Abstract
The enediyne natural products have been explored for their phenomenal cytotoxicity. The development of enediynes into anticancer drugs has been successfully achieved through the utilization of polymer- and antibody–drug conjugates (ADCs) as drug delivery systems. An increasing inventory of enediynes would benefit current application of ADCs in many oncology programs. Innovations in expanding the enediyne inventory should take advantage of the current knowledge of enediyne biosynthesis and post-genomics technologies. Bioinformatics analysis of microbial genomes reveals that enediynes are underexplored, in particular from Actinomycetales. This digest highlights the emerging opportunities to explore microbial genomics for the discovery of novel enediyne natural products.
Keywords: Enediyne polyketide synthase, Biosynthetic gene cluster, Genome mining, ADC payload, Natural products
Graphical abstract
The enediyne natural products are the most cytotoxic molecules in existence today, and their use as anticancer drugs has been demonstrated clinically.1–6 The natural enediynes have seen limited use as clinical drugs mainly because of substantial toxicity, however, various polymer-based delivery systems or antibody–drug conjugates (ADCs) have shown great clinical success or promise in anticancer therapy.7–12 Indeed, the poly(styrene-co-maleic acid)-conjugated neocarzinostatin (SMANCS®) has been marketed since 1994 for use against hepatoma.2 Various ADCs have been developed or are in varying stages of development, including a CD33 mAB-calicheamicin (CAL) conjugate (i.e., Mylotarg®) for acute myeloid leukemia (AML), a CD22 mAB-CAL conjugate (inotuzumab ozogamicin) for non-Hodgkin lymphoma,10,11 as well as, several mAB-C-1027 conjugates for hepatoma3,13 and mAB-uncialamycin (UCM) conjugates for selected tumors.14 (Pfizer voluntarily withdrew Mylotarg® from the market in 2010; however, significant survival benefits observed in recent phase III trials suggest that Mylotarg® may have an important future role in treating patients with good- or intermediate-risk AML.15) These examples clearly demonstrate that the enediynes can be developed into powerful drugs when their extremely potent cytotoxicity is harnessed and delivered to tumor cells. It is remarkable that among the 11 enediynes known to date, two, neocarzinostatin (NCS) and CAL, have been developed into clinical drugs and one, C-1027, is in clinical trials, representing an astonishing ~30% success rate with the enediyne class of natural products (Fig. 1). Developing innovative methods to discover new enediynes therefore holds a great promise for anticancer drug discovery.
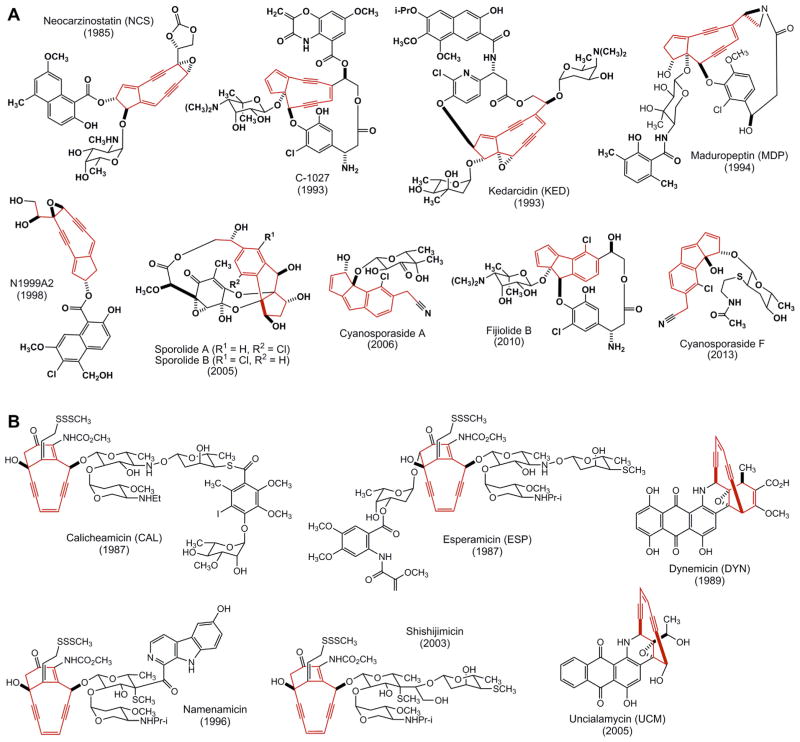
Figure 1.
