Abstract
Cystic fibrosis (CF) is a lethal genetic disorder most commonly caused by the F508del mutation in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. It is not readily amenable to gene therapy because of its systemic nature and challenges including in vivo gene delivery and transient gene expression. Here, we use triplex-forming PNA molecules and donor DNA in biodegradable polymer nanoparticles to correct F508del. We confirm modification with sequencing and a functional chloride efflux assay. In vitro correction of chloride efflux occurs in up to 25% of human cells. Deep sequencing reveals negligible off-target effects in partially homologous sites. Intranasal application of nanoparticles in CF mice produces changes in nasal epithelium potential differences consistent with corrected CFTR, with gene correction also detected in lung tissue. This work represents facile genome engineering in vivo with oligonucleotides using a nanoparticle system to achieve clinically relevant levels of gene editing without off-target effects.
INTRODUCTION
Cystic fibrosis (CF) is an autosomal recessive, multi-system disease caused by defects in the cystic fibrosis transmembrane conductance regulator (CFTR), an ion channel that mediates chloride transport. Lack of CFTR function causes obstructive lung disease, intestinal obstruction syndromes, liver dysfunction, exocrine and endocrine pancreatic dysfunction, and infertility. Since the sequencing and cloning of the CFTR gene in 1989 1–3, numerous mutations resulting in CF have been identified 2, 4. The most common mutation in CF is a three base-pair deletion (F508del) on chromosome 7, which results in the loss of a phenylalanine residue, causing increased degradation of the CFTR protein before it can reach the cell surface.
Although CF is one of the most rigorously characterized genetic diseases, current treatment of patients with CF focuses on symptomatic management rather than correction of the genetic defect. Some studies have demonstrated increased F508del activity with agents such as curcumin 5, 6 or histone deacetylase inhibitors 7; VX-770 increases the activity of the CFTR protein in patients who have the less common G551D mutation. Gene therapy has remained unsuccessful in CF, due to challenges including in vivo delivery to the lung and other organ systems. In recent years, there have been many advances in gene therapy for treatment of diseases involving the hematolymphoid system, where harvest and ex vivo manipulation of cells for autologous transplantation is possible: examples include the use of zinc finger nucleases targeting CCR5 to produce HIV-1 resistant cells8, correction of the ABCD1 gene by lentiviral vectors9, and correction of SCID using retroviral gene transfer10. In contrast, harvest and autologous transplant is not a readily available option in CF, due to the involvement of the lung and other internal organs.
As one approach, the UK Cystic Fibrosis Gene Therapy Consortium is testing liposomes to deliver plasmids containing cDNA encoding CFTR to the lung. Other clinical trials have used viral vectors for delivery of the CFTR gene with limited success (reviewed here11), or CFTR expression plasmids that are compacted by polyethylene glycol-substituted lysine 30-mer peptides 12. Delivery of plasmid DNA for gene addition without targeted insertion does not correct the endogenous gene and is not subject to normal CFTR gene regulation, while virus-mediated integration of the CFTR cDNA could introduce the risk of non-specific integration into important genomic sites. New gene delivery vectors include a chimeric Ad5F35 vector that showed much higher efficiency than traditional Ad5 vectors 13. Researchers have demonstrated that treatment with the microRNA miR-138 leads to improved synthesis of CFTR-F508del 14, and have also shown that lentiviruses can be used for gene transfer to porcine airways 15. Other current gene and cell therapy strategies have been recently reviewed 16.
We reasoned that an attractive approach for CF gene therapy would be site-specific gene editing and correction of F508del. Unlike the delivery of plasmids containing CFTR, which would be transiently expressed, or non-specific viral integration, site-specific gene editing would correct CFTR at its endogenous site, resulting in permanent gene modification under normal regulatory control. Current approaches for site-specific gene editing include short fragment homologous recombination using DNA fragments containing the correct CFTR sequence that can recombine with F508del CFTR genomic DNA, resulting in gene correction 17–19, including introduction of the F508del mutation into normal mouse lung18. Zinc finger nucleases (ZFNs20) have recently been used to insert a CFTR transgene at the CCR5 locus 21 and for modification of F508del at levels <1% in vitro 22. CRISPR/Cas-9 technology has been used to correct F508del in intestinal organoids from CF patients in culture 23, but with high off-target effects (one out of twenty-five surveyed genes in a single analyzed clone). In addition, the efficiency of gene modification was low: approximately 0.3% of treated organoids (3 to 6/1400) had the desired modification 23. In vivo delivery is an important challenge, which was not attempted in this prior work with CRISPR/Cas9 or ZFNs.
In our approach, we use triplex-forming peptide nucleic acids (PNAs). PNAs are synthetic oligonucleotide analogs, resistant to protease and nuclease degradation, that can induce DNA repair upon sequence-specific triplex formation at targeted genomic sites 24, 25. Targeted genome engineering using this technique has historically been at low frequencies. Modification of PNA molecules with cell-penetrating peptides allowed for direct in vivo delivery in a supF reporter transgenic mouse, resulting in gene correction up to 0.06% in vivo. Our previous work has shown that triplex-forming PNAs and short donor DNA fragments can be used to edit the human β-globin gene 26 and the CCR5 gene 27. Work by Schleifman et al 27 has demonstrated that tail-clamp triplex-forming PNAs, containing an extended Watson-Crick strand invasion domain for the formation of PNA/DNA/PNA triplexes, have increased activity for gene modification.
Nanoparticles are an attractive delivery tool 28 because of the numerous avenues for improving and fine-tuning delivery, including decoration with targeting ligands or cell penetrating peptides 29, use of surface modifiers to avoid uptake by phagocytes 30, and exploration of polymer compositions and formulations to create vehicles for effective clinical application31. Recently, we have developed biodegradable poly(lactic-co-glycolic) acid (PLGA) nanoparticles for PNA delivery. Using this approach, we were able to achieve increased in vitro modification of human hematopoietic stem cells (up to 0.9%) 32. Application of these nanoparticles for modification of the human CCR5 gene resulted in in vivo gene modification of up to 0.5% in the spleen 33. Our group also developed a new method for surface-modifying PLGA nanoparticles with cell-penetrating peptides29, and found that this class of nanoparticles could deliver PNA molecules for inhibition of microRNAs in a lymphoma model34. This surface modification strategy was combined with a polymer blend strategy for enhanced delivery of plasmid DNA to primary human lung cells in vitro 35 and importantly in vivo to epithelial airway cells in mice36. Enhanced delivery of nanoparticles to lung type I epithelium and macrophages was seen using this polymer blend.
Here, we use tail-clamp PNAs and donor DNA molecules delivered in polymer nanoparticles to correct the F508del CFTR mutation both in vitro in human bronchial epithelial cells and in vivo in a CF murine model. By combining our recently developed tail-clamp PNA and surface-modified polymer blend nanoparticle technologies, we achieve in vitro and in vivo gene correction an order of magnitude higher than previously achieved.
RESULTS
Design of novel triplex-forming PNA molecules targeting F508del CFTR
We designed a donor DNA molecule homologous to the targeted region containing the F508del sequence, and three tail-clamp PNA molecules that bind near this site at homopurine/homopyrimidine stretches (Fig 1a,b, Supplementary Fig 1). A gel shift binding assay was used to confirm binding of these PNA molecules to the desired targets – successful binding is indicated by presence of a DNA band more proximally on the gel as the bound triplex-forming PNA molecule slows down the transit of the complex (Fig 1c, Supplementary Fig 1). Several bands may be present due to different binding configurations, as previously described 37. We also designed an AS-PCR assay to differentiate between the integrated donor sequence and the endogenous F508del sequence, using plasmids for optimization and validation of the allele specificity of the PCR reaction (Supplementary Fig 2). Primers specific to the donor DNA selectively amplified the plasmid containing the donor sequence, whereas primers specific to the F508del sequence only amplified the plasmid containing the F508del sequence. Importantly, spiking of the PCR reaction on genomic DNA with excess donor DNA or excess PNA did not lead to a false positive PCR artifact. However, spiking of the PCR reaction with donor DNA and PNA at high doses did result in inhibition of the PCR reaction (Supplementary Fig 2 D), suggesting that our AS-PCR may not pick up all samples with corrected genomes. We also noted occasional amplification of the F508del sequence with donor primers, which would not lead to false positives or negatives when trying to detect the donor sequence. Because of these limitations, AS-PCR was only used as an initial screening tool to identify active molecules before moving to sequencing and functional studies.
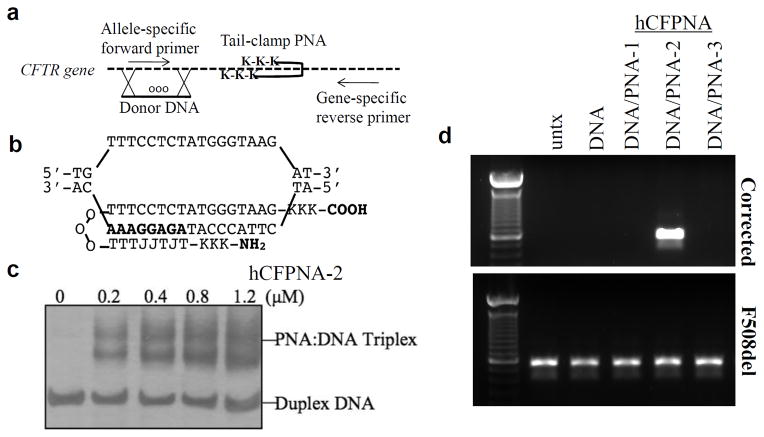
Figure 1. Tail-clamp hCFPNA2 mediates correction of F508DEL in the CFTR gene.
(a) Overall strategy for PNA-induced recombination and gene correction, and detection of modification by AS-PCR. (b) Schematic of hCFPNA2 forming a PNA/DNA/PNA triplex. J represents pseudoisocytosine, a C analog for improved triplex formation at physiologic pH. (c) Gel shift assay to test the binding of each designed PNA to the respective target site in CFTR: results for hCFPNA2. PNA was incubated with plasmid DNA with the targeted binding site, the binding region excised, and the product analyzed on PAGE gel with silver stain. (d) Human CFBE cells were treated with 2 mg/mL nanoparticles for 24 hours, then harvested for genomic DNA extraction. AS-PCR was performed to detect the corrected sequence.
To screen PNA molecules for gene editing activity, PLGA nanoparticles were loaded with PNAs and donor DNAs using a double emulsion solvent evaporation technique as previously described32. Nanoparticles with the donor DNAs alone, or DNAs and the various PNA molecules, were then tested on CF bronchial epithelial (CFBE) cells containing F508del (CFBE41o-)38. AS-PCR showed that F508del cells treated with PLGA nanoparticles containing both donor DNAs and hCFPNA2 had the desired modification present (Fig 1d). Nanoparticles with donor DNA alone or with donor DNA plus either hCFPNA1 or hCFPNA3 were not effective.
Confirmation of gene modification in isolated clones
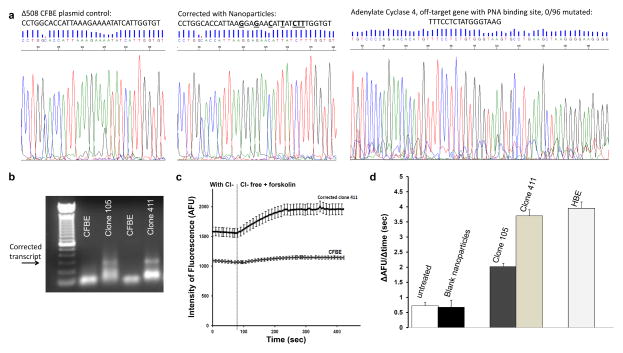
The nanoparticle-treated cell populations were seeded at limiting dilution into 96-well plates, and expanded to isolate clones positive for the modification (Supplementary Fig 3). The frequency of modification in cells treated once with PLGA nanoparticles containing hCFPNA2 and the donor DNA, as calculated by limiting dilution analysis 39, was 0.5–0.96%. Populations positive for CFTR gene correction were expanded by repeated limiting dilution to create more homogeneous clones, with the modification persisting over months of cell expansion. A 700 base-pair region around the modification site was amplified by PCR and sequenced, confirming the presence of the corrected sequence in clone 411 (Fig 2a), and regular sequencing with limited PCR cycles revealed heterozygosity of the sample (Supplementary Fig 3c) although with low sequencing quality. Higher quality reads were obtained by deep-sequencing, which revealed that clone 411 was indeed heterozygous. Clone 411 was found to have 15897/35178 (45%) of alleles with the modified sequence, implying a heterozygous population with possibly a few contaminating unmodified cells (which may have remained even after the limiting dilution cell isolation process).
Figure 2. Confirmation of CFTR modification in isolated clones.
After human CFBE cells were treated with nanoparticles, modified clones were isolated by limiting dilution. (a) A 712 bp region around the modification site was amplified by PCR and sent for direct sequencing. A plasmid control with the uncorrected site, and cells corrected with nanoparticles (clone number 411 from limiting dilution cloning procedure) are shown. In addition, 96 clones of cells which were positive for the CF modification were analyzed for off-target modification in the adenylate cyclase gene which has partial homology to the PNA binding site. No mutations were found. (b) Reverse transcriptase, allele-specific PCR using primers designed such that one binds to the corrected transcript, and the other within the adjacent exon. The presence of a band indicates a transcript with the corrected sequence conferred by the donor DNA in corrected clone 411. Lower bands indicate non-specific primer-dimer amplification. (c) Chloride efflux was measured using N-[ethoxycarbonylmethyl]-6-methoxy-quinolinium bromide (MQAE), a fluorescent indicator dye. Example traces from untreated CFBE41o- cells (n=23) (bottom) and a corrected CFBE clone (n=26) (top) are shown. Error bars = standard error of the mean. (d) Summary of chloride efflux: cell-averaged arbitrary fluorescence units per minute (AFU/min) for untreated CFBE cells (n=138), blank treated cells (n=168), modified clones (n=108 for clone 105, n=100 for clone 411), and wild type 16HBE14o- cells (n=113). Error bars = standard error of the mean. Efflux rates of HBE cells (p<0.0001) and clone 105 (p=0.0061) and clone 411 (p<0.0001) were significantly different from that of untreated CF cells. There was no difference in chloride efflux between untreated cells and those treated with blank particles. We used one way ANOVA with multiple comparisons to analyze chloride efflux in untreated CF cells, blank particle treated CF cells, clone 105, clone 411 and normal human bronchial epithelial cells (16HBE14o-).
A region of an unrelated gene that has homology to the hCFPNA2 binding site, except for one base-pair mismatch, adenylate cyclase type 4 on chromosome 14, was also sequenced, and no mutations were identified in 96 sequenced clones. While this regular sequencing in clones would not be able to identify mutations at a frequency lower than 1/96, additional experiments to ascertain off-target effects were performed in treated cells (see below). Correction of the CFTR gene was also confirmed using reverse transcriptase, allele-specific PCR on RNA extracted from a positive clone, as seen by the band corresponding to the modified sequence (Fig 2b).
Chloride efflux in the positive clones was quantified using MQAE, a fluorescent indicator dye, and perfusate solutions that switched from chloride containing solutions to chloride free solutions in the presence of forskolin and IBMX to maximally activate functional CFTR at the cell surface 40, 41. While untreated cells had minimal chloride efflux (flat line), the positive clones had increased chloride efflux in individually tested cells (Fig 2c). The increased chloride efflux was calculated by measuring the rate of change in fluorescence over time (ΔAFU/Δsec) as perfusate solutions were changed from chloride containing to chloride free solutions in the presence of a CFTR stimulating cocktail. Chloride efflux was found to be significantly increased in the positive clones (Fig 2d). Chloride efflux in clones was found to be similar to efflux in wild-type human bronchial epithelial (HBE) cells, although there was some variation between clones. For instance, “clone” 105, which had lower response, was found to have 350/8346168 of alleles modified in one deep sequencing run, suggesting a heterogeneous population with variable expansion of modified cells.
Improved in vitro activity of PLGA/PBAE/MPG nanoparticles
Nanoparticles were then formulated from a blend of PLGA and 15% (wt%) poly (beta amino ester) (PBAE), surface modified with the nuclear-localization sequence-containing cell-penetrating peptide MPG (modified PLGA/PBAE/MPG nanoparticles)35. Particles exhibited uniform size and morphology on SEM, and released most of their contents quickly, within the first 6–12 hours of incubation in PBS at 37°C, although there was more sustained release of nucleic acid cargo using the modified nanoparticles (Supplementary Fig 4). Increased uptake of fluorescently-labeled PNA molecules was seen when PLGA/PBAE/MPG nanoparticles were used on human CFBE cells (Fig 3).
Figure 3. Modified nanoparticles for increased PNA uptake.
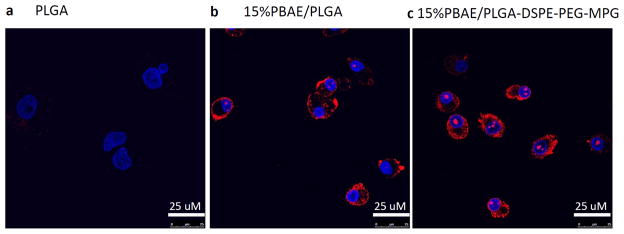
CFBE cells were plated on slides and treated with (a) 0.5 mg/mL PLGA nanoparticles, (b) 0.5 mg/mL 15% PBAE/PLGA nanoparticles, or (c) 0.5 mg/mL 15% PBAE/PLGA nanoparticles with MPG surface modification. All nanoparticles contained tamra-labeled PNA (red) and unlabeled DNA. Cells were stained with DAPI (blue) to track nuclear uptake. The Leica TCS SP5 confocal microscope was used to image cells.
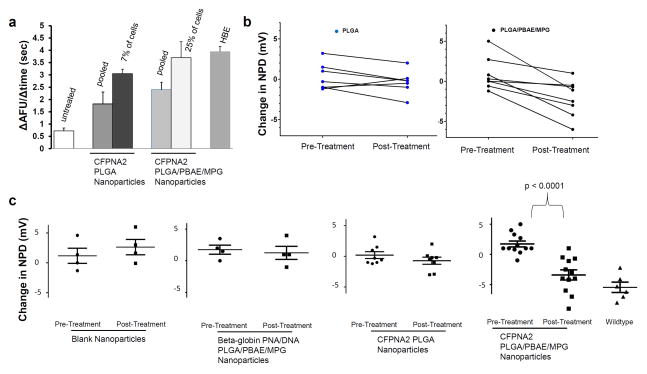
Change in chloride efflux was seen in CFBE cells serially treated three times with nanoparticles, without isolation of positive cells (Fig 4a). Of note, interrogation of individual cells in these studies allowed us to quantify the absolute number of cells with functional chloride efflux. Approximately 7% of the PLGA-nanoparticle treated cells demonstrated efflux similar to positive controls, and when CFBE cells were treated repeatedly with modified PLGA/PBAE/MPG nanoparticles 25% of cells demonstrated efflux equivalent to positive controls; this difference in modification efficiency was statistically significant (p=0.003 two-tailed Fisher’s exact test). Cells treated similarly with PNA-carrying nanoparticles targeting a non-related genomic target or hCFPNA2 with a different donor DNA targeting a non-related genomic target did not have any change in chloride efflux (Supplementary Fig 5). Our previous work has suggested that this modified nanoparticle formulation is also optimal for in vivo delivery of cargo to the respiratory epithelium. We have recently shown that PLGA/PBAE/MPG nanoparticles are taken up by both macrophages and lung epithelial cells in mice36.
Figure 4. Functional correction of CFTR with modified nanoparticles in vitro and in vivo. (a).
Summary of chloride efflux: cell-averaged arbitrary fluorescence units per minute (AFU/min) for untreated CFBE cells (n=138), treated cells (n=150), and wildtype 16HBE14o- cells (n=113). F508del CFBE cells were plated at 10% confluence, then treated 3 times with 2 mg/mL particles over 7 days. They were then replated on slides and allowed 7–10 days to grow to confluence before the MQAE assay was performed to determine chloride efflux. CFPNA2 NPs = cell population treated with PLGA nanoparticles containing hCFPNA2 and donor DNA. 7% of these cells exhibited efflux similar to positive HBE controls. CFPNA2 Modified NPs = cell population treated with PLGA/PBAE/MPG nanoparticles containing hCFPNA2 and donor DNA. 25% of these cells exhibited efflux similar to positive HBE controls. p=0.003 two-tailed Fisher’s exact test between PLGA and PBAE/PLGA/MPG treated cells. Error bars show the SD. (b) Mice were treated by intranasal infusion with nanoparticles. Nasal potential difference measurements were assessed prior to nanoparticle treatment, and subsequent to treatment. The response to a 0Cl + amiloride + forksolin perfusate after nanoparticle treatment was compared to the response prior to treatment. Each data point represents one mouse, with a line connecting pre and post-treatment values. Mice treated with PLGA (left panel) or PLGA/PBAE/MPG nanoparticles (right panel) containing PNA/DNA are shown. Pre and post treatment changes in NPD were compared using paired t tests for each mouse. Treatment with PLGA nanoparticles demonstrated no difference in NPD; however treatment with PLGA/PBAE/MPG nanoparticles demonstrated a significant change in NPD (p=0.004). (c) Nasal potential difference changes in functional and control nanoparticle treated CF mice. Each mouse is represented with an individual data point; in addition, the mean is shown with a horizontal line, surrounded by error bars showing the standard error of the mean. Pre and post treatment changes in NPD were compared using unpaired t tests for each group. In the last panel, nasal potential difference changes in wild type mice are shown for comparison.
Correction of murine F508del in vivo
To account for slight sequence variation between the mouse and human CFTR genes, new donor DNAs and PNAs were designed to target the mouse gene and correct the mouse F508del mutation. Binding of the mouse-specific PNA to the target DNA was confirmed by gel shift assay (Supplementary Fig 1). PLGA and PLGA/PBAE/MPG nanoparticles were formulated to contain the mouse-specific triplex-forming PNA and donor DNA, and CF mice42 were treated with the nanoparticle suspension by intranasal application on days 1, 3, 6, and 9. Four days after the last treatment (day 14), correction of the mouse CFTR mutation in the nasal epithelium was assayed by measuring the nasal potential difference, a non-invasive assay used to detect chloride transport in vivo. Normally, CF nasal epithelia (human and mice) exhibit a large lumen negative nasal potential that is amiloride sensitive as well as a lack of activation of cyclic AMP stimulated chloride efflux. This can be contrasted with a more modest amiloride sensitive response and the presence of robust cyclic AMP stimulated chloride efflux in non-affected tissue. The lack of activation of cyclic AMP stimulated chloride flux is due directly to CFTR dysfunction and serves as a surrogate of CFTR activity.
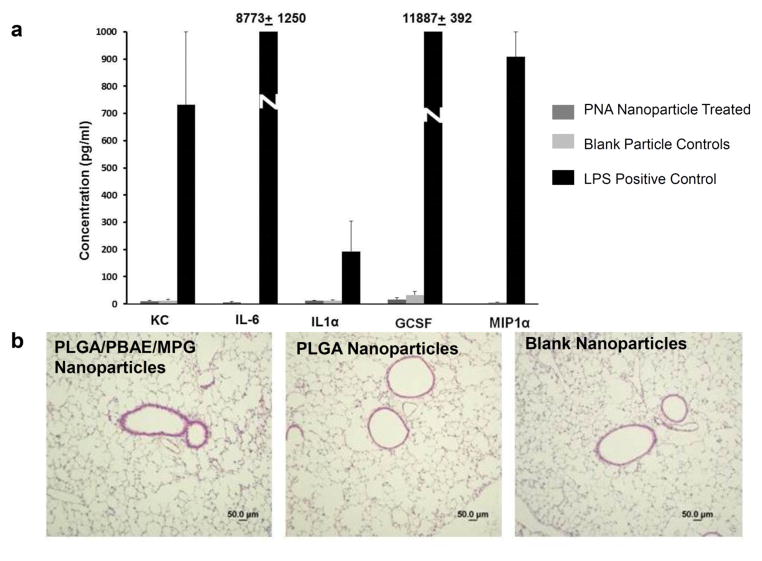
After intranasal delivery of mCFPNA2/donor DNA containing nanoparticles, we found the impaired response to cyclic AMP stimulation was partially corrected, with mice exhibiting nasal potential differences that hyperpolarized in response to forskolin, which is more characteristic of wild-type mice (Fig 4b). The degree of hyperpolarization in mice treated with unmodified PLGA nanoparticles containing mCFPNA2/donor DNA was modest and did not reach statistical significance. However, after intranasal delivery with PLGA/PBAE/MPG nanoparticles, we found that the response to cyclic AMP stimulation was much more robust, with mice exhibiting a significant increase in their response to forskolin (Fig 4c). No significant change was seen in mice treated in parallel with blank nanoparticles, or in mice treated in parallel with PNA/DNA containing PLGA/PBAE/MPG nanoparticles targeting an unrelated genomic target but with similar base composition (Fig 4c). In these control experiments, additional CF mice were treated identically to the experimental group with PLGA/PBAE/MPG nanoparticles containing either no nucleic acid cargo, or with PNA and DNA targeting human β-globin; these PNA had similar base composition as the CF-targeted DNA but with 12 mismatches out of 17 in the Watson-Crick domain. The β-globin PNA was shown to be functionally active for inducing gene editing in β-globin33 but had no effect on the CFTR gene. For comparison, cyclic AMP responses of the nasal potential difference assays in wild-type mice were more robust (Fig 4c); this is expected given that wild-type mice have a homogenous population of wild-type CFTR-containing cells. In addition to the partial correction of the impaired cyclic AMP response we observed a significant reduction in the large lumen negative nasal potential in CF mice after treatment with PLGA/PBAE/MPG nanoparticles. This amiloride-sensitive portion of NPD was significantly reduced post treatment and similar in magnitude to that observed in wild-type mice (Supplementary Figure 6). Finally, there was no increased production of inflammatory cytokines in bronchoalveolar lavage fluid of treated mice (Fig 5a), and lungs showed normal histology (Fig 5b). Histology of limited nasal epithelial samples showed no obvious differences between treated and untreated mice (Supplementary Figure 7). There was a reduction in inflammatory cells in the bronchoalveolar lavage (BAL) of CF mice treated with PLGA/PBAE/MPG nanoparticles when compared to untreated CF mice: for n=4 mice in each group, average BAL cell counts were 1.24 × 105 in untreated CF mice, 0.4 × 105 in treated CF mice, and 0.32 × 105 for wildtype mice, p=0.03 for untreated versus treated CF mice.
Figure 5. Minimal inflammatory response with nanoparticle treatment.
(a) Cytokine production in bronchoalveolar lavage fluid of treated and control mice, with BAL from LPS treated control mice shown as a positive control, using Luminex bead-based assay. (b) Histology of treated and untreated mouse lungs, paraffin embedded and stained with H&E.
Confirmation of modification with deep sequencing
Modification was further confirmed in nanoparticle-treated human CFBE cells and in nanoparticle-treated mouse nasal epithelium and lung by deep sequencing, which allows for sequencing of millions of individual CFTR gene alleles in populations of cells (Table 1). In human CFBE cells treated in vitro serially three times with PLGA/PBAE/MPG particles, targeted modification frequency approached 10%. Increased efficiency of PLGA/PBAE/MPG nanoparticles over PLGA nanoparticles was also confirmed. In mice treated serially with PLGA/PBAE/MPG nanoparticles as described above, modification in the nasal epithelium was more than 5%, and more than 1% in the lung (Table 1); modification was not detected in vivo when plain PLGA nanoparticles were used. In addition, deep sequencing of cDNA amplicons produced from lung mRNA detected at least greater than 80-fold higher expression of corrected CFTR RNA in a treated mouse (PLGA/PBAE/MPG particles) versus untreated, demonstrating that the modification was present at the mRNA level, consistent with our findings of functional correction.
Table 1. Deep sequencing confirms efficient modification with low off-target effects.
F508del CFBE cells were plated at 10% confluence, then treated 3 times with 2 mg/mL particles over 7 days. Mice were treated with intranasal infusion of nanoparticles as described above. Controls were treated with blank nanoparticles without nucleic acid cargo. Primers with sample-specific barcodes were used to amplify 100 bp regions of the targeted region in mouse and human CFTR, as well as in a partially homologous site in human chromosome 4 or a partially homologous site in the murine X chromosome (similarity to the donor DNA sequence). Samples were analyzed by deep sequencing of alleles in the population using Illumina HiSeq sequencer. Sequences were aligned and enumerated to quantify modified sequences to determine percentage modification. For off-target sites, sequences with any mutations in the central region of interest above the machine error rate were enumerated.
| Sample | Modified CFTR sequences | %Correction | Modified off-target sequences | %Off-Target | |
|---|---|---|---|---|---|
| in vitro human CFBE cells | Control CFBE | 0/1894182 | <0.0005% | 0/1102030 | <0.00009% |
| PLGA Nanoparticles | 1502/1016551 | 0.15% | 0/236874 | <0.0004% | |
| PBAE/PLGA/MPG Nanoparticles | 947458/10279296 | 9.2% | 0/10304922 | <0.00001% | |
| in vivo CF mouse model | Control nasal epithelium | 0/46633 | <0.002% | 0/517496 | <0.0002% |
| Control lung | 0/1385709 | <0.0001% | 0/121970 | <0.001% | |
| PLGA Nanoparticles – Nasal Epithelium | 0/406270 | <0.00025% | |||
| PBAE/PLGA/MPG Nanoparticles – nasal epithelium | 31092/547521 | 5.7% | 0/1380607 | <0.0001% | |
| PBAE/PLGA/MPG Nanoparticles - lung | 9052/732024 | 1.2% | 0/1385709 | <0.0001% |
Off-Target Effects
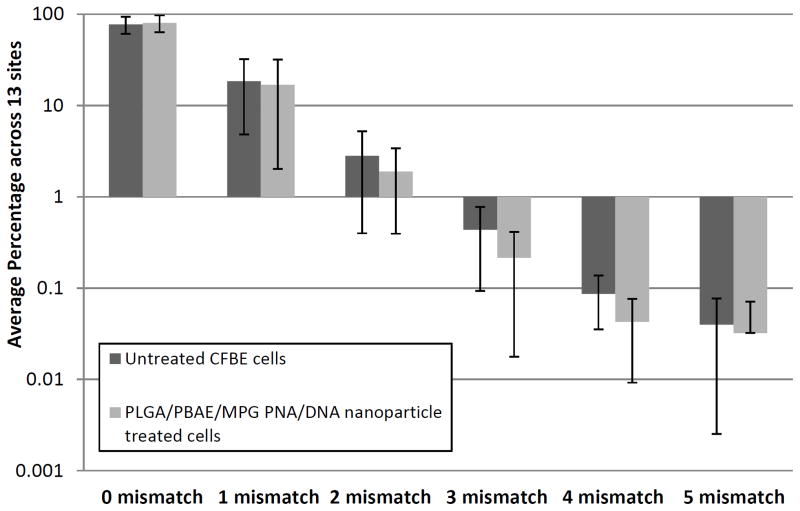
In addition, off-target modification in sites partially homologous to CFTR was examined. A section of chromosome 4 with 80% homology to the human donor DNA was queried in human cells (flanking features included a type II inositol-3,4-bisphosphate 4-phosphatase and a ubiquitin carboxyl-terminal hydrolase), and a similar section of the X chromosome with 50% homology to the donor DNA sequence (uncharacterized proteins) was queried in mice. In millions of sequenced alleles at these sites, there were no detected mutations above the machine-specific error rate (Table 1). In addition, thirteen additional off-target sites in the human genome with partial homology (>14 bp) to hCFPNA2 were queried in treated CFBE cells by deep sequencing. In these thirteen additional sites, off-target mutation/error rates were similar to untreated controls (Fig 6, Supplementary Table 1). For instance, for both untreated and treated cells, approximately 80 +/- 15% (average across the 13 sites) of queried sequences had zero mismatches (no difference above the machine-specific error rate between samples), and similarly there were no differences in the number of sequences with one to five mismatches in the queried sites. No differences in mutation frequencies above the error rate were seen at the individual sites. Finally, a single-cell gel electrophoresis assay (comet assay) was used to assess for the presence of DNA double-stranded breaks (Supplementary Fig 8). In this assay, electrophoresis of lysed cells results in migration of fragmented DNA, producing images that resemble comets when observed by fluorescent microscopy, with the length of the comet “tail” corresponding to the number of DNA breaks. No difference was seen between cells treated with DNA-containing and PNA/DNA-containing nanoparticles. In contrast, there was a slight but statistically significant increase in comet tail moments in cells treated with a human codon-optimized Cas9 expression plasmid43, which is designed to express CRISPR associated protein 9, the DNA nuclease used in CRISPR-based gene editing technologies that induces double stranded breaks.
Figure 6. Deep sequencing in additional human genomic sites showed no increased mutation rates in comparison to untreated controls.
200 bp regions of DNA with partial homology to the PNA binding site (>14 bps) in 13 disparate genomic sites were amplified by PCR, and PCR amplicons were sent for deep sequencing using the HiSeq (Illumina) 75 bp paired-end reads. CFBE cells treated 3 times with 2 mg/mL PLGA/PBAE/MPG particles with PNA/DNA were compared to untreated CFBE cells. The total number of aligned sequences were queried, and the number of sequences with 0 to 5 mismatched based pairs. At each of the 13 off-target sites, the percentage of sequences that had 0 to 5 mismatched base pairs was calculated. The average and standard deviation across the 13 genomic sites for 0 to 5 mismatches are shown. Mismatch percentages broken down by site are shown in Supplementary Table 1.
DISCUSSION
We have developed a new approach for the treatment of CF, which combines novel PNA/DNA molecules targeting the common F508del mutation in CFTR both in vitro in human bronchial epithelial cells and in vivo in a CF mouse, with enhanced gene correction using modified nanoparticles. Three PNA molecules were designed to bind to the human CFTR gene at sites within 350 base pairs of the F508del mutation. These sites were chosen because homopurine/homopyrimidine sites are required for Hoogsteen binding and triple helix formation, and previous studies indicate triplex-forming PNAs can increase levels of gene recombination at sites up to 750 base pairs (bp) from the target, with drop-off when the target is further than 400 bp away 44. While all three PNA molecules we designed were found to bind to their respective targeted sites in CFTR, only hCFPNA2 was able to induce consistent gene modification in CFTR in conjunction with the donor DNA as detected by AS-PCR. In prior work, we have also noticed some variability in the ability of certain triplex-forming PNA molecules to induce gene modification 26, 44. Factors which may contribute to differences in PNA intracellular activity include accessibility of the binding site in the cellular chromatin, folding dynamics of the molecules being used, and strength of binding in intracellular conditions.
Cloning by limiting dilution of nanoparticle-treated cells allowed interrogation of gene correction at the level of individual cells. Modification was passed on to cell progeny through months of cloning, demonstrating heritability. Gene modification in these positive clones was further confirmed by direct sequencing, and deep sequencing. CFTR gene correction in the positive clones was also confirmed by the presence of sequence-corrected mRNA and by functional analysis in an MQAE chloride flux assay. Positive clones had increased chloride flux in comparison to untreated F508del CFBE cells.
Modified PLGA/PBAE/MPG nanoparticles loaded with the PNA and donor DNA showed improved activity over PLGA nanoparticles, as demonstrated by MQAE chloride flux and deep sequencing both in vitro and in vivo. Of note, PLGA/PBAE/MPG nanoparticles carrying PNA/DNA cargo targeting an unrelated genomic site did not produce changes in chloride flux either in vitro or in vivo, suggesting that the observed effects are due to gene modification rather than a non-specific physiologic effect. In other work, using nanoparticles loaded with fluorescent dyes as tracers, we demonstrated that intranasal administration of PLGA/PBAE/MPG nanoparticles produced significantly greater nanoparticle association with airway epithelial cells than PLGA nanoparticles36. After multiple in vitro treatments with PLGA/PBAE/MPG nanoparticles, chloride efflux of CFBE cells approached that of normal human bronchial epithelial cells, and modification frequencies of up to 25% based on functional chloride efflux. However, it is possible that some individual cells with increased efflux may have had enhanced chloride transport due to a bystander effect from being adjacent to corrected cells, and not from direct modification. In addition, it is possible that corrected cells have a selection advantage, resulting in their preferential expansion. Deep sequencing showed modification up to 10% and no off-target effects above background mutation/read errors rates in untreated cells as assessed in 13 sites with partial homology for possible off-target binding of the PNA. In addition, no increased DNA damage was detected in treated cells by comet assay. This is expected, given that tail-clamp PNA molecules have very low levels of binding to mismatched sites and do not have any intrinsic nuclease activity. Unlike nuclease-based approaches to gene editing like zinc-finger nuclease and CRISPR, PNAs do not directly make strand breaks but instead provoke endogenous DNA repair pathways in the cell to mediate sequence conversion and gene correction that is templated by the co-introduced donor DNA. A low frequency of off-target effects will be of utmost importance for gene editing in this chronic, systemic disease.
In addition, surface-modified PLGA/PBAE/MPG nanoparticles showed greater genome engineering capacity after direct in vivo administration. Multiple intranasal treatments with PLGA/PBAE/MPG nanoparticles containing the murine CFTR-specific triplex-forming PNAs and donor DNAs were found to significantly modify the characteristic nasal potential difference defect in CF mice. Modification frequencies were greater than 5% in the nasal epithelium and 1% in the lung, with no detectable off-target mutations in a partially homologous site. There was no enhanced inflammatory cytokine production or changes in lung histology, highlighting the low immunogenicity of our approach. Because of this low toxicity, we speculate that longer courses of treatment are feasible and should enhance gene modification. Since correction of only one defective allele is required for restoration of chloride flux in cells, and studies have indicated that as little as 6–10% of cells need to be corrected for normal levels of ion transport in culture 45, our system has the potential to achieve gene correction at a clinically relevant level.
While the work presented here provides a foundation for PNA-based gene editing of CFTR, there are still many avenues left unexplored for further improvement of this in vivo genome engineering strategy. Future PNA designs to explore may include gamma-PNA molecules with a stereogenic mini-PEG group at the gamma position to enhance binding, use of guanidine-G-clamp PNA monomers to enhance PNA binding, or PNA/peptide conjugates for increased nuclear PNA uptake. Improvements in nanoparticle formulation may include use of surface modifiers targeting specific cell types, exploration of new polymers developed by our lab and others for oligonucleotide delivery, and formulation techniques to vary nanoparticle size. While we have shown that the combination of tail-clamp PNAs and PLGA/PBAE/MPG nanoparticles improves gene modification with triplex-forming oligonucleotides by an order of magnitude compared to previous approaches, there are still many possibilities for further enhancement of in vivo gene modification.
In addition, for translation to clinical use, several avenues of study must be further explored, including inflammatory response in CF mice animal models with more robust respiratory phenotypes46. Although the CF mouse model used in these studies has a mild respiratory phenotype which increases with age, the robust spontaneous pathology observed in the human disease process is not seen. One might consider using the ENaC-Tg overexpressing mouse to model the ability of nanoparticles to penetrate thickened airway mucus however given there is no underlying CFTR defect the efficacy of gene editing in such a model would be limited. In addition the properties of CF mucus may be quite different than that of the ENaC-Tg model due to alterations in airway surface liquid pH and bicarbonate concentrations that have been reported in CF47, 48. Alternatively, future studies could use house dust mite extracts to potentially induce mucus hyperproduction which may better approximate the airway pathology observed in advanced CF lung disease. Other potential models to explore include the CF pig model49, which most closely mirrors human disease. Modification of lung epithelial progenitors would also be required for more long-term correction of phenotype; while permanent genomic change using PNA/DNA is less transient than plasmid-based approaches and the changes will be passed on to daughter cells, some modified cells may be lost over time with regular turnover of the respiratory epithelium.
In summary, we have shown that PNA/DNA-encapsulating nanoparticles can correct the F508del mutation in human CFBE cells, resulting in the appearance of CFTR dependent chloride transport. We have also shown that intranasal treatment with PLGA/PBAE/MPG nanoparticles can correct chloride flux as measured by changes in nasal potential difference measurements in a CF mouse model, and leads to gene modification in the lung. This is the first demonstration of the use of triplex-mediated recombination for gene correction in CFTR. Modified PLGA/PBAE/MPG nanoparticles showed enhanced efficacy over PLGA nanoparticles, allowing us to achieve site-specific gene modification at levels an order of magnitude higher than previously possible with the PNA technology. While ZFNs and CRISPR/Cas9 systems have previously been used for CFTR gene editing, gene modification has been at lower frequencies with higher off-target effects, and without a system for in vivo translation. The delivery vehicle presented here could be used for direct in vivo gene editing using other nucleic-acid based approaches, such as short fragment homologous recombination and CRISPR/Cas9 plasmid systems.
AND METHODS
Oligonucleotides
PNAs with an 8-amino-2,6-dioxaoctanoic acid linker were purchased from Bio-Synthesis (Lewisville TX) or Panagene (Daejeon, Korea) and purified by HPLC. Donor oligonucleotides 50 nt in length were synthesized by Midland Certified Reagent (Midland TX), 5′- and 3′-end protected by three phosphorothioate internucleoside linkages at each end and purified by reversed phase-HPLC. Sequences of PNA molecules used are given in Figure 1 and Supplementary Figure 1.
Human donor DNA sequence: 5′TTCTGTATCTATATTCATCATAGGAAACACCAAAGATAATGTTCTCCTTAATGGTGCCAGG3′
Mouse donor DNA sequence: 5′TCTTATATCTGTACTCATCATAGGAAACACCAAAGATAATGTTCTCCTTGATAGTACCCGG3′
In our mismatched PNA control experiments, a PNA molecule targeting the human β-globin gene was used with 12 mismatches in the Watson Crick domain relative to the CF PNA2:
JTTTJTTTJTJT-OOO-TCTCTTTCTTTCAGGGCA β-globin-targeted PNA
TJTJJTTT-OOO-TTTCCTCTATGGGTAAG CFTR-targeted PNA
The β-globin-targeted PNA has 8 T, 5 C, 2 A, 3 G in the Watson-Crick domain and 8 T and 4 J in the Hoogsteen domain.
The CFTR-targeted PNA has 7 T, 3 C, 3 A, 4 G in the Watson-Crick domain and 5 T and 3 J in the Hoogsteen domain.
Gel Shift Assays for PNA Binding
To test the binding of candidate tail-clamp PNA molecules to the targeted site in the CFTR gene, PNA was incubated with plasmid DNA containing the target site at 37°C overnight, with 10 μM KCl in TE at final volume of 10 μL. Samples were digested with restriction enzymes flanking the binding site (EcoRI and BamHI), and the products run on an 8% non-denaturing PAGE gel. A silver stain was used to visualize the products. Uncropped gel images are included in Supplementary Figure 9.
Nanoparticle Formulation and Characterization
PLGA nanoparticles loaded with PNA and DNA were formulated and characterized using a double-emulsion solvent evaporation technique as previously described32. Instead of 1:1 PNA:DNA, 1:2 PNA:DNA was loaded in each batch in initial screening studies (20 μL of 2 mM donor, 20 μL of 1 mM PNA per 80 mg of PLGA). For particles in subsequent studies, 80 nmole (40 uL of 2 mM solution) of PNA and 40 nmole (20 uL of 2 mM solution) of DNA were used per 80 mg particle batch (scaled up or down accordingly). Briefly, 80 mg of polymer was dissolved in 160 uL dichloromethane overnight. PNA and DNA were dissolved in RNase/DNase free water. The PNA and DNA were then added dropwise into the dissolved polymer while vortexing, then sonicated for 10 seconds three times (Tekmar Probe Sonicator, Cincinatti, Ohio). The polymer solution was then added dropwise to 3.2 mL of 5% poly (vinyl alcohol) (PVA) while vortexing, then sonicated for 10 seconds three times using a probe sonicator on ice. The emulsion was then transferred to 20 mL of 0.3% PVA in a beaker with a stir bar, and left for 3 hours to let the solvent evaporate. This solution was then washed three times by ultracentrifugation with 10 mL of water, then resuspended in 2.5 mL of water and transferred into to Eppendorf tubes. Eppendorfs were frozen at −80*C for at least 2 hours, then transferred to a lyophilizer for 3 days.
Poly(beta amino ester) (PBAE) was synthesized by a Michael addition reaction of 1,4-butanediol diacrylate (Alfa Aesar Organics, Ward Hill, MA) and 4,4′-trimethylenedipiperidine (Sigma, Milwaukee, WI) as previously reported50. DSPE-PEG(2000)-maleimide was purchased from Avanti Polar Lipids (Alabaster, AL). MPG peptides were purchased from Keck (Yale University). CPPs were covalently linked to DSPE-PEG-maleimide as previously reported35. PLGA/PBAE particles contained 15% PBAE (wt%), and solvent from these particles was evaporated overnight in PVA instead of for three hours as above. To make surface-modified particles, DSPE-PEG-MPG was added to the 5.0%PVA solution during formation of the second emulsion at a 5 nmol/mg ligand-to-polymer ratio. In subsequent studies, particles were loaded as indicated. SEM imaging and controlled release studies were performed as before32. Briefly, for SEM imaging, particles were sputter coated with gold prior to imaging. For controlled release studies, particles were dissolved in 600 uL of DNase/RNase free water, put in a 37°C shaker, and at set timepoints centrifuged at 13000 RPM in a microfuge; at each timepoint, the supernatant was examined using a NanoDrop 8000 for nucleic acid content.
Cell Culture
CFBE cells (CFBE41o-) and human bronchial epithelial cells (16HBE14o-)38 were grown with LHC-8 media (Invitrogen) with 10% FBS, 1X antibiotic antimycotic (Gibco), and tobramycin 40 mg per 500 mL (Sigma). Once grown to confluence, cells were trypsinized by first washing with 0.05% trypsin, then adding 0.25% trypsin for 5 minutes, and harvesting with RPMI medium with 10% FBS. Cells were frozen in 5% DMSO in culture medium as necessary. Nanoparticles were resuspended in culture media by vigorous vortexing and water sonication, then added directly to cells at concentrations of 2 mg/mL/1×10^6 cells (corresponding to approximately 10^9 PNA/DNA molecules delivered to each cell assuming 100% efficiency).
To test primers, a 712 base pair region of the CFTR gene, with either the F508DEL or corrected sequence (including our silent modifications), was cloned into plasmids. PCR reactions were first tested on plasmids. Gradient and step-down PCR at varying conditions was performed to ensure that F508del primers only amplified the F508del plasmid, and the donor-specific primers only amplified the donor-sequence-containing plasmids. Primers are included in Supplementary Table 2.
Genomic DNA extraction and AS-PCR
Genomic DNA was harvested from cells and purified using the Wizard Genomic DNA Purification kit (Promega, Madison WI). Equal amounts of genomic DNA from each sample were subjected to allele-specific PCR, with a gene-specific reverse primer, and an allele-specific forward primer in which the 3′ end corresponds to the 6 bp modified sequence. Quantitative PCR was performed using a Stratagene Mx 3000P cycler. 0.2 μM donor DNA was used in spiking experiments. Copy numbers of DNA in the PCR reaction were approximately 10^14 copies of genomic DNA and 10^12 copies of spiked donor DNA. PCR products were separated on a 1% agarose gel and visualized using a gel imager. Relative gene modification was calculated using the 2−ΔΔCt method, with the average of the untreated controls used as the reference groups51.
AS-PCR conditions are as follows. Platinum Taq polymerase (Invitrogen, Carlsbad CA) was used for PCR reactions: 5 uL betaine, 4.25 uL water, 2.5 uL 10x Platinum Taq PCR buffer, 1.25 uL 50 mM MgCl2, 0.5 uL dNTPs, 0.5 uL each primer at 10 uM, 0.5 uL Platinum Taq polymerase, and 10 uL of genomic DNA at 40 ng/uL. PCR cycler conditions for human CFTR were as follows: 95°C 2 min, 94°C 30 sec, 69°C 1min, 72°C 1 min, 94°C 30 sec, 68°C 1min, 72°C 1 min, 94°C 30 sec, 67°C 1min, 72°C 1 min, 94°C 30 sec, 66°C 1min, 72°C 1 min, 94°C 30 sec, 65°C 1min, 72°C 1 min, [94°C 30 sec, 65°C 1min, 72°C 1 min] x 35 cycles, 72°C 2 min, hold at 4°C 1. PCR cycler conditions for mouse CFTR were as follows: 94°C for 5 min, [94°C 30 sec 66.9°C (for detection of F508del) or 68.3°C (for detection of modification) 45 sec, 72°C 1 min] x 40, 72°C 6 min, hold at 4°C. Conditions were optimized using plasmids containing the target sequences as indicated above. Of note, our donor sequences contain an additional 4 base-pairs of silent mutations distinguishing the donor sequence from wild-type CFTR, to ensure that contaminating wild-type cells (environmental or from other cell cultures) do not appear as false-positives. Uncropped gel image is included in Supplementary Figure 10.
For regular sequencing, High Fidelity Platinum Taq Polymerase (Invitrogen, Carlsbad CA) was used. PCR conditions for production of amplicons for regular sequencing were as follows: 0.5 uL dNTPs, 2.5 uL 10x HiFi Buffer, 1.5 uL 50 mM MgCl2, 14.1 uL water, 0.4 uL Taq HiFi, 0.5 uL each primer at 10 uM, 5 uLgenomic DNA at 80 ng/uL. PCR cycler conditions were as follows: 94°C 2 min, [94°C 30 sec 55°C 45 sec 68°C 1 min] x 35, 68°C 1 min, hold at 4°C.
RNA extraction and Reverse-Transcription AS-PCR
RNAeasy Plus Qiagen Kit (Gaithersburg, MD) was used to extract RNA, and Invitrogen superscript III kit (Carlsbody, CA) was used to make cDNA. PCR reactions contained cDNA, 20% Betaine, 0.2 mM dNTPs, Advantage 2 Polymerase Mix, 0.2 μM of each primer, and 2% platinum taq.
Gene-specific reverse primer:
5′ CCTAGTTTTGTTAGCCATCAGTTTACAGAC 3′
F508DEL CF primer: 5′GCCTGGCACCATTAAAGAAAATATCATTGG3′
Primer for corrected/donor: 5′CCTGGCACCATTAAGGAGAACATTATCTT 3′
PCR cycler conditions were as follows: 95°C 5 min, [95°C 30 sec 65°C 1 min 72°C 1min]x35, 72°C 5 min, hold at 4°C.
Deep Sequencing
Genomic DNA was isolated from treated cells or mouse tissue, and PCR reactions performed with high fidelity TAQ polymerase. Each PCR tube consisted of 28.2 μL dH2O, 5 μL 10x HiFi Buffer, 3 μL 50mM MgCl2, 1 μL DNTP, 1 μL each of forward and reverse primer, 0.8 μL HiFi Platinum Taq and 10 μL DNA template. Separate barcoded primers (6 bp barcode plus primer) were used for each sample. PCR conditions were as follows: For regular sequencing, High Fidelity Platinum Taq Polymerase (Invitrogen, Carlsbad CA) was used. PCR conditions for production of amplicons for regular sequencing were as follows: 0.5 uL dNTPs, 2.5 uL 10x HiFi Buffer, 1.5 uL 50 mM MgCl2, 14.1 uL water, 0.4 uL Taq HiFi, 0.5 uL each primer at 10 uM, 5 uLgenomic DNA at 80 ng/uL. PCR cycler conditions were as follows: 94°C 2 min, [94°C 30 sec 55°C 45 sec 68°C 1 min] x 35, 68°C 1 min, hold at 4°C. PCR products were prepared by end-repair and adapter ligation according to Illumina protocols (San Diego, CA), and pooled samples sequenced by the Illumina HiSeq with 75 paired-end reads at the W.M. Keck Facility at Yale University. Sequences have been uploaded to http://www.ncbi.nlm.nih.gov/bioproject/276283.
Analysis was performed using software designed by Nicole McNeer and Ken Hui (Yale School of Medicine), PERL file available as Supplementary Software online, and using software designed by Yong Kong (associate director Yale Bioinformatics Resource Center), code available at http://graphics.med.yale.edu/trim/. The program Btrim was used to trim off low-quality regions of each read and to assign the trimmed reads to each barcode52. The number of reads with modified sequence or original sequence were also searched by using Btrim. For off-target sites, the trimmed reads were mapped using program bowtie253. Primers are listed in Supplementary Table 2.
MQAE Assay for Chloride Flux
N-[ethoxycarbonylmethyl]-6-methoxy-quinolinium bromide (MQAE) is a chloride sensitive fluorescent dye we used to assess chloride flux in plated CFBE cells as previously described40. Cells were grown to confluence directly on coverslips. Then, cells were placed in Cl- containing solution (135 mM NaCl, 5 mM KCl, 1 mM CaCl2, 1.2 mM MgSO4, 2 mM NaH2PO4, 2 mM HEPES, and 10 mM glucose), then moved to a chloride free solution (135 mM NaCyclamate, 3 mM KGluconate, 0.5 mM CaCyclamate, 1.2 mM MgSO4, 2 mM KH2PO4, 2 mM HEPES, 10 mM glucose). Finally, chloride flux was assessed in solution with forskolin (10 μM) and IBMX (100 μM) added. MQAE experiments were performed on an Olympus IX-71 inverted microscope, with MQAE excited at 354 nm and fluorescence measured at 460 nm every 5 s. Fluorescence was measured on a cell-by-cell basis, with 30 to 100 cells catalogued per slide. The rate of change in MQAE fluorescence (arbitrary fluorescence units AFU/time) was graphed, and AFU/min was compared between groups. Graphs shown are normalized to background.
Animal Model
We used a mouse model homozygous for the F508DEL mutation on a fully backcrossed C57/BL6 background 42. Mice were between 12 and 40 weeks of age (the majority between 3 and 6 months of age), an equal mix of male and female. Nanoparticles were resuspended at 1 mg in 50 μL PBS, sonicated and administered to mice by intranasal instillation. Mice were treated with a total of 7 mg of nanoparticles over a course of 2 weeks (one treatment every other day) – this corresponds to a total of approximately 3.5 nmoles of donor DNA (~10^15 copies) and 7 nmoles of PNA per mouse (~2×10^15 copies). Estimating about 400 million cells/mouse lung, this corresponds to approximately 5 million PNA and 2.5 million DNA molecules per murine lung cell, if delivery to the lung is 100% efficient. Control mice were treated identically with either blank nanoparticles without nucleic acid cargo, or with nanoparticles containing PNA/DNA targeting human β-globin. While a scrambled PNA would provide the most closely matched molecular control, this off-target PNA provides a control of effects from non-specific PNA activity. Each independently performed experiment included at least one CF-targeted PNA/DNA treated mouse and one control mouse. All procedures were performed in compliance with relevant laws and institutional guidelines, and were approved by the Yale University Institutional Animal Care and Use Committee.
Nasal potential differences (NPDs) were measured as previously described5. Briefly, mice were anesthetized with ketamine/xylazine, and one electrode probe placed into one nostril, with a reference electrode with 3% agar in Ringer’s solution placed subcutaneously. A microperfusion pump was used to flow solution through the electrode probe at 0.2 mL/hour. Potential differences were measured first with a control Ringer’s solution, then with Ringer’s solution containing 100 μM amiloride, then a chloride-free solution with amiloride, and then chloride-free solution with amiloride and forskolin/IBMX. NPDs were measured prior to and after the nanoparticle treatment.
Bronchoalveolar lavage (BAL) fluid analysis and lung histology
BAL fluid was collected by standard protocols as previously described54, and cytokines measured using a microsphere-based multiplex assay per manufacturer instructions (Luminex; Millipore, Billerica, MA). To collect the lungs for histopathology, a midline incision from sternum to diaphragm was performed and, to remove blood from the pulmonary circulation, PBS was perfused via the right ventricle using a 20g needle. Lungs were inflated with 0.5% low melt agarose at constant pressure, then removed from the chest and placed in fixative. Paraffin embedded tissues were stained with hemotoxylin and eosin stain for imaging.
Comet Assay
300000 CFBE cells/well were pated on 6-well plates in 1 mL media, then treated with 2mg/mL of PBAE/MPG/PLGA nanoparticles either with DNA alone or both DNA and PNA, or with lipofectamine to deliver 2 ug of human cas9 plasmid #41815 (Addgene, Cambridge, MA)43. After 24 hours, cells were scraped and harvested, and prepared using the Trevigen CometAssay kit per manufacturer protocol (Trevigen, Gaithersburg, MD). Briefly, cells were suspended in agarose, added to comet slides, allowed to set, incubated 1 hr in lysis solution, placed in electrophoresis solution for 30 min, then run at 21 V for 45 min, placed in acetate solution for 30 min, 70% ethanol solution for 30 min, dried, stained with Sybr Green for 30 min, then visualized using an EVOS microscope. TriTek Comet Score freeware was used to analyze images.
Data Analysis
Data is represented as individual data points or as mean and standard error of the mean unless otherwise indicated. Data analysis was performed with Excel and Prism commercial software. Statistical significance was determined using student’s t-test, two-tailed, unpaired, or Fisher’s exact test. Code used for deep sequencing analysis is available at http://graphics.med.yale.edu/trim/.
Supplementary Material
Acknowledgments
We thank Faye Rogers for her technical assistance and helpful discussions. We are grateful to the Yale Center for Genome Analysis core facility and Y.K., Fransesc Lopez, Ken Hui, Michael Zhao, and Gregory Breuer for assistance of analysis of deep sequencing data presented in this work. This work was supported by the NIGMS Medical Scientist Training Program T32GM07205 (to N.A.M.), the Hartwell Foundation (to M.E.E.) and the National Institute of Health grants R01HL082655 and R01AI112443 (to P.M.G) and R01EB000487 (to W.M.S.). This work was also supported by F30HL110372 from the National Heart, Lung, and Blood institute (to N.A.M.) and Grant Number TL 1 RR024137 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH) and NIH Roadmap for Medical Research (to R.J.F.), and the Yale Pediatric Basic Science Training Program T32HD068201 from the National Child Health and Human Development Institute (to L.P.). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other funding organizations.
Footnotes
Authors contributions
Experiments were designed by N.A.M., K.A., C.C., P.M.G., W.M.S., and M.E.E. Experiments were performed and data analyzed by N.A.M., K.A., R.J.F., C.C., S.K., A.G., E.Q, L.P., Y.K., and M.E.E. Important reagents were prepared and provided by E.Q., R.B., and J.G. Written manuscript was prepared by N.A.M., K.A., E.Q., P.M.G., W.M.S., and M.E.E.
Conflict of financial interests: The authors have no conflicts of interest to disclose.
Accession codes: The sequences generated by deep sequencing have been deposited in SRA database under the accession codes (awaiting upload). Sequences have been uploaded to http://www.ncbi.nlm.nih.gov/bioproject/276283.
References
- 1.Riordan JR, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 2.Kerem B, et al. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 3.Rommens JM, et al. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- 4.Goetzinger KR, Cahill AG. An update on cystic fibrosis screening. Clinics in laboratory medicine. 2010;30:533–543. doi: 10.1016/j.cll.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Egan ME, et al. Curcumin, a major constituent of turmeric, corrects cystic fibrosis defects. Science. 2004;304:600–602. doi: 10.1126/science.1093941. [DOI] [PubMed] [Google Scholar]
- 6.Cartiera MS, et al. Partial correction of cystic fibrosis defects with PLGA nanoparticles encapsulating curcumin. Mol Pharm. 2010;7:86–93. doi: 10.1021/mp900138a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hutt DM, et al. Reduced histone deacetylase 7 activity restores function to misfolded CFTR in cystic fibrosis. Nature chemical biology. 2010;6:25–33. doi: 10.1038/nchembio.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holt N, et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cartier N, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 10.Aiuti A, et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N Engl J Med. 2009;360:447–458. doi: 10.1056/NEJMoa0805817. [DOI] [PubMed] [Google Scholar]
- 11.Griesenbach U, Alton EW. Gene transfer to the lung: lessons learned from more than 2 decades of CF gene therapy. Adv Drug Deliv Rev. 2009;61:128–139. doi: 10.1016/j.addr.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 12.Konstan MW, et al. Compacted DNA nanoparticles administered to the nasal mucosa of cystic fibrosis subjects are safe and demonstrate partial to complete cystic fibrosis transmembrane regulator reconstitution. Hum Gene Ther. 2004;15:1255–1269. doi: 10.1089/hum.2004.15.1255. [DOI] [PubMed] [Google Scholar]
- 13.Granio O, et al. Adenovirus 5-fiber 35 chimeric vector mediates efficient apical correction of the cystic fibrosis transmembrane conductance regulator defect in cystic fibrosis primary airway epithelia. Hum Gene Ther. 2010;21:251–269. doi: 10.1089/hum.2009.056. [DOI] [PubMed] [Google Scholar]
- 14.Ramachandran S, et al. A microRNA network regulates expression and biosynthesis of wild-type and DeltaF508 mutant cystic fibrosis transmembrane conductance regulator. Proc Natl Acad Sci U S A. 2012;109:13362–13367. doi: 10.1073/pnas.1210906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinn PL, et al. Lentiviral vector gene transfer to porcine airways. Molecular therapy Nucleic acids. 2012;1:e56. doi: 10.1038/mtna.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oakland M, Sinn PL, McCray PB., Jr Advances in cell and gene-based therapies for cystic fibrosis lung disease. Mol Ther. 2012;20:1108–1115. doi: 10.1038/mt.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goncz KK, Kunzelmann K, Xu Z, Gruenert DC. Targeted replacement of normal and mutant CFTR sequences in human airway epithelial cells using DNA fragments. Hum Mol Genet. 1998;7:1913–1919. doi: 10.1093/hmg/7.12.1913. [DOI] [PubMed] [Google Scholar]
- 18.Goncz KK, et al. Expression of DeltaF508 CFTR in normal mouse lung after site-specific modification of CFTR sequences by SFHR. Gene Ther. 2001;8:961–965. doi: 10.1038/sj.gt.3301476. [DOI] [PubMed] [Google Scholar]
- 19.Bruscia E, et al. Isolation of CF cell lines corrected at DeltaF508-CFTR locus by SFHR-mediated targeting. Gene Ther. 2002;9:683–685. doi: 10.1038/sj.gt.3301741. [DOI] [PubMed] [Google Scholar]
- 20.Beumer K, Bhattacharyya G, Bibikova M, Trautman JK, Carroll D. Efficient gene targeting in Drosophila with zinc-finger nucleases. Genetics. 2006;172:2391–2403. doi: 10.1534/genetics.105.052829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramalingam S, et al. Generation and Genetic Engineering of Human Induced Pluripotent Stem Cells Using Designed Zinc Finger Nucleases. Stem cells and development. 2012 doi: 10.1089/scd.2012.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee Ciaran M, RF, Hollywood Jennifer A, Scallan Martina F, Harrison Patrick T. Correction of the ΔF508 Mutation in the Cystic Fibrosis Transmembrane Conductance Regulator Gene by Zinc-Finger Nuclease Homology-Directed Repair. BioResearch Open Access. 2012;1:99–108. doi: 10.1089/biores.2012.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwank G, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell stem cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 24.Rogers FA, Vasquez KM, Egholm M, Glazer PM. Site-directed recombination via bifunctional PNA-DNA conjugates. Proc Natl Acad Sci U S A. 2002;99:16695–16700. doi: 10.1073/pnas.262556899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majumdar A, et al. Targeted gene knockout mediated by triple helix forming oligonucleotides. Nature genetics. 1998;20:212–214. doi: 10.1038/2530. [DOI] [PubMed] [Google Scholar]
- 26.Chin JY, et al. Correction of a splice-site mutation in the beta-globin gene stimulated by triplex-forming peptide nucleic acids. Proc Natl Acad Sci U S A. 2008;105:13514–13519. doi: 10.1073/pnas.0711793105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schleifman EB, et al. Targeted Disruption of the CCR5 Gene in Human Hematopoietic Stem Cells Stimulated by Peptide Nucleic Acids. Chem Biol. 18:1189–1198. doi: 10.1016/j.chembiol.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hubbell JA, Chilkoti A. Chemistry. Nanomaterials for drug delivery. Science. 2012;337:303–305. doi: 10.1126/science.1219657. [DOI] [PubMed] [Google Scholar]
- 29.Cheng CJ, Saltzman WM. Enhanced siRNA delivery into cells by exploiting the synergy between targeting ligands and cell-penetrating peptides. Biomaterials. 2011;32:6194–6203. doi: 10.1016/j.biomaterials.2011.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez PL, et al. Minimal “Self” peptides that inhibit phagocytic clearance and enhance delivery of nanoparticles. Science. 2013;339:971–975. doi: 10.1126/science.1229568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hrkach J, et al. Preclinical development and clinical translation of a PSMA-targeted docetaxel nanoparticle with a differentiated pharmacological profile. Sci Transl Med. 2012;4:128ra139. doi: 10.1126/scitranslmed.3003651. [DOI] [PubMed] [Google Scholar]
- 32.McNeer NA, et al. Nanoparticles deliver triplex-forming PNAs for site-specific genomic recombination in CD34+ human hematopoietic progenitors. Mol Ther. 2011;19:172–180. doi: 10.1038/mt.2010.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McNeer NA, et al. Systemic delivery of triplex-forming PNA and donor DNA by nanoparticles mediates site-specific genome editing of human hematopoietic cells in vivo. Gene Ther. 2013;20:658–659. doi: 10.1038/gt.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Babar IA, et al. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1201516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fields RJ, et al. Surface modified poly(beta amino ester)-containing nanoparticles for plasmid DNA delivery. J Control Release. 2012 doi: 10.1016/j.jconrel.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fields RJ, Quijano E, McNeer NA, Caputo Barone C, Bahal R, Anandalingam K, Egan ME, Saltzman WM. Modified Poly(lactic-co-glycolic acid) Nanoparticles for Enhanced Cellular Uptake and Gene Editing in the Lung. Advanced Healthcare Materials. 2014;2014 doi: 10.1002/adhm.201400355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nielsen PE. Peptide nucleic acids : protocols and applications. 2. Horizon Bioscience; Wymondham: 2004. [Google Scholar]
- 38.Gruenert DC, Willems M, Cassiman JJ, Frizzell RA. Established cell lines used in cystic fibrosis research. Journal of cystic fibrosis : official journal of the European Cystic Fibrosis Society. 2004;3 (Suppl 2):191–196. doi: 10.1016/j.jcf.2004.05.040. [DOI] [PubMed] [Google Scholar]
- 39.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 40.Shenoy A, et al. Calcium-modulated chloride pathways contribute to chloride flux in murine cystic fibrosis-affected macrophages. Pediatric research. 2011;70:447–452. doi: 10.1203/PDR.0b013e31822f2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Egan ME, et al. Calcium-pump inhibitors induce functional surface expression of Delta F508-CFTR protein in cystic fibrosis epithelial cells. Nat Med. 2002;8:485–492. doi: 10.1038/nm0502-485. [DOI] [PubMed] [Google Scholar]
- 42.Zeiher BG, et al. A mouse model for the delta F508 allele of cystic fibrosis. The Journal of clinical investigation. 1995;96:2051–2064. doi: 10.1172/JCI118253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knauert MP, et al. Distance and affinity dependence of triplex-induced recombination. Biochemistry. 2005;44:3856–3864. doi: 10.1021/bi0481040. [DOI] [PubMed] [Google Scholar]
- 45.Johnson LG, et al. Efficiency of gene transfer for restoration of normal airway epithelial function in cystic fibrosis. Nature genetics. 1992;2:21–25. doi: 10.1038/ng0992-21. [DOI] [PubMed] [Google Scholar]
- 46.Wilke M, et al. Mouse models of cystic fibrosis: phenotypic analysis and research applications. Journal of cystic fibrosis : official journal of the European Cystic Fibrosis Society. 2011;10 (Suppl 2):S152–171. doi: 10.1016/S1569-1993(11)60020-9. [DOI] [PubMed] [Google Scholar]
- 47.Quinton PM. Cystic fibrosis: impaired bicarbonate secretion and mucoviscidosis. Lancet. 2008;372:415–417. doi: 10.1016/S0140-6736(08)61162-9. [DOI] [PubMed] [Google Scholar]
- 48.Quinton PM. Role of epithelial HCO3(-) transport in mucin secretion: lessons from cystic fibrosis. American journal of physiology Cell physiology. 2010;299:C1222–1233. doi: 10.1152/ajpcell.00362.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ostedgaard LS, et al. The DeltaF508 mutation causes CFTR misprocessing and cystic fibrosis-like disease in pigs. Sci Transl Med. 2011;3:74ra24. doi: 10.1126/scitranslmed.3001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akinc A, Anderson DG, Lynn DM, Langer R. Synthesis of poly(beta-amino ester)s optimized for highly effective gene delivery. Bioconjug Chem. 2003;14:979–988. doi: 10.1021/bc034067y. [DOI] [PubMed] [Google Scholar]
- 51.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 52.Kong Y. Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics. 2011;98:152–153. doi: 10.1016/j.ygeno.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 53.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bruscia EM, et al. Macrophages directly contribute to the exaggerated inflammatory response in cystic fibrosis transmembrane conductance regulator-/- mice. American journal of respiratory cell and molecular biology. 2009;40:295–304. doi: 10.1165/rcmb.2008-0170OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.