Abstract
Identification and isolation of hematopoietic stem cells (HSC) in mice is most commonly based on the expression of surface molecules Kit and Sca-1 and the absence of markers of mature lineages. However, Sca-1 is absent or weakly expressed in hematopoietic progenitors in many strains including NOD, BALB/c, C3H and CBA mice. In addition, both Kit and Sca-1 levels are modulated following bone marrow injury. In these cases, other markers and dye exclusion methods have been employed to identify HSC, yet there is no antibody-based stain that enables identification of HSC and early progenitors when Kit and Sca-1 are inadequate. CD201 is a marker that is highly restricted to HSC and progenitors, and CD27 is expressed at moderate to high levels on HSC. We show here that combining CD201 and CD27 enables highly efficient isolation of long-term HSC in NOD mice, as well as in other strains including SJL, FVB, AKR, BALB/c, C3H and CBA. We also find that HSC appear to maintain expression of CD201 and CD27 after hematopoietic injury, when Kit expression is downregulated. These results suggest a widely applicable yet simple alternative for HSC isolation in settings where Kit and Sca-1 expression are insufficient.
Introduction
Hematopoietic stem cells (HSC) are defined by their ability to durably give rise to all lineages of the blood and immune system; they are essential for bone marrow (BM) transplantation, and they are usually isolated based on their expression of unique combinations of cell surface proteins. Studies of HSC in wild type C57BL/6 (B6) mice have predominantly used Kit and Sca-1 (also called Ly-6A/E) as well as the absence of markers of lineage committed cells (lin−), to identify HSC, termed KLS (or LSK) staining1-4. Although additional markers including CD34, CD150 and CD48 can be used to further enrich HSC, they are commonly used in combination with the KLS stain4-6. However, Sca-1 is not robustly expressed in all mouse strains, hindering the application of this stain to diverse model systems7. Mice of the Ly6.1 haplotype, including BALB/c, C3H and CBA strains, express very low levels of Sca-17,8.
In addition, both Kit and Sca-1 expression levels are dynamically regulated in response to hematopoietic injury9. Isolation of HSC in these cases has been enabled by the Hoechst dye exclusion approach, but this method is more technically challenging than antibody staining and thus alternative antibody staining approaches are needed to facilitate studies in which Kit and Sca-1 are insufficient10.
One model system where an alternative HSC stain is needed is the non-obese diabetic (NOD) mouse, which is the predominant mouse model of spontaneous autoimmune diabetes. Several studies have reported on the ability of HSC transplantation to prevent, halt or reverse progression of diabetes in NOD mice11,12. Although Sca-1 is used as an identifying marker for HSC in some of these transplantation studies, NOD HSC fail to express high levels of Sca-1 (despite the fact that NOD have the Ly6.2 haplotype), suggesting that these studies may have been impacted by transplantation of progenitor populations that were poorly enriched for HSC7,13.
We investigated the use of alternative markers that could identify HSC in NOD mice. CD201, a type I transmembrane receptor, is expressed at high levels on murine HSC14. Although CD201 is a highly specific marker for HSC, it is still used in combination with Sca-1 and SLAM-family markers CD150 and CD48 to identify a more enriched HSC population14-16. CD27 is another marker that is expressed on HSC and downstream progenitors17,18. Although it has been proposed that the CD27 positive subset of hematopoietic progenitors does not contain long-term HSC, other studies suggest that most CD34− long-term HSC express CD27 at moderately high levels17,19.
We show here that CD27 and CD201 identify HSC independently of Sca-1 in NOD mice. This identification method was applicable in several other strains, including C57B/6, SJL, FVB/N, AKR, BALB/c, C3H/He and CBA. In addition, these markers identify HSC and progenitors in mice that have downregulated Kit as a result of hematopoietic injury. CD27 and CD201 therefore enable identification and isolation of highly enriched hematopoietic stem and progenitor cells in models where Sca-1 and Kit are unable to identify a distinct progenitor population.
Methods
Mice
C57BL/6J (stock no. 000664), NOD/ShiLtJ (001976), SJL/J (000686), FVB/NJ (001800), AKR/J (000648), BALB/cJ (000651), C3H/HeJ (000659) and CBA/J (000656) mice were purchased from Jackson Laboratories. NOD-mRaspberry (mRasp) transgenic mice were provided by Dr. Jason Gaglia. NOD, NOD-mRaspberry transgenic, B6-GFP transgenic and Rag−/− transgenic mice were bred at the Joslin Diabetes Center Animal Facility. Ages of donor and recipient mice ranged from 4 -12 weeks at time of initial treatment and sacrifice.
All strains were maintained at the Joslin Diabetes Center Animal Facility and fed with standard mouse chow and water. All animal procedures were approved by the Joslin IACUC.
Isolation and staining of bone marrow
Bone marrow (BM) was harvested from donor mice by flushing contents of both tibias and femurs into staining buffer (2% filtered FBS, 1mM EDTA in PBS). Red blood cell lysis buffer was added for 1 minute, debris was removed by filtration and bone marrow cellularity was assessed on a hemocytometer.
Antibodies and flow cytometry
For bone marrow, cells were stained with antibodies to (hybridoma names and vendors in parentheses): Kit-APC/Cy7 (2B8, Becton Dickinson), CD27-FITC (LG.3A10, Biolegend), CD201-APC (RCR-16, Biolegend), Sca-1 (D7, BV605, BD; and E13-161-7, Pacific Blue, Biolegend), CD150-Biotin (TC15-12F12.2; with streptavidin-Brilliant Violet 421, BD), CD4 (GK1.5), CD8 (53-6.7), CD3ε (145-2C11), Ter119 (TER-119), B220 (RA3-6B2), Gr1 (RB6-8C5), Mac-1 (M1/70) were used. Antibodies to CD4, CD8, CD3ε, Ter119, B220, Gr-1 and Mac-1 were purchased as conjugates to PE/Cy5 (Biolegend). For peripheral blood, cells were stained with antibodies to: CD3ε-Pacific Blue (17A2, Biolegend), CD3ε-PE (145-2C11, Biolegend) CD45-PECy5 (30-F11, Biolegend), Mac-1-PECy7 (M1/70, Biolegend), Gr1-Alexa Fluor 680 (in house), B220-Pacific Blue (RA3-6B2, Biolegend), B220-FITC (in house). Cells were stained on ice in staining buffer for 1 hour, washed, and resuspended in staining buffer with propidium iodide (0.25ug/ml). Analysis and sorting of stained cells were performed on an LSRII (BD Biosciences) or on a FACSAria (BD Biosciences) at the Joslin Diabetes Center Flow Core.
Irradiation and transplantation of sorted bone marrow populations
Recipient mice were lethally-irradiated with 2 treatments of 4.75 Gray at 0 and 4 hours and transplanted immediately following the second dose; irradiated mice were provided with antibiotic water (sulfamethoxazole 3%) for two weeks. For transplantation, donor BM was stained with anti-lineage antibodies (CD4, CD8, CD3ε, Ter119, B220, Gr1, Mac1; unconjugated, Bio X Cell) and depleted using Dynabeads (M-450 Sheep anti-Rat IgG, Invitrogen). Following lineage depletion, the remaining BM was stained with antibodies to: CD27, CD201, and other HSC markers as specified in the results, and specified populations were sorted into staining media using a FACSAria (BD). Recipient mice were anesthetized in an isoflurane chamber, and sorted populations were subsequently transplanted by retro-orbital injection.
For the BM injury model, 10 week old C57BL/6 mice were sublethally irradiated with 3 or 6 Gray. Irradiated mice were sacrificed 48hrs post-irradiation and BM was analyzed by flow cytometry.
Statistical analysis
Statistical analysis was performed with GraphPad Prism 5 software (La Jolla, CA).
Results
NOD HSC lack Sca-1 expression
In order to characterize the hematopoietic stem and progenitor population in NOD BM, we stained whole BM from B6 and NOD mice using the KLS method. In contrast to the distinct progenitor population identified by this stain in B6 bone marrow, we found that NOD bone marrow did not contain a distinct Kit+Sca-1+ population (Figure 1A, top panels). We further tested two commonly used monoclonal antibodies to Sca-1: D7 and E13.1, to determine if either of these anti-Sca-1 antibodies would enable identification of a distinct Kit+Sca-1+ progenitor population in NOD mice20,21. Although both clones stained a subset of the Kit+ population in B6 bone marrow, they failed to mark a substantial subset of Kit+ cells in NOD bone marrow (Figure 1A, top panels).
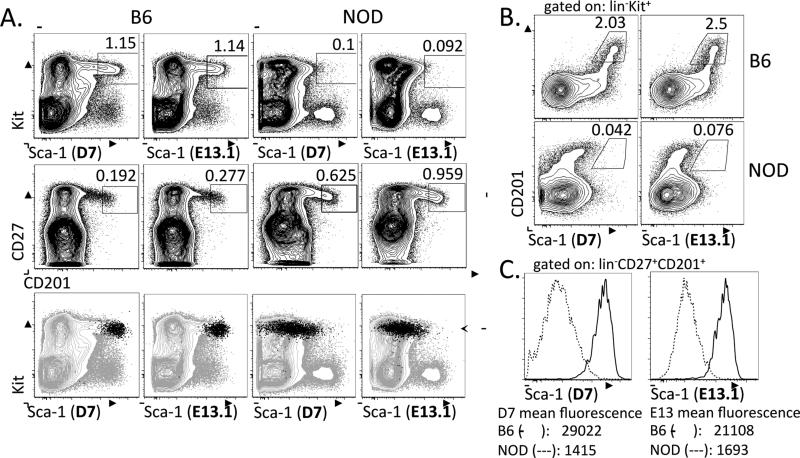
Figure 1. CD201 and CD27 are expressed in both B6 and NOD bone marrow, and stain a subset of the Kit + progenitor population.
A. NOD and B6 bone marrow cells were stained for Kit, Sca-1, CD27 and CD201. Each plot shows pre-gated live, lin− cells, and each column of 3 plots is from an individual stained sample. Both the D7 and E13.1 clones of Sca-1 stain a distinct progenitor population in B6 BM, but not in NOD BM (top panels). In contrast, a distinct CD27+CD201+ population is present in both B6 and NOD BM (middle panels). The CD27+CD201+ population is overlaid onto the Kit vs. Sca-1 plot (bottom panels). B. Plots of CD201 versus Sca-1 staining in B6 and NOD BM progenitors show that the CD201+ progenitor population is high for Sca-1 in B6 and low for Sca-1 in NOD BM. Plots are pre-gated on live, lin−Kit+ cells. C. Comparison of mean fluorescence of Sca-1 staining in the lin−CD27+CD201+ BM populations from B6 and NOD mice with both the D7 and E13 clones.
We stained bone marrow from B6 and NOD mice with CD27 and CD201 to determine whether these two markers exhibited similar staining across both strains of mice. We found that CD27 and CD201 identified a distinct population of cells from the BM of both strains (Figure 1A, middle panels). In the B6 BM, the CD27+CD201+ population uniformly overlapped with the tip of the KLS population, consistent with this population representing early stem and progenitor cells as would be predicted by previous studies using CD20114 (Figure 1A, bottom panels). In addition, the entire CD27+CD201+ population expressed high levels of Kit in both B6 and NOD (Figure 1A, bottom panels). Whereas the CD201+ population in B6 mice was universally high for Sca-1, the CD201+ population in NOD mice showed little, if any Sca-1 expression (Figure 1B, Table 1). When we gated on the CD27+CD201+ population from both B6 and NOD BM, the Sca-1 levels were 12-21 fold higher in B6 (Figure 1C, Table 1). Taken together, these results indicate that CD27 and CD201 stain a subset of hematopoietic stem and progenitor cells in B6 and NOD bone marrow.
Table 1.
Quantification of Sca-1 (D7) expression in B6 and NOD bone marrow populations.
| % Sca-1+ |
Sca-1 mean fluorescence |
|||||
|---|---|---|---|---|---|---|
| Gated BM population | B6 (n=3) | NOD (n=3) | p value | B6 (n=3) | NOD (n=3) | p value |
| lin-Kit+ | 4.3 ± 0.2 | 1.7 ± 0.2 | 0.0007 | 5800 ± 180 | 4500 ± 150 | 0.0057 |
| lin-CD27+CD201+ | 95 ± 0.6 | 1.2 ± 0.2 | <0.0001 | 8100 ± 250 | 610 ± 10 | <0.0001 |
The vast majority of HSC in NOD mice are found within the CD27+ CD201+ population
We sought to determine whether we could use CD201 and CD27 to isolate a progenitor population containing HSC in NOD mice. Because HSC identity can only be confirmed by transplantation and long-term repopulating ability, we performed transplantation assays.
NOD bone marrow was depleted of downstream lineages and the remaining cells were sorted into two populations, a CD27+CD201+ fraction and the remaining fraction that was excluded by the CD27+CD201+ gate, termed the “not” population. To compare the HSC content of these separate populations, we co-transplanted the reciprocal populations from NOD and mRaspberry-transgenic NOD (mRasp) mice into irradiated recipients (Figure 2A).
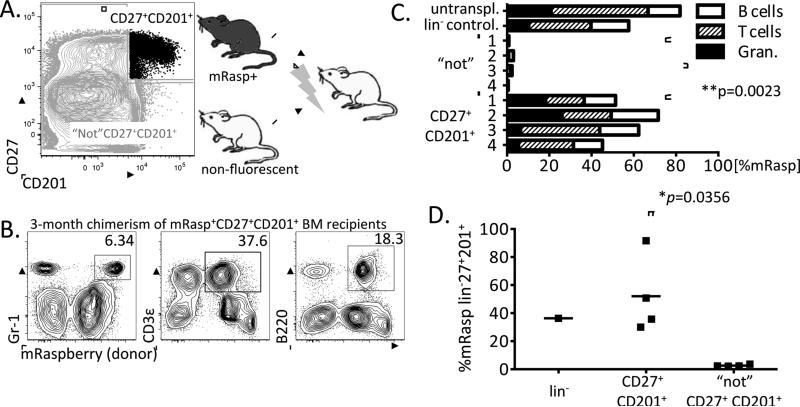
Figure 2. The lin−CD27+CD201+ population contains the vast majority of HSC in NOD BM.
A. Lin− NOD BM was sorted into two fractions, a CD27+CD201+ population and the “not” CD27+CD201+ population, which contains all the remaining cells. These populations were co-transplanted from mRaspberry and non-fluorescent NOD donors into lethally-irradiated non-fluorescent NOD recipients. B. Three months post-transplantation, peripheral blood donor chimerism was measured. FACs plots show chimerism of one representative recipient of mRasp+CD27+CD201+ BM. C. Peripheral blood chimerism at 3 months post-transplantation within each transplanted mouse (*p=0.0023, unpaired student's t-test). Peripheral blood of an untransplanted mRasp mouse (untranspl.), and one mouse receiving 0.5 BM equivalents of lin− mRasp BM cells (lin− control) are shown as controls. D. CD27+CD201+ cells self-renew: three month chimerism of donor derived lin−CD27+CD201+ cells in the BM of mice transplanted with mRasp+CD27+CD201+ (n=4) or mRasp+ “not” CD27+CD201+ BM progenitors (n=4) (*p=0.0356, unpaired student's t-test).
We analyzed T cell, B cell and granulocyte chimerism from peripheral blood at 3 months post-transplantation, and found that mice transplanted with mRasp CD27+CD201+ progenitors exhibited high levels of mRasp chimerism (45.2% - 71.6%) in all downstream lineages (Figure 2B and 2C). This engraftment level was similar to a control mouse that had received whole bone marrow from an mRasp donor. In contrast, the mice that received the “not” CD27+ CD201+ mRasp donor cells showed very little mRasp contribution to downstream lineages (<4%) (Figure 2C).
To determine the self-renewal capability of the CD27+CD201+ population, we analyzed the bone marrow of the transplanted mice at 3 months post-transplantation. In mice that received mRasp CD27+CD201+ cells (n=4), we found a similar CD27+CD201+ population 3 months later in the bone marrow. In contrast, recipient mice that had received the mRasp “not” CD27+CD201+ population (n=4) exhibited virtually no mRasp chimerism (<3%) in the CD27+CD201+ gate, and almost no other bone marrow chimerism at three months post-transplantation (Figure 2D). Notably, the peripheral blood chimerism roughly matched the chimerism level of the CD27+CD201+ population in the bone marrow, consistent with CD27+CD201+ cells giving rise to durable chimerism of all hematopoietic lineages.
These results indicate that the distinct lin−CD27+CD201+ population in NOD mice contains the vast majority of long-term HSC, both giving rise to myeloid and lymphoid lineages, and also self-renewing.
CD201 and CD27 mark HSC and progenitors in many mouse strains
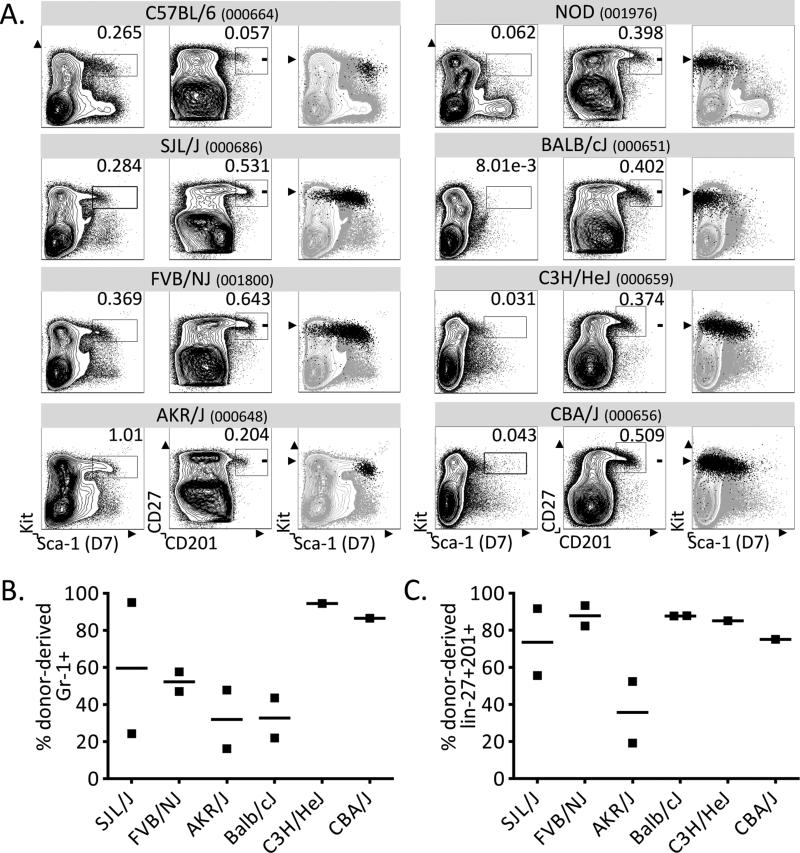
We next tested whether CD201 and CD27 marked HSC in strains other than NOD mice. We selected eight commonly used mouse strains and found that four of these lacked high levels of Sca-1 in their bone marrow (NOD, BALB/c, C3H/HeJ, CBA/J). All eight strains displayed a distinct CD27+CD201+ population regardless of Sca-1 expression levels (Figure 3A). In order to test whether these subsets contained functional and long-term HSC, we sorted CD27+CD201+ progenitors from six strains and co-transplanted each population with Rag-2-deficient (Rag−/−) GFP-transgenic (GFP+) helper bone marrow into lethally-irradiated Rag−/− GFP+ recipients. At 2 months post-transplantation, we analyzed cells from the bone marrow and spleen from each recipient and found substantial donor chimerism from the transplanted CD27+ CD201+ populations (Figure 3B and 3C). In the three strains that expressed Sca-1, we confirmed the presence of donor-derived HSC that expressed not only Sca-1, but also the additional HSC markers Kit and CD150 (data not shown)5. These findings indicate that CD27 and CD201 mark progenitors and HSC across several mouse strains. Of particular significance, this presents a widely applicable alternative to identify and isolate HSC in mice that do not express Sca-1.
Figure 3. CD201 and CD27 stain progenitors independently of variation in Sca-1 expression across strains.
A. The Kit+Sca-1+ stains are shown for 8 strains of mice (left panels). While the KLS population is only apparent in some strains, all tested mouse strains exhibit a distinct CD27+CD201+ population (middle panels), which completely or partially overlaps with the Kit+Sca-1+ population in mice that express Sca-1 (right panels). Plots are pre-gated on live, lin− BM. B. CD27+CD201+ progenitors were transplanted into lethally-irradiated Rag−/−GFP+ recipients and the percentage of donor-derived GFP− granulocytes (CD45+CD3−CD19-Gr1+Mac1+) was assessed by flow cytometry of splenocytes at 8 weeks post-transplantation. C. Bone marrow was assessed for donor-derived GFP− HSC and progenitor chimerism (lin-Kit+CD27+CD201+) at 8 weeks post-transplantation by flow cytometry. In all strains, sorted CD27+CD201+ BM cells resulted in substantial HSC and progenitor chimerism. In both B and C, Rag−/−GFP+ whole BM was co-transplanted to ensure survival.
Expression of CD27 and CD201, but not Sca-1 or Kit, is maintained following hematopoietic injury
Although B6 mice express Sca-1 and Kit under normal circumstances, certain conditions lead to aberrant expression of these markers. For example, previous studies demonstrated that irradiation of bone marrow leads to upregulation of Sca-1 and downregulation of Kit9. Identification of the HSC and progenitor populations with the KLS method is therefore hindered even in B6 bone marrow under conditions of hematopoietic stress and injury. Given that CD27 and CD201 can be used to isolate HSC independently of Sca-1 and Kit, we tested whether these two markers were maintained after hematopoietic stress and injury.
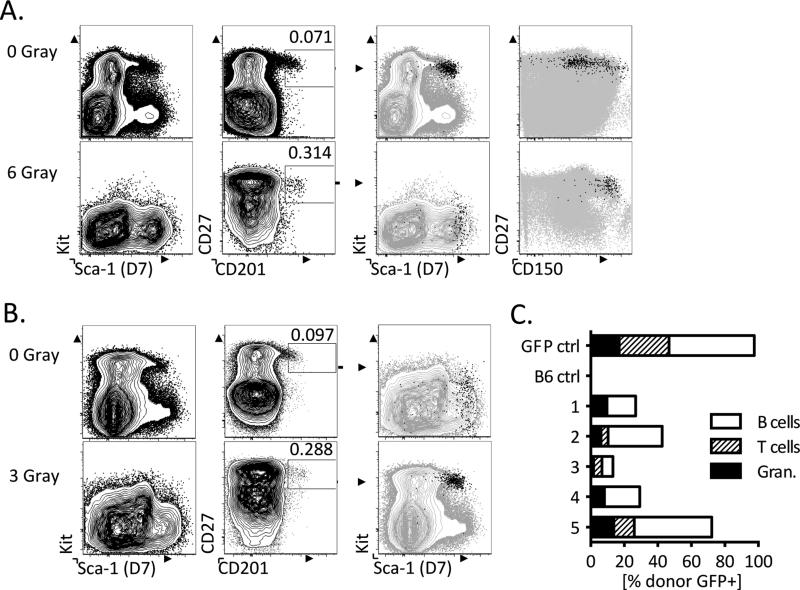
We sublethally irradiated B6 mice and analyzed their bone marrow 48 hours later to test whether expression of CD27 and CD201 is maintained following irradiation-induced hematopoietic stress. Consistent with previous findings, Sca-1 and Kit did not identify a distinct HSC or progenitor population (Figure 4A, bottom left panels). In contrast, we found a distinct CD27+ CD201+ population that exhibited increased levels of Sca-1 and was enriched for the additional HSC marker CD150 (Figure 4A, bottom right panels)5. This staining pattern was consistent in mice that had received 6 Gray (Figure 4A) and 3 Gray (Figure 4B) doses of irradiation (Figure 4A and 4B).
Figure 4. CD27 and CD201 identify progenitors in a post-irradiation setting of altered Kit and Sca-1 expression.
A and B. Hematopoietic progenitors do not maintain expression of Kit 48 hours after sublethal 6 Gray (A) and 3 Gray (B) irradiation in B6 mice (n=2) as compared to a non-irradiated control (left panels). Within the same stained samples CD27 and CD201 levels are maintained and identify a clear progenitor population (second panels). This CD27+CD201+ progenitor population exhibits increased Sca-1 and decreased Kit expression (third panels). The CD27+CD201+ progenitor population expresses high levels of CD150, a hematopoietic stem and progenitor marker (fourth panels). All plots are pre-gated on live, lin− BM. C. CD27+CD201+GFP+ progenitors were isolated from the BM of irradiated mice 48 hours post irradiation (3 Gray) and transplanted competitively with 250,000 WBM from non-irradiated B6 mice (n=5). Peripheral blood was assessed for donor granulocyte, T cell and B cell chimerism at 8 weeks post transplant.
The resistance of the CD27+CD201+ population to irradiation is consistent with studies indicating that long-term HSCs have increased radioresistance due to cell cycle phase9. In order to confirm that this population contained functional HSC, we performed competitive transplants into lethally-irradiated recipient B6 mice. We isolated the CD27+ CD201+ population from the bone marrow of mice that had been sublethally irradiated (3 Gray) 48hrs prior and co-transplanted with 250,000 whole bone marrow from non-irradiated B6 mice. 8 weeks after transplantation, we found substantial granulocyte, T-cell and B-cell chimerism from the CD27+ CD201+ population, clearly indicating that HSC continue to express CD27 and CD201 after irradiation (Figure 4C). Taken together, our results show that CD27 and CD201 are robust markers for HSC and hematopoietic progenitors across a variety of mouse strains and under conditions of hematopoietic injury.
Discussion
CD201 and CD27 as HSC surface markers
CD201, also referred to as Endothelial Protein C Receptor, or EPCR, is a type I transmembrane receptor that binds and activates protein C by the thrombin-thrombomodulin complex22. Its role in HSC function is unclear, given that adult EPCR knockout mice show no obvious hematopoietic defects23,24. However, fractional transplantations in wild type B6 mice have suggested that HSC activity is always associated with expression of CD201, and therefore CD201 is a potentially highly specific marker for isolation of murine HSC14.
CD27 is a member of the tumor necrosis factor (TNF) receptor family, and together with its ligand CD70, is thought to play a role in maintenance of HSC phenotype and function by preventing differentiation of HSC toward the myeloid lineage when activated25. CD27 has not been extensively utilized as an HSC marker, and it was previously suggested that CD27 expression was low or absent in long-term HSC19. However, our analysis, supported by a screen for HSC markers conducted by Morita et al, instead indicates that the long-term HSC are slightly lower for CD27 than downstream progenitors, but still express robust levels17. Consistent with this idea, our data show that the hematopoietic progenitors expressing the highest surface levels of CD201, which are highly enriched for HSC14, have only slightly lower CD27 levels (Figure 1A).
Dynamic regulation of Kit and Sca-1
In light of the growing potential of HSC transplantation for treatment of chronic diseases, the ability to sort and transplant an HSC-enriched population in a variety of mouse models is becoming increasingly important. However, Kit and Sca-1, the most commonly used markers for hematopoietic stem and progenitors, are dynamically regulated in response to hematopoietic stress and injury. Even at sublethal doses of irradiation, murine bone marrow exhibits significant downregulation of Kit and a moderate increase in Sca-1 levels, hindering the identification of an HSC-containing population by KLS staining9. Further examples of dynamic regulation of Kit and Sca-1 can be found following antibody-mediated blockade of Kit signaling, a method that facilitates engraftment of donor HSC in some mouse strains26, and in response to interferon-α and interferon-γ–induced proliferation, which activates Sca-1 expression on hematopoietic progenitors27-29.
Identification methods such as Hoechst dye exclusion, also termed “side population”, have previously been used to identify HSC under conditions of dynamically regulated or entirely absent Kit and Sca-1 expression10. Although this method is independent of Kit and Sca-1, it requires more time and resources for isolation than antibody staining approaches. In contrast to Kit and Sca-1, CD27 and CD201 identify a distinct population of hematopoietic stem and progenitor cells across multiple strains of mice, and also after irradiation. Thus, this antibody-based stain provides a reliable solution for the isolation of murine HSC in cases where the standard KLS stain is not applicable.
Interestingly, while CD27 and CD201 identify a population of hematopoietic stem and progenitor cells in all strains tested within this study, the frequency of this population can vary by more than 10-fold between strains. This is somewhat similar to variations seen in the KLS population in strains that express Sca-1; in this study, we observed a 4-fold difference in the frequency of KLS cells between strains. Other studies have functionally determined HSC and early progenitor frequencies in multiple mouse strains and have also found substantial differences, and in some cases, have identified genetic polymorphisms that account for these differences30-33. It is possible that some of these same polymorphisms contribute to the differences reported here in the frequencies of the CD27+CD201+ populations.
HSC in autoimmunity and type 1 diabetes
Autoimmune disease, in particular, is a growing target for HSC-mediated therapies, given that HSC differentiate down all lineages of the immune system and can induce tolerance and reverse autoimmunity34. BALB/c mice are often utilized as models of induced autoimmunity, but universally lack Sca-1 expression7. Similarly, the NOD mouse model is well known for developing spontaneous autoimmune diabetes, but has decreased Sca-1 expression on hematopoietic progenitors13. Our data support these findings suggesting that NOD HSC express lower levels of Sca-1, and thus cannot be robustly identified and isolated with the KLS staining method. Taken together, these findings have impeded the development of strategies for HSC-mediated therapies in the NOD model.
Nevertheless, transplantation of disease-resistant NOD Kit+ Sca-1+ cells, usually in combination with myeloablative conditioning, has resulted in prevention of disease in NOD mice11,12. Although NOD HSC do not express high levels of Sca-1, it is likely that the Kit+ Sca-1+ population contains a small fraction of HSC in NOD, given that the CD27+ CD201+ contains a fraction of cells that express moderate and low levels of Sca-1 (Figure 1A and B). Thus, although studies conducting tolerance induction by transplanting the Kit+ Sca-1+ population may not have transplanted the entire HSC population, it is likely that the small fraction of HSC contained in the Kit+ Sca-1+ population was sufficient to enable long-term HSC engraftment and chimerism. In the future, HSC identification and transplantation studies in the NOD mouse model and in other Sca-1 deficient strains will be facilitated by the Sca-1-independent staining method described here.
Highlights.
- CD27 and CD201 specifically mark murine hematopoietic stem and progenitor cells
- These markers enable isolation of HSC in many mouse strains, including NOD mice
- CD27 and CD201 are especially useful when HSC lack Sca-1 or Kit expression
Acknowledgements
This study was funded by grants from the Peabody Foundation, The A&M Stewart Trust, The Fleisher Family Foundation, and the Pittsburgh Foundation to TS. SEV was funded by a fellowship from the Harvard Stem Cell Institute Internship Program. The authors gratefully acknowledge the Joslin Diabetes Center Flow Cytometry Core, supported by an NIH Diabetes Research Center grant (NIH award P30DK036836) and the Harvard Stem Cell Institute. The authors acknowledge the excellent support of the Joslin Animal Facility. The authors thank Leo Wang for helpful advice, and MiJeong Kim and Martin Thelin for comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authorship Contributions
SEV designed and performed research and wrote the paper. TS designed and performed research and wrote the paper. MAI designed research.
Disclosure of Conflicts of Interest
The authors have no conflicts of interest to disclose.
References
- 1.Ogawa M, Matsuzaki Y, Nishikawa S, et al. Expression and function of c-kit in hemopoietic progenitor cells. Journal of Experimental Medicine. 1991;174(1):63–71. doi: 10.1084/jem.174.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okada S, Nakauchi H, Nagayoshi K, et al. Enrichment and characterization of murine hematopoietic stem cells that express c-kit molecule. Blood. 1991;78(7):1706–1712. [PubMed] [Google Scholar]
- 3.Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241(4861):58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 4.Mayle A, Luo M, Jeong M, Goodell MA. Flow cytometry analysis of murine hematopoietic stem cells. Cytometry A. 2013;83(1):27–37. doi: 10.1002/cyto.a.22093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiel MJ, Yilmaz ÖH, Iwashita T, et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121(7):1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 6.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273(5272):242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 7.Spangrude GJ, Brooks DM. Mouse strain variability in the expression of the hematopoietic stem cell antigen Ly-6A/E by bone marrow cells. Blood. 1993;82(11):3327–3332. [PubMed] [Google Scholar]
- 8.Lee PY, Wang J-X, Parisini E, Dascher CC, Nigrovic PA. Ly6 family proteins in neutrophil biology. J. Leukoc. Biol. 2013;94(4):585–594. doi: 10.1189/jlb.0113014. [DOI] [PubMed] [Google Scholar]
- 9.Simonnet AJ, Nehmé J, Vaigot P, et al. Phenotypic and functional changes induced in hematopoietic stem/progenitor cells after gamma-ray radiation exposure. Stem Cells. 2009;27(6):1400–1409. doi: 10.1002/stem.66. [DOI] [PubMed] [Google Scholar]
- 10.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. Journal of Experimental Medicine. 1996;183(4):1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steptoe RJ, Ritchie JM, Harrison LC. Transfer of hematopoietic stem cells encoding autoantigen prevents autoimmune diabetes. J. Clin. Invest. 2003;111(9):1357–1363. doi: 10.1172/JCI15995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beilhack GF, Scheffold YC, Weissman IL, et al. Purified allogeneic hematopoietic stem cell transplantation blocks diabetes pathogenesis in NOD mice. Diabetes. 2003;52(1):59–68. doi: 10.2337/diabetes.52.1.59. [DOI] [PubMed] [Google Scholar]
- 13.Chilton PM, Rezzoug F, Ratajczak MZ, et al. Hematopoietic stem cells from NOD mice exhibit autonomous behavior and a competitive advantage in allogeneic recipients. Blood. 2005;105(5):2189–2197. doi: 10.1182/blood-2004-07-2757. [DOI] [PubMed] [Google Scholar]
- 14.Balazs AB, Fabian AJ, Esmon CT, Mulligan RC. Endothelial protein C receptor (CD201) explicitly identifies hematopoietic stem cells in murine bone marrow. Blood. 2006;107(6):2317–2321. doi: 10.1182/blood-2005-06-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inlay MA, Serwold T, Mosley A, et al. Identification of Multipotent Progenitors that Emerge Prior to Hematopoietic Stem Cells in Embryonic Development. Stem Cell Reports. 2014;2(4):457–472. doi: 10.1016/j.stemcr.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kent DG, Copley MR, Benz C, et al. Prospective isolation and molecular characterization of hematopoietic stem cells with durable self-renewal potential. Blood. 2009;113(25):6342–6350. doi: 10.1182/blood-2008-12-192054. [DOI] [PubMed] [Google Scholar]
- 17.Morita Y, Ema H, Nakauchi H. Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. J. Exp. Med. 2010;207(6):1173–1182. doi: 10.1084/jem.20091318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serwold T, Ehrlich LIR, Weissman IL. Reductive isolation from bone marrow and blood implicates common lymphoid progenitors as the major source of thymopoiesis. Blood. 2009;113(4):807–815. doi: 10.1182/blood-2008-08-173682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiesmann A, Phillips RL, Mojica M, et al. Expression of CD27 on murine hematopoietic stem and progenitor cells. Immunity. 2000;12(2):193–199. doi: 10.1016/s1074-7613(00)80172-7. [DOI] [PubMed] [Google Scholar]
- 20.Aihara Y, Bühring HJ, Aihara M, Klein J. An attempt to produce “pre-T” cell hybridomas and to identify their antigens. Eur. J. Immunol. 1986;16(11):1391–1399. doi: 10.1002/eji.1830161113. [DOI] [PubMed] [Google Scholar]
- 21.Ortega G, Korty PE, Shevach EM, Malek TR. Role of Ly-6 in lymphocyte activation. I. Characterization of a monoclonal antibody to a nonpolymorphic Ly-6 specificity. J. Immunol. 1986;137(10):3240–3246. [PubMed] [Google Scholar]
- 22.Fukudome K, Esmon CT. Identification, cloning, and regulation of a novel endothelial cell protein C/activated protein C receptor. J. Biol. Chem. 1994;269(42):26486–26491. [PubMed] [Google Scholar]
- 23.Esmon CT. The endothelial cell protein C receptor. Thromb. Haemost. 2000;83(5):639–643. [PubMed] [Google Scholar]
- 24.Li W, Zheng X, Gu J-M, et al. Extraembryonic expression of EPCR is essential for embryonic viability. Blood. 2005;106(8):2716–2722. doi: 10.1182/blood-2005-01-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nolte MA, Arens R, van Os R, et al. Immune activation modulates hematopoiesis through interactions between CD27 and CD70. Nat Immunol. 2005;6(4):412–418. doi: 10.1038/ni1174. [DOI] [PubMed] [Google Scholar]
- 26.Czechowicz A, Kraft D, Weissman IL, Bhattacharya D. Efficient transplantation via antibody-based clearance of hematopoietic stem cell niches. Science. 2007;318(5854):1296–1299. doi: 10.1126/science.1149726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao X, Ren G, Liang L, et al. Brief report: interferon-gamma induces expansion of Lin(−)Sca-1(+)C-Kit(+) Cells. Stem Cells. 2010;28(1):122–126. doi: 10.1002/stem.252. [DOI] [PubMed] [Google Scholar]
- 28.Dumont FJ, Palfree RG, Coker LZ. Phenotypic changes induced by interferon in resting T cells: major enhancement of Ly-6 antigen expression. J. Immunol. 1986;137(1):201–210. [PubMed] [Google Scholar]
- 29.Essers MAG, Offner S, Blanco-Bose WE, et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458(7240):904–908. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- 30.de Haan G, Nijhof W, Van Zant G. Mouse strain-dependent changes in frequency and proliferation of hematopoietic stem cells during aging: correlation between lifespan and cycling activity. Blood. 1997;89(5):1543–1550. [PubMed] [Google Scholar]
- 31.de Haan G, Van Zant G. Intrinsic and extrinsic control of hemopoietic stem cell numbers: mapping of a stem cell gene. Journal of Experimental Medicine. 1997;186(4):529–536. doi: 10.1084/jem.186.4.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morrison SJ, Qian D, Jerabek L, et al. A genetic determinant that specifically regulates the frequency of hematopoietic stem cells. J. Immunol. 2002;168(2):635–642. doi: 10.4049/jimmunol.168.2.635. [DOI] [PubMed] [Google Scholar]
- 33.Henckaerts E, Geiger H, Langer JC, et al. Genetically determined variation in the number of phenotypically defined hematopoietic progenitor and stem cells and in their response to early-acting cytokines. Blood. 2002;99(11):3947–3954. doi: 10.1182/blood.v99.11.3947. [DOI] [PubMed] [Google Scholar]
- 34.Li HW, Sykes M. Emerging concepts in haematopoietic cell transplantation. Nat Rev Immunol. 2012;12(6):403–416. doi: 10.1038/nri3226. [DOI] [PMC free article] [PubMed] [Google Scholar]