Structures of the 11 enediynes, (A) five nine-membered and (B) six 10-membered ones, known to date with the enediyne cores highlighted in red. The sporolides, cyanosporasides, and fijiolides were proposed to be derived from nine-membered enediyne precursors based on their chlorine substitution pattern after cycloaromatization. Given in parentheses are the years when each of the enediyne structures was established. CAL and NCS have been developed into clinical drugs, C-1027 is currently in phase II trials, and several analogues of UCM have been evaluated as ADC payloads in preclinical studies.
Inspired by the recent success of two ADCs (Adcetris® in 2011 and Kadcyla® in 2013), interests in anticancer ADCs continue to grow, and virtually every major pharmaceutical company with an oncology program now has an initiative on ADCs.9–11 Among the 30+ ADCs currently in development, however, the majority of them use one of the few available cytotoxic drugs (auristatin, CAL, maytansinoid, doxorubicin, camptothecin, duocarmycin, and C-1027), all of which are of natural product origin.9–11 The ADC field is in critical need of new, highly potent cytotoxic payloads (IC50s at 1 nM–10 pM), active in many tumor types, with improved physical, chemical, and biological properties. Therefore, new enediynes would be extremely valuable assets in the development of safer, more effective ADCs with a validated mode of action. This digest highlights the emerging opportunities to explore microbial genomics for the discovery of novel enediyne natural products, in particular, those from the Actinomycetales.
Enediyne discovery, structures, and activities
Enediyne natural products are very rare. Since the NCS chromophore structure was first elucidated in 1985,16 only 11 enediynes are structurally characterized to date, with an additional four isolated in the cycloaromatized form (Fig. 1).17,18 Discovery of a new enediyne natural product is therefore a significant event in natural products chemistry, biology, and drug discovery.1–6 The enediyne natural products are classified into two subcategories according to the size of the enediyne cores. Members of the nine-membered enediynes are commonly chromoproteins, consisting of an apoprotein and the enediyne chromophore, including NCS from Streptomyces carzinostaticus, 16 C-1027 from Streptomyces globisporus,19 kedarcidin (KED) from a Streptoalloteichus sp.,20 maduropeptin (MDP) from Actinomadura madurae,21 N1999A2 from Streptomyces sp. AJ9493,22 the sporolides (SPOs) from Salinispora tropica,23 the cyanosporasides (CYAs) from Salinispora pacifica CNS-143,24 the cyanosporasides (CYNs) Streptomyces sp. CNT-179,25 and the fijiolides from Nocardiopsis sp. CNS-653,26 with the latter four isolated in the absence of an apoprotein. 10-Membered enediynes are discrete small molecules, including the CALs from Micromonospora echinospora,27 dynemicin (DYN) from Micromonospora chersina,28 the esperamicins (ESPs) from Actinomadura verrucosospora,29 namenamicin from Polysyncraton lithostrotum,30 and UCM from Streptomyces uncialis.31
While members of the enediyne family of natural products exhibit remarkable structural diversities, all enediynes contain a core consisting of two acetylenic groups conjugated to a double bond or incipient double bond within the nine- or 10-membered carbocycle (Fig. 1). As a consequence of this structural feature, these compounds share a common mode of action. Electronic rearrangement (Bergman or Myers-Saito rearrangement) of the enediyne carbocycle produces a transient benzenoid diradical.1–6 When positioned within the minor groove of DNA, the diradical abstracts hydrogen atoms from the deoxyribose backbone of duplex DNA; the DNA-centered radicals can then cause interstrand crosslinks (ICLs), react with molecular oxygen leading ultimately to DNA double-strand breaks (DSBs), or both, resulting in the phenomenal cytotoxicity known for the enediynes.4–6,17,18,30,31
Enediynes as payload candidates for anticancer ADCs
ADCs provide the possibility of selectively ablating cancer cells by combining the specificity of a mAB for a target antigen with the delivery of a highly potent cytotoxic agent.7–12 The ideal number of drug molecules per mAB for most current ADCs appears to be about four.12 Underconjugation can decrease potency of the resultant ADCs, whereas overconjugation can lead to decreased circulation half-life, reduced tolerability, and impaired antigen binding.12 The preferred payload molecules therefore have to be highly cytotoxic (IC50s at 1 nM–10 pM) and ideally active in many tumor types. The enediynes represent some of the most cytotoxic molecules in existence today (for example, the IC50s of CAL and C-1027 towards selected cancer cell lines are in the range of 10 pM–10−3 pM).1–6,30,31 While the enediynes are mostly known for their activity through DNA DSBs, ICL is an alternative mode of action.30–33 The ICL property of the enediynes can be exploited to target solid tumors or other cancer cells under hypoxic environments, which do not respond well to enediynes that predominantly induce oxygen-dependent DSBs.32,33 The exquisite potency and mechanisms of action of the enediynes make them ideal payload candidates for ADCs. The utilities of enediynes as payloads of anticancer ADCs have been demonstrated in clinical therapy.7–12
Enediyne biosynthesis and engineering
The enediyne natural products present an outstanding opportunity to (i) decipher the genetic and biochemical basis for the biosynthesis of complex natural products,17,18 (ii) explore ways to make novel analogues by manipulating genes governing their biosynthesis,30,31,34–36 and (iii) discover new enediyne natural products by mining microbial genomes for the trademark enediyne biosynthetic machineries. 37,38 Significant progress has been made in enediyne biosynthesis and engineering in the last decade. Highlights include: (i) cloning and characterization of the C-1027,39 NCS,40 MDP,41 KED,42 SPO,43 CYA and CYN25 biosynthetic gene clusters as examples of nine-membered enediyne natural products, (ii) cloning and characterization of the CAL,44 ESP (partial),37,38 and DYN45 biosynthetic gene clusters as examples of 10-membered enediyne natural products (Fig. 2A), (iii) establishment of the polyketide origin for all enediynes and discovery of the enediyne PKS as an acyl carrier protein (ACP)-dependent, self-phosphopantetheinylating, iteratively acting, type I polyketide synthase (PKS) that produces a linear polyene to initiate both nine- and 10-membered enediyne core biosynthesis (Fig. 2B),46,47 (iv) characterization of numerous enzymes from the enediyne biosynthetic machineries that catalyze novel chemistries,17,18 (v) engineered production of novel enediyne analogues with altered modes of action,30,31 and exploitation of the ICL property of the enediynes to target solid tumors or other cancer cells under hypoxic environments,32,33 (vi) manipulation of the enediyne biosynthetic machinery for titer improvement and production of selected enediynes in sufficient quantities to support further mechanistic studies and preclinical development,35,36 (vii) development of methodologies to screen microorganisms for the discovery of new enediyne natural products, 37,38 and (viii) discovery that, in spite of the fact that only 11 enediyne natural products (14 if including the ones isolated in the cycloaromatized form) have been isolated to date (Fig. 1), the biosynthetic potential of Actinomycetales to produce enediynes is greatly underappreciated.37,38 These findings have laid the foundation to explore microbial genomics for the discovery of new enediyne natural products. Thus, (i) genome survey of microbial strain collections for the hallmark enediyne PKS gene cassettes could allow rapid identification of potential enediyne producers, (ii) genome sequencing of the potential producers for the enediyne biosynthetic gene clusters could allow accurate prediction of the structural novelty of the new enediynes, and (iii) genetic manipulation and fermentation optimization of the most promising producers would ultimately allow efficient production, isolation, and structural and biological characterization of new enediyne natural products.
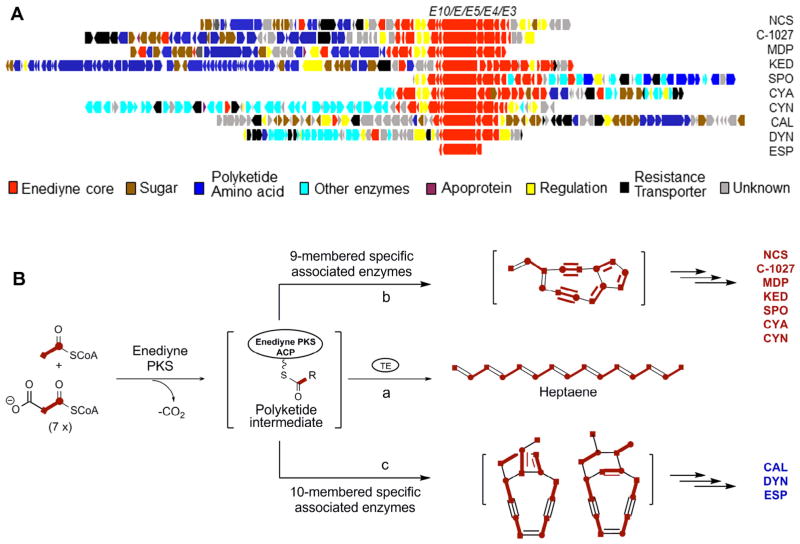
Figure 2.
Characterization of the selected nine- and 10-membered enediyne biosynthetic machinery laying the foundation to explore microbial genomics for the discovery of new enediyne natural products. (A) Alignment of the 10 known enediyne biosynthetic gene clusters, highlighting the enediyne PKS gene cassettes (i.e., E10, E, E5, E4, and E3) common to all enediynes and varying open reading frames (color-coded) for the diverse peripheral moieties, as well as for pathway regulation and self-resistance. (B) A unified model for enediyne core biosynthesis, featuring the production of heptaene by PKS and TE in the absence of associated enzymes (path a), and functional interactions between PKS-TE and nine- or 10-membered specific associated enzymes differentiating nine- (path b) or 10-membered (path c) core biosynthesis.
Strain prioritization for novel natural product discovery
Traditional microbial natural product discovery programs begin by fermenting each strain individually, often in various media, followed by preparation of crude extracts. Two fundamental methods to search for novel natural products from extracts are: bioassay-guided fractionation and chemical profiling of compounds possessing unique structural novelty.48,49 The desired molecule of interest must be produced in sufficient amounts to facilitate isolation and characterization on a reasonable timeframe. The discovery of a new natural product typically requires three principal steps: dereplication of known compounds to avoid redundancies, isolation of the targeted molecules, and structural elucidation of the purified natural products. Indeed, discovery of the known enediynes relied mainly on bioassay-guided, extensive natural product dereplication.16–31 The biochemical induction assay (BIA) that detects DNA damaging agents was most commonly used to selectively assay for enediyne production and subsequent isolation.37,50 While successful, only 14 enediyne natural products are known to date. This tedious and laborious traditional process could be significantly shortened if the biosynthetic potential of a strain collection is known in advance. Resources could then be devoted preferentially to only interrogate strains that hold the highest promise in producing novel natural products, thereby accelerating their discovery, isolation, and structural characterization.
Complementary to traditional approaches, the progress made in the last two decades in connecting natural products to the genes that encode their biosynthesis has fundamentally changed the landscape of natural products research and sparked the emergence of a suite of contemporary approaches to natural product discovery.48,49,51–53 Thus, genes are now as important as chemistry in categorizing known natural products and identifying new ones. Advances in microbial genomics have unequivocally demonstrated that we are missing ~90% of the natural product biosynthetic capacity of even the workhorse producers, the Actinobacteria.52,53 To access this untapped reservoir of potentially new natural products, two principal strategies have been applied to induce these ‘cryptic biosynthetic pathways’. The ‘epigenetic’-related approaches include challenging the microorganisms through culture conditions, nutritional or environmental factors, external cues, and stress, as well as, exploiting interspecies crosstalk.54 The genomics-based approaches include mining the genomes to predict metabolite structures, engineering the pathways by manipulating pathway-specific or global regulators, and expressing the cryptic pathways in selected heterologous hosts.51–53,55 While each of the various approaches has different strengths and weaknesses, they have been successful in yielding cryptic natural products but only on a case-by-case basis and are far from being of practical use for natural product discovery. Thus, in spite of the rapid advances in DNA sequencing technologies and bioinformatics, it is still impractical to sequence and annotate all strains within a large collection as a practical means to discover new natural products. High-throughput methods, rapidly surveying the biosynthetic potential of a strain collection so that strains that harbor the highest biosynthetic potential can then be identified, prioritized, and subjected to epigenetic- and/or genomics-based approaches, could fundamentally change how natural products are discovered.48,49
Enediyne PKS gene cassettes as a beacon for enediyne producers
In spite of their remarkable structural diversity, the 14 known enediyne natural products all feature a nine- or 10-membered enediyne core (Fig. 1). Comparative bioinformatics analyses of the seven nine- (NCS, C-1027, MDP, KED, SPO, CYA, and CYN)25,39–43 and three 10-membered (CAL, DYN, and ESP)37,38,44,45 enediyne PKS loci revealed a set of five genes common to all enediynes (i.e., the enediyne PKS gene cassette consisting of E10/E/E5/E4/E3) (Fig. 2A); no apparent conservation was observed beyond the enediyne PKS gene cassettes, accounting for the structural diversity characteristic for the periphery moieties of the enediynes (Fig. 1). This remarkable sequence homology has resulted in a unified model for the enediyne PKS cassette to catalyze the formation of both nine- and 10-membered enediyne cores. This model has been further supported by the fact that: (i) PKS-TE (i.e., E–E10) catalyze the biosynthesis of heptaene as a shunt metabolite (Fig. 2B) in the absence of the associated enzymes,46,47 (ii) heptaene is co-produced in fermentations of known enediyne producers, and (iii) heptaene production is PKS-dependent—inactivation of the pksE gene abolished heptaene production and complementation of the ΔpksE mutation with a functional pksE restored its production.47 These observations prompted us to select genes within the enediyne PKS cassettes as probes to survey genomes for the presence of enediyne biosynthetic machinery.37,38 Once the new enediyne producers are identified, heptaene production, complementary to BIA for enediyne production,37,50 could be used as a sensitive phenotypic indicator to follow enediyne production upon fermentation optimization.35,36,47
Actinomycetales as the most prolific enediyne producers
It is now clear that the biosynthetic potential of natural products is significantly underestimated based on the natural products isolated to date.51–53 This is also true for the enediyne natural products,37,38 in spite of the fact that enediyne natural products are extremely rare and only 14 enediyne natural products are known to date (Fig. 1). A great challenge is to rapidly identify the most promising enediyne producers, from a large strain collection, so that all resources could be devoted to interrogate only the strains that hold the highest potential in producing novel enediyne natural products. To validate the utility of the selected genes within the enediyne PKS cassette as a beacon to search for new enediyne producers from a large strain collection, we carried out a virtual survey of the entire GenBank, using each of the five genes within the enediyne PKS cassette, alone or in combination, as queries, for genes encoding enediyne biosynthetic machineries. We were able to identify all 10 confirmed enediyne biosynthetic machineries (Fig. 2A), validating the utility and specificity of the genes within the enediyne PKS cassette as probes. While each of the five genes alone yields essentially the same outputs, E5, E, or E10 are preferred, and the combination of E5/E or E/E10 afford the most specific results. Most significantly, we identified 51 additional enediyne PKS cassettes from 47 organisms (four strains contain two enediyne PKS cassettes each) not known as enediyne producers. Together with the 10 known enediyne biosynthetic machineries, the 61 total enediyne PKS cassettes support our early findings that the biosynthetic potential of enediynes is significantly underappreciated37,38 (i.e., a total of 61 enediyne biosynthetic gene clusters from the GenBank database as of September 23, 2014). All of the 61 gene clusters are of bacterial origin (out of 4889), and most strikingly, 54 of the 61 clusters are in the order of Actinomycetales (out of 796). While the Actinomycetales are well known as the most prolific producers of biologically active secondary metabolites, 56 this is the first time revealing the Actinomycetales as the most prolific producers of enediyne natural products.
Enediyne structural diversity as predicted by microbial genomics
The new enediyne biosynthetic gene clusters can be readily differentiated from the known ones upon phylogenetic analysis on the basis of the enediyne PKS cassettes. As exemplified by the phylogenetic analysis in Figure 3A, which is based on the amino acid sequences of the E enzymes, the 10 known enediyne PKS cassettes are well dispersed, and the 51 newly identified enediyne PKS cassettes are distinct from the 10 known enediyne PKS cassettes. Most significantly, many of the new enediyne PKS cassettes fell into clades that were not represented by any of the 10 known enediyne natural products, a provocative suggestion for novel enediyne structures (Fig. 3A).
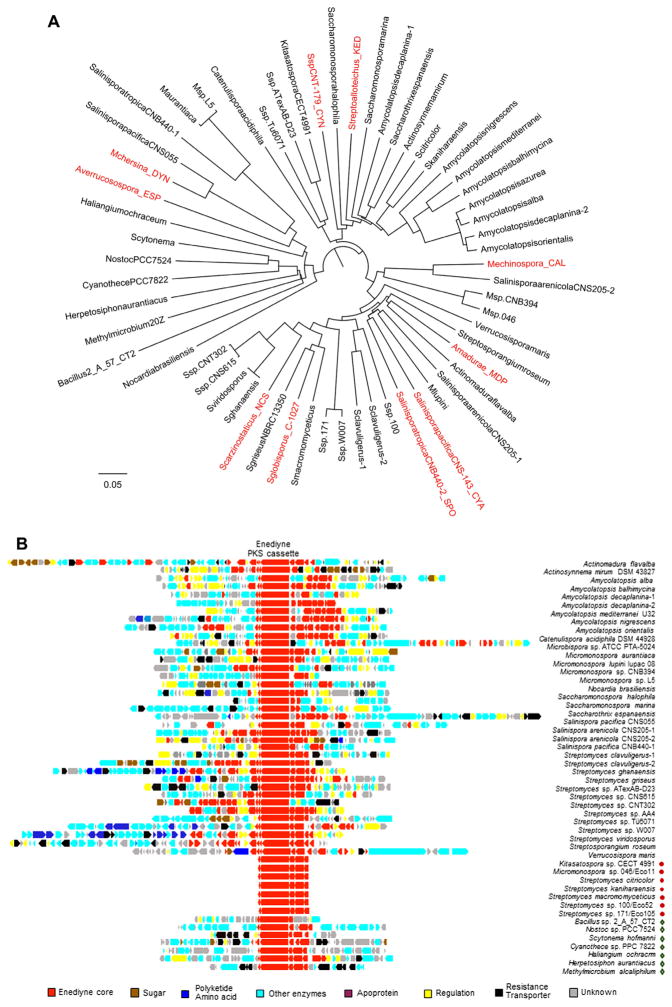
Figure 3.
Exploration of microbial genomics to discover new enediyne natural products. (A) Phylogenetic analysis of enediyne PKS cassettes, as exemplified with the amino acid sequences of Es, from the 49 putative enediyne gene clusters (two Es excluded due to sequence gaps) and the 10 known enediyne gene clusters (in red). (B) Annotation and alignment of 44 new enediyne gene clusters, as well as seven additional enediyne PKS cassettes, in comparison with the 10 known gene clusters in Figure 2A. While all clusters feature the conserved enediyne PKS cassette (highlighted in red), they also possess other open reading frames (color-coded) that are unprecedented in gene clusters that encode production of the known enediynes, indicative of novel structural features and functional groups for the new enediyne natural products. Of the total 57 enediyne producers (i.e., 47 new producers with four containing two enediyne clusters each and the 10 known enediyne producers), 50 are actinomycetales. (◇) Non-actinomycetales, (●) enediyne PKS cassettes only, and GenBank accession numbers for the new enediyne gene clusters are given in reference.57
While the hallmark nine- or 10-membered enediyne cores are essential for their phenomenal cytotoxicity, the enediyne natural products are rich in peripheral moieties, and it is the diverse peripheral chemistry displayed for each of the enediyne natural products that fine-tunes their biological activities. We have annotated the 51 new enediyne gene clusters, though the boundaries of the clusters are hypothetical, and they appear to be true enediyne biosynthetic clusters, based on features common to the biosynthesis of the known enediynes. As highlighted in Figure 3B, all the clusters feature the conserved enediyne PKS cassettes indicating the hosts are true enediyne producers, and the gene clusters are rich in other open reading frames that are unprecedented in gene clusters that encode the production of the known enediyne natural products. The extraordinary number of novel open reading frames is indicative of novel structural features and peripheral moieties, which holds promise for the discovery of new enediyne natural products.
Conclusions and prospects
Given the extreme potencies of the enediyne family of natural products,1–6 their proven utility as cytotoxic payloads of anticancer ADCs, and the astonishing success rate of these rare natural products in the clinic,7–12 the discovery of new enediynes, as well as alternative producers of known enediynes with high titers or better growth characteristics, is a critical step in the development of enediynes as novel anticancer drugs. Thorough bioinformatics analysis of microbial genomes reveals that enediyne natural product gene clusters are widely distributed among Actinomycetales (50 out of 796 sequenced bacterial genomes, accounting for 6.3%), and yet remain underexplored, as the newly identified gene clusters are non-redundant and highly unique.37,38 The abundant and highly diverse enediyne gene clusters found within public databases should provoke the interests of scientific communities in pursuing novel enediyne natