774 (Figure 6B). The formation of H-bonds with Ser774 of PI3Kα might contribute to the selectivity of 7 and 14. Similarly to the PI3Kα-selective 10, 14 forms a H-bond with Tyr836 of PI3Kα (Figure 6B).
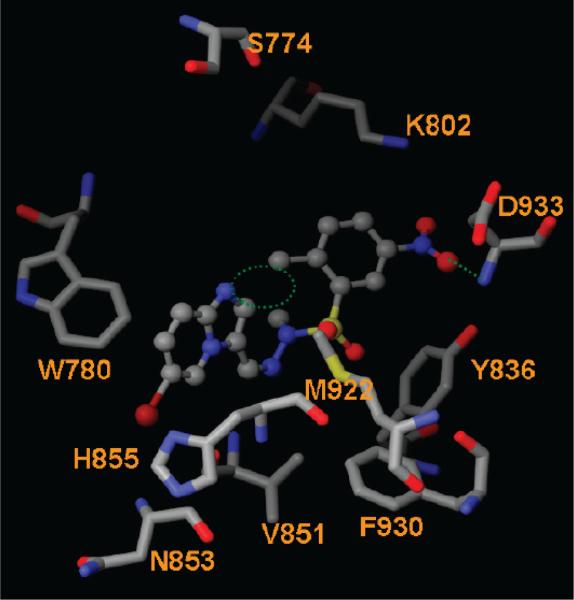
Similarly to the 3-phenol functional group in 14, the meta NO2 group of 15 (Figure 2) appears to be critical. Removal of the ortho methyl and the meta nitro groups and introduction of a para fluoro yielded compound 16, which is a 216-fold weaker inhibitor of PI3Kα than 15. This indicates that the ortho methyl and the meta NO2 groups are critical for PI3Kα inhibition. These two functional groups were maintained in compounds 17–29. Compounds 17–19 form H-bonds with Val851, Asp933, and/or Ser854 of the α isoform. However, none of these three compounds interacts with Val882 of the γ isoform; instead, they form H-bonds with Asp964 and Ser806. The binding affinity appears to relate to the van der Waals interactions between the imidazo[1,2-α]pyridines and the phenyl group of compound 17. Removal of the methyl group at the R1 position of 17 reduced the steric clash between R1 and the ortho methyl group on the phenyl ring of 19 (dotted circle in Figure 7). The decreased steric hindrance allows 19 to adopt conformations that allow for H-bond formation with Val851 and Asp933 (Figure 7). The introduction of a methyl group at the R3 position allows 19 to fit into a lipophilic cavity formed by Phe930, Met922, and Tyr836 (Figure 7). All of these factors might help to explain the potency and high selectivity index of 133 for 19. GSK2126458 inhibits PI3Kα, H1047R mutant, and PI3Kγ with Ki values of 0.019, 0.06, and 0.009 nM, respectively.42 The X-ray cocrystal structure of PI3Kγ/ GSK2126458 shows that it forms H-bonds with Val882, Tyr867, and Lys833 of the γ isoform.42 The corresponding residues in the α isoforms (both wt and H1047R) are Val851, Tyr836, and Lys802. H-bond formation with these residues is a characteristic of many PI3Kα-active ligands. This might explain the weaker selectivity of this compound.
Figure 7.
Interactions between PI3Kα and compound 19. The H-bond interactions are depicted as green dotted lines. The potential steric clash between R1 and the methyl group on the phenyl ring of 19 is highlighted as a green dotted circle.
3.4 Binding Mode of the PI3Kα Mutants/Ligand Interactions
It might be tempting to assume that an H1047R mutation, being distant from the binding pocket of PI3Kα, might not have a significant effect on ligand binding. The superposition of PI3Kα (3RD0) and its mutant (3HHM) reveals that this is not the case. Two noticeable differences are observed in loop 1 and loop 2 between the wt and H1047R PI3Kα forms. Loop 1 is the activation loop Arg770–Pro778 (yellow loop1), and loop 2 is the C2 domain loop (Val346–Lys353, pink loop 2, Figure 4A). The native PI3Kα apoprotein adopts a more open activation loop in the Met772–Arg777 region (red loop 1, Figure 4A). This loop in the H1047R mutant moves 1.33 Å toward the ligand, forming a slightly narrower binding pocket. The snapshot structures collected from our MD simulations, as well as those in Han and Zhang's experiments, also reveal a more closed conformation of the activation loop in the mutant.43 Histidine1047 of PI3Kα forms a H-bond with the main-chain carbonyl of Leu956, a residue within the activation loop. The mutation of His1047 to an arginine orients the positively charged arginine side chain toward the negatively charged membrane phospholipids and 90° away from the original position of the native histidine, disrupting the H-bond with Leu956.23 The C2 domain of the H1047R mutant moves toward the membrane by 7.30 Å (pink loop 2, Figure 4A). These structural changes in the H1047R mutant might alter the interaction of PI3Kα with the cell membrane, an interaction vital to its lipid kinase activity. Indeed, biochemical assays show that the enzymatic activity of the PI3Kα H1047R mutant is differentially regulated by lipid membrane composition.10
Interestingly, the wild-type PI3Kγ protein contains residue Arg1076, corresponding to Arg1047 in the PI3Kα H1047R mutant. Structural inspection reveals that the activation loop (residues Met804–Lys809) of PI3Kγ (gray loop 1, Figure 4B) adopts a closed conformation similar to the corresponding loop conformation on the PI3Kα mutant (residues Met772–Arg777 of PI3Kα, yellow loop 1, H1047R mutant; red loop1, native PI3Kα; Figure 4B). The conformation of loop residues His962–His967 of PI3Kγ (gray loop 2, Figure 4B) moves toward the ligand binding site, making the binding pocket smaller for the γ isoform. Structural differences in the kinase domains of wt PI3Kα, PI3Kα H1047R mutant, and PI3Kγ present a unique opportunity to design selective PI3Kα inhibitors.
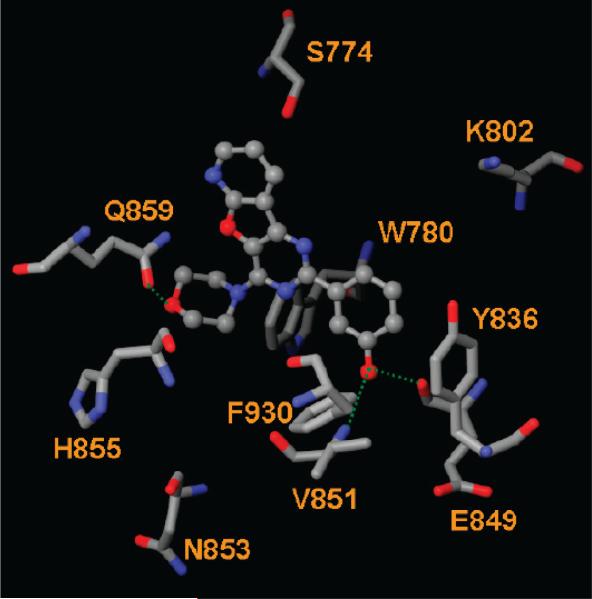
Our docking studies show that the docking scores of most PI3K inhibitors are comparable against both the wt and H1047R forms of PI3Kα. This indicates that an inhibitor effective against the wt PI3Kα might also be active against the H1047R mutant. For example, the docking scores of 9 against wt and mutant PI3Kα are −13.48 and −12.99 kcal/ mol, respectively, consistent with the very similar IC50 values of 4.0 and 4.6 nM for 9 against PI3Kα and the PI3Kα H1047R mutant, respectively.14 Compound 9 forms H-bonds with Val851 and Gln859 of both the wt and H1047R mutant. Some compounds such as 14, however, have a higher docking score for the H1047R mutant. Inspection of interacting residues shows that 14 forms H-bonds with Val851, Ser774, and Tyr836 of the wt and with Val851, Glu849, and Gln859 of the H1047R mutant (Figure 8). Table 4 shows that nine ligands were identified to form H-bonds with Gln859 of the H1047R mutant whereas only four formed H-bonds with Gln859 of the wt. This suggests that residue Gln859 might be critical for ligand binding to the PI3Kα H1047R mutant. Finally, many ligands form H-bonds with Ser854 and Ser774 of the H1047R mutant (Table 4), suggesting an important role for these two residues.
Figure 8.
Interactions between PI3Kα H1047R mutant and 14. The H-bond interactions are depicted as green dotted lines.
3.5 Effect of Protein Flexibility on Ligand Binding
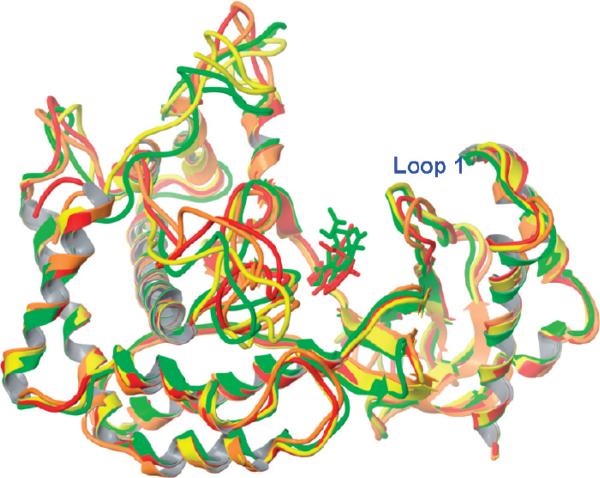
To investigate the role of protein flexibility in ligand binding, we carried out a 2-ns MD simulation on both the native and mutant PI3Kα. The native 2RD0 is an apo crystal structure, and therefore, the conformation of wortmannin in 3HHM was used for 2RD0 after structural alignment. The collected trajectories were compared with the crystal structure, and the stable rmsd values (Figure S1, Supporting Information) indicate that our systems were well-equilibrated. The 2000thps snapshots from both model systems were compared to their crystal structures (Figure 9). Figure 9 shows that the α-helices and β-sheets overlap well, whereas the loop regions are quite flexible. The activation loop 1 in the wt form adopts a more open conformation (orange loop) at the 2000th snapshot in comparison to that of the crystal structure (red loop, Figure 9); this is in good agreement with Han and Zhang's observation.43 The conformation for the same loop in the mutant protein, however, experienced little change. The widened flap (loop 1) in the wt makes it possible for a ligand to move deeper into the binding pocket. This is exactly what we observed in the docked pose of wortmannin (red ligand, Figure 9) in the MD-generated model. Loop 2 (residues Glu849–His855, Figures 9 and 10), however, remains quite stable during the MD simulations. This explains why Val851 is able to form H-bonds with many ligands in both the wt and mutant MD-generated models. The conformations of loop 3 (residues Gly912–Asn920) and loop 4 (residues Asp933–Arg951) (Figure 10) exhibited high flexibility, similar to Han and Zhang's observation.43
Figure 9.
Kinase domains of 2RD0 and 3HHM before and after MD simulations. Color code: 2RD0 crystal, red; 2RD0 MD model, orange; 3HHM crystal, yellow; 3HHM MD model, green. Wortmannin: red, docked pose from the 2RD0 MD model; green, docked pose from the 3HHM MD model.
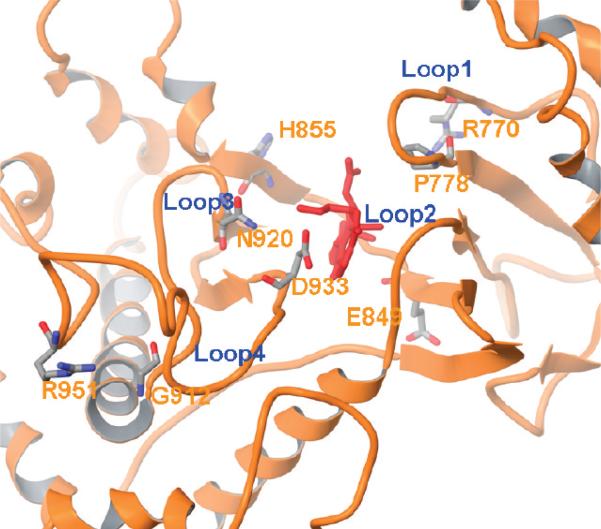
Figure 10.
Loop conformations of the 2RD0 MD model. Wortmannin is depicted in red. Residues denoting the start and end of the loop are highlighted and labeled in a stick model.
Docking of all 33 ligands to the MD-generated 2RD0 and 3HHM proteins was carried out, and the docking scores and residues forming H-bonds with ligands are listed in Tables 1 and 4, respectively. In comparison to the crystal structures, many ligands have better docking scores when binding to the MD-generated models, presumably because of the movement of loops 3 and 4, as well as loop 1 (for the wild-type model, Figure 10). Residues providing H-bonds to ligands vary according to their positions in the loops. Residues in and around loop 2 (Glu849–His855) experience less conformational movement and, therefore, are more frequently observed to form H-bonds with ligands; these residues include Tyr836, Glu849, Val851, Ser854, His855, and Gln859. Ser854 of both the wt and mutant models appear to form H-bonds with ligands. More ligands form H-bonds with Gln859 of the H1047R mutant models (both the crystal and MD-generated structures) suggesting that Gln859 of the H1047R mutant might play a more important role in ligand binding. The importance of Gln859 in ligand binding to PI3Kα and H1047R mutant has been confirmed by both other computational21 and experimental23 work. Residue Ser774 of 3HHM (MD model) forms H-bonds with eight ligands, whereas no H-bonds with Ser774 were observed in the MD-generated 2RD0 model. The outward movement of loop 1 of 2RD0 during MD simulations oriented the side-chain hydroxyl group away from the binding pocket. In this conformation, the main-chain carbonyl of Ser774 was a H-bond donor (Figure 11A). In contrast, the relatively closed activation loop positioned the Ser774 hydroxy group toward the binding pocket, making it available as a H-bond donor and acceptor in the mutant model (Figure 11B). Hence, ligands able to form H-bonds with Ser774 could be H1047R-mutant-specific.
Figure 11.
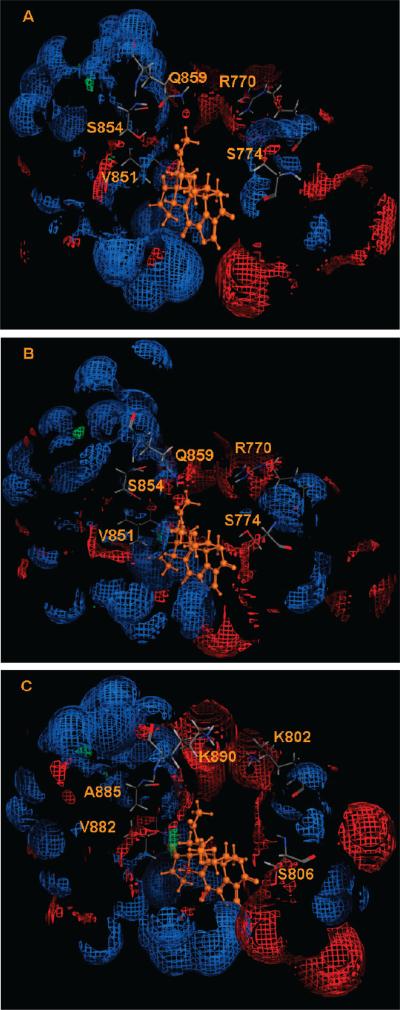
Electrostatic surface of the binding pockets of (A) wt PI3Kα (2RD0), (B) H1047R mutant (3HHM), and (C) PI3Kγ (3IBE). The hydrophobic region is depicted as green; H-bond acceptor, red; and H-bond donor, blue. The electrostatic surfaces were made with the MOE program.26
Residues in loop 3 (Gly912-Asn920) and loop 4 (Asp933-Arg951) were very flexible during MD simulations (Figures 9 and 10). The flexibility of loops 3 and 4 might explain the disappearance of H-bonds with Asp933 in the MD models in both the wt and mutant R isoforms. As illustrated in both our and Han and Zhang's MD simulations,43 Asp933 might be an important residue in providing a required structural scaffold to the kinase domain by forming a H-bond with Lys776; however, its location in the highly flexible loop might reduce its importance in ligand binding.
The electrostatic map of PI3Kγ shows that the hydroxyl group of Ser806 (equivalent to Ser774 of PI3Kα) was positioned toward the ligand binding site and served as a H-bond acceptor (Figure 11C). This agrees with the observation that both H1047R mutant and PI3Kγ adopt a slightly closed activation loop. The more noticeable difference among the three proteins lies in Gln859; it is positioned deeper in the binding pocket in the H1047R mutant, making it more likely to form favorable H-bonds with ligands, an observation consistent with our docking results. The corresponding residue in the γ isoform is Lys890. Because of its extended side-chain structure, Lys890 flips out of the binding pocket (Figure 11C), making it less likely to interact with incoming ligands. This explains why there are only three ligands that form H-bonds with Lys890 of PI3Kγ (Table 4). Ser854 from both the wt and mutant PI3Kα forms is able to form H-bonds with ligands as either a H-bond donor or acceptor, whereas the corresponding residue in PI3Kγ is Ala885, lacking the H-bond forming side chain. The electrostatic maps surrounding Val851 for the three proteins are very similar; this residue participates in H-bond interactions with ligands through its main-chain NH group. This provides a reasonable explanation that many ligands form H-bonds with Val851 of the wt and mutant PI3Kα and Val882 of the γ isoform. Our docking results suggest that interactions with residues Ser774, Ser854, and Gln859 might be exploited for isoform- and/or mutant-specific ligand design. The inhibition specificity mediated by Gln859 and Ser774 was also observed in Frédérick and Denny's experiments.20
4. CONCLUSIONS
The prevalence of PI3K signaling abnormalities in human cancer cells has made PI3K an attractive target for anticancer drug discovery. The fact that oncogenic mutations such as H1047R increase kinase activity by affecting loop confirmations in the activation and C2 domains has provided the opportunity to develop potent and selective inhibitors targeting both the wt PI3Kα and its H1047R mutant. Structural differences in the kinase loop between the wt PI3Kα and H1047R mutant and PI3Kγ could be exploited in the development of a new generation of inhibitors that target both the isoform-specific and/or mutant-active PI3Kα. Several observations have arisen from this investigation. First, as exemplified by 7, 14, and 19, 3-aryl phenol and nitro functional groups appear to increase selectivity toward PI3Kα. Inspection of ligand/PI3Kα interactions shows that these functional groups form H-bonds to PI3Kα through the residues Ser774, Tyr836, Val851, and Gln859. The respective residues in PI3Kγ are Ser806, Tyr867, Val882, and Lys890. Second, selective and potent inhibition of PI3Kα seems to require a strong interaction with residues Ser774, Tyr836, Ser854, and Gln859. Third, the conformations of Ser774, Ser854, and Gln859 might be used to design PI3Kα H1047R-mutant-specific inhibitors. Fourth, formation of H-bonds with Ser806 and Lys802 of PI3Kγ is likely to direct γ-isoformspecific inhibition. Fifth, size differences in the binding pocket due to the plasticity of the loop structures could be exploited to design inhibitors that are active against both the PI3Kα wt and H1047R mutant.
ACKNOWLEDGMENT
The work was supported in part by the Research Corporation for Science Advancement. D.A.S. acknowledges Al-Zaytoonah Private University of Jordan for financial support.
Footnotes
Supporting Information Available: Results from the sequence alignments and docking scores of the drug-like molecules and the rmsd values of MD trajectories. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES AND NOTES
- 1.Bader AG, Kang S, Zhao L, Vogt PK. Oncogenic PI3K deregulates transcription and translation. Nat. Rev. Cancer. 2005;5:921–929. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- 2.Chang HW, Aoki M, Fruman D, Auger KR, Bellacosa A, Tsichlis PN, Cantley LC, Roberts TM, Vogt PK. Transformation of chicken cells by the gene encoding the catalytic subunit of PI 3-Kinase. Science. 1997;276:1848–1850. doi: 10.1126/science.276.5320.1848. [DOI] [PubMed] [Google Scholar]
- 3.Katso R, Okkenhaug K, Ahmadi K, White S, Timm J, Waterfield MD. Cellular function of phosphoinositide 3-kinases: Implications for development, homeostasis and cancer. Annu. Rev. Cell. Dev. Biol. 2001;17:615–675. doi: 10.1146/annurev.cellbio.17.1.615. [DOI] [PubMed] [Google Scholar]
- 4.Vanhaesebroeck B, Waterfield MD. Signaling by distinct classes of phosphoinositide 3-kinases. Exp. Cell Res. 1999;253:239–254. doi: 10.1006/excr.1999.4701. [DOI] [PubMed] [Google Scholar]
- 5.Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, Kinzler KW, Vogelstein B, Gabelli SB, Amzel LM. The structure of a human p110α/p85α complex elucidates the effects of oncogenic PI3Kα mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 6.Perrone F, Lampis A, Orsenigo M, Di Bartolomeo M, Gevorgyan A, Losa M, Frattini M, Riva C, Andreola S, Bajetta E, Bertario L, Leo E, Pierotti MA, Pilotti S. PI3KCA/PTEN deregulation contributes to impaired responses to cetuximab in metastatic colorectal cancer patients. Ann. Oncol. 2009;20:84–90. doi: 10.1093/annonc/mdn541. [DOI] [PubMed] [Google Scholar]
- 7.She QB, Solit D, Basso A, Moasser MM. Resistance to gefitinib (ZD1839, Iressa) in PTEN null HER overexpressing tumor cells can be overcome through restoration of PTEN function or pharmacologic modulation of constitutive PI3K/Akt pathway signaling. Clin. Cancer Res. 2003;9:4340–4346. [PubMed] [Google Scholar]
- 8.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, Kinzler KW, Vogelstein B, Velculescu VE. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554–554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 9.Liu I, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discovery. 2009;8:627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carson JD, Aller GV, Lehr R, Sinnamon RH, Kirkpartick RB, Auger KR, Dhanak D, Copeland RA, Gontarek RR, Tummino PJ, Luo L. Effects of oncogenic p110α subunit mutations on the lipid kinase activity of phosphoinositide 3-kinase. Biochem. J. 2008;409:519–524. doi: 10.1042/BJ20070681. [DOI] [PubMed] [Google Scholar]
- 11.Zask A, Kaplan J, Toral-Barza L, Hollander I, Young M, Tischler M, Gaydos C, Cinque M, Lucas J, Yu K. Synthesis and structure-activity relationships of ring-opened 17 hydroxywortmannins: Potent phosphinositide 3-kinase inhibitors with improved properties and anticancer efficacy. J. Med. Chem. 2008;51:1319–1323. doi: 10.1021/jm7012858. [DOI] [PubMed] [Google Scholar]
- 12.Hayakawa M, Kaizawa H, Moritomo H, Koizumi T, Ohishi T, Okada M, Ohta M, Tsukamoto S, Parker P, Workman P, Waterfield M. Synthesis and biological evaluation of 4-morpholino-2-phenylquinazolines and related derivatives as novel PI3 kinase p110α inhibitors. Bioorg. Med. Chem. 2006;14:6847–6858. doi: 10.1016/j.bmc.2006.06.046. [DOI] [PubMed] [Google Scholar]
- 13.Zvelebil MJ, Waterfield MD, Shuttleworth SJ. Structural analysis of PI3-kinase isoforms: Identification of residues enabling selective inhibition by small molecule ATP-competitive inhibitors. Arch. Biochem. Biophys. 2008;477:404–410. doi: 10.1016/j.abb.2008.06.024. [DOI] [PubMed] [Google Scholar]
- 14.Maira SM, Stauffer F, Brueggen J, Furet P, Schnell C, Fritsch C, Brachmann S, Chène P, De Pover A, Schoemaker K, Fabbro D, Gabriel D, Simonen M, Murphy L, Finan P, Sellers W, García-Echeverría C. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol. Cancer Ther. 2008;7:1851–1863. doi: 10.1158/1535-7163.MCT-08-0017. [DOI] [PubMed] [Google Scholar]
- 15.Marion F, Williams DE, Patrick BO, Hollander I, Mallon R, Kim SC, Roll DM, Feldberg L, Soest RV, Andersen RJ. Liphagal, a selective inhibitor of PI3 kinase α isolated from the sponge Aka coralliphaga: Structure elucidation and biomimetic synthesis. Org. Lett. 2006;8:321–324. doi: 10.1021/ol052744t. [DOI] [PubMed] [Google Scholar]
- 16.Gale S, Croasdell G. 28th Annual JPMorgan Healthcare Conferences—Exelixis and Nektar Therapeutics. IDrugs. 2010;13(3):139–141. [PubMed] [Google Scholar]
- 17.Markman B, LuRusso PM, Patnaik A, Heath E, Laird AD, van Leeuwen B, Papadopoulos KP, Baselga JA. phase I dose-escalation study of the safety, pharmacokinetics and pharmacodynamics of XL-765, a novel inhibitor of PI3K and mTOR, administered orally to patients with solid tumors. Eur. J. Cancer Suppl. 2008;6(12):68–69. [Google Scholar]
- 18.Hayakawa M, Kawaguchi K, Kaizawa H, Koizumi T, Ohishi T, Yamano M, Okada M, Ohata M, Tsukamoto S, Reynaud F, Parker P, Workman P, Waterfield M. Synthesis and biological evaluation of sulfonylhydrazone-substituted imidazole[1,2-a]pyridines as novel PI3 kinase p110α inhibitors. Bioorg. Med. Chem. 2007;15:5837–5844. doi: 10.1016/j.bmc.2007.05.070. [DOI] [PubMed] [Google Scholar]
- 19.Kendall JD, Rewcastle GW, Frederick R, Mawson C, Denny WA, Marshall ES, Baguley BC, Chaussade C, Jackson SP, Shepherd PR. Synthesis, biological evaluation and molecular modelling of sulfonohydrazides as selective PI3K p110α inhibitors. Bioorg. Med. Chem. 2007;15:7677–7687. doi: 10.1016/j.bmc.2007.08.062. [DOI] [PubMed] [Google Scholar]
- 20.Frédérick R, Denny WA. Phosphoinositide-3-kinases (PI3Ks): Combined comparative modeling and 3D-QSAR to rationalize the inhibition of p110α. J. Chem. Inf. Model. 2008;48:629–638. doi: 10.1021/ci700348m. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Wang Y, Zhang F. Pharmacophore modeling and 3D-QSAR analysis of phosphoinositide 3-kinase p110α inhibitors. J. Mol. Model. 2010;9:1449–1460. doi: 10.1007/s00894-010-0659-y. [DOI] [PubMed] [Google Scholar]
- 22.Zask A, Verheijen JC, Curran K, Kaplan J, Richard DJ, Nowak P, Malwitz DJ, Brooijmans N, Bard J, Svenson K, Lucas J, Toral-Barza L, Zhang WG, Hollander I, Gibbons JJ, Abraham RT, Ayral-Kaloustian S, Mansour TS, Yu K. ATP-competitive inhibitors of the mammalian target of rapamycin: Design and synthesis of highly potent and selective pyrazolopyrimidines. J. Med. Chem. 2009;52:5013–5016. doi: 10.1021/jm900851f. [DOI] [PubMed] [Google Scholar]
- 23.Mandelker D, Gabelli S, Schmidt-kittler O, Zhu J, Cheong I, Huang CH, Kinzler K, Vogelstein B, Amzel M. A frequent kinase domain mutation that changes the interaction between PI3K α and the membrane. Proc. Natl. Acad. Sci. U.S.A. 2009;106:16996–17001. doi: 10.1073/pnas.0908444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holm L, Park J. DaliLite workbench for protein structure comparison. [Jan 28, 2010];Bioinformatics. 2000 16:566–567. doi: 10.1093/bioinformatics/16.6.566. available online at http://www.ebi.ac.uk/Tools/dalilite/index.html. [DOI] [PubMed] [Google Scholar]
- 25.Walker EH, Pacold ME, Perisic O, Stephen L, Hawkins PT, Whymann MP, Williams RL. Structural determinations of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin and staurosporine. Mol. Cell. 2000;6:909–919. doi: 10.1016/s1097-2765(05)00089-4. [DOI] [PubMed] [Google Scholar]
- 26.The Molecular Operating Environment (MOE) Chemical Computing Group Inc.; Montreal, Quebec, Canada: 2009. [Google Scholar]
- 27.Thoden JB, Miran SG, Phillips JC, Howard AJ, Raushel FM, Holden HM. Carbomoyl phosphate synthetase: Caught in the act of glutamine hydrolysis. Biochemistry. 1998;37:8825–8831. doi: 10.1021/bi9807761. [DOI] [PubMed] [Google Scholar]
- 28.Sheng Y, Laister R, Lemak A, Wu B, Tai E, Duan S, Lukin J, Sunnerhagen M, Srisailam S, Karra M, Benchimol S, Arrowsmith CH. Molecular basis of Pirh2-mediated p53 ubiquitylation. Nat. Struct. Mol. Biol. 2008;15:1334–1342. doi: 10.1038/nsmb.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu JW, Cocina AE, Chai J, Hay BA, Shi Y. Structural analysis of a functional DIAP1 fragment bound to grim and hid peptides. Mol. Cell. 2001;8:95–104. doi: 10.1016/s1097-2765(01)00282-9. [DOI] [PubMed] [Google Scholar]
- 30.Milburn MV, Hassell AM, Lambert MH, Jordan SR, Proudfoot AE, Graber P, Wells TN. A novel dimer configuration revealed by the crystal structure at 2.4 Å resolution of human interleukin-5. Nature. 1993;363:172–176. doi: 10.1038/363172a0. [DOI] [PubMed] [Google Scholar]
- 31.Protein Preparation Wizard, Maestro, MacroModel, and Glide. Schrödinger, LLC; Portland, OR: 2009. [Google Scholar]
- 32.Pineda-Lucena A, Liao J, Wu B, Yee A, Cort JR, Kennedy MA, Edwards AM, Arrowsmith CH. NMR Structure of the Hypothetical Protein Encoded by the YjbJ gene from Escherichia coli. Proteins: Struct. Funct. Bioinf. 2002;47:572–574. doi: 10.1002/prot.10120. [DOI] [PubMed] [Google Scholar]
- 33.NCI Open Database Compounds, release 3. National Cancer Institute, National Institutes of Health; Bethseda, MD: Sep, 2003. [Aug 18, 2008]. Available online at: http://cactus.nci.nih.gov/download/nci. [Google Scholar]
- 34.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 35.Zhong HZ, Carlson HA. Computational studies and peptidomimetic design for the human p53-MDM2 complex. Proteins: Struct. Funct. Bioinf. 2005;58:222–234. doi: 10.1002/prot.20275. [DOI] [PubMed] [Google Scholar]
- 36.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 37.Darden T, York D, Pedersen L. Particle mesh Ewald: An N · log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 38.Case DA, Darden DA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, Merz KM, Wang B, Hayik S, Roitberg A, Seabra G, Kolossvàry I, Wong KF, Paesani F, Vanicek J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Yang L, Mongan J, Hornak V, Kollman PA. AMBER 10. University of California; San Francisco, CA: 2008. [Google Scholar]
- 39.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling CL. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins: Struct. Funct. Bioinf. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Norman BH, Shih C, Toth JE, Ray JE, Dodge JA, Johnson DW, Rutherford PG, Schultz RM, Worzalla JF, Vlahos CJ. Studies on the mechanism of phosphatidylinositol 3-kinase inhibition by wortmannin and related analogs. J. Med. Chem. 1996;39:1106–1111. doi: 10.1021/jm950619p. [DOI] [PubMed] [Google Scholar]
- 41.Hayakawa M, Kaizawa H, Moritomo H, Koizumi T, Ohishi T, Yamano M, Okada M, Ohta M, Tsukamoto S, Raynaud FI, Workman P, Waterfield M, Parker P. Synthesis and biological evaluation of pyrido[3′,2′:4,5]furo [3,2-d]pyrimidine derivatives as novel PI3 kinase p110α inhibitors. Bioorg. Med. Chem. Lett. 2007;17:2438–2442. doi: 10.1016/j.bmcl.2007.02.032. [DOI] [PubMed] [Google Scholar]
- 42.Knight SD, Adams ND, Burgess JL, Chaudhari AM, Darcy MG, Donatelli CA, Luengo JI, Newlander KA, Parrish CA, Ridgers LH, Sarpong MA, Schmidt SJ, Van Aller GS, Carson JD, Diamond MA, Elkins PA, Gardiner CM, Garver E, Gilbert SA, Gontarek RR, Jackson JR, Kershner KL, Luo L, Raha K, Sherk CS, Sung C-M, Sutton D, Tummino PJ, Wegrzyn RJ, Auger KR, Dhanak D. Discovery of GSK2126458, a highly potent inhibitor of PI3K and the mammalian target of rapamycin. ACS Med. Chem. Lett. 2010;1:39–43. doi: 10.1021/ml900028r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han M, Zhang JZH. Class I phosphor-inositide-3-kinase (PI3Ks) isoform-specific inhibition study by the combination of docking and molecular dynamics simulation. J. Chem. Inf. Model. 2010;50:136–145. doi: 10.1021/ci900175n. [DOI] [PubMed] [Google Scholar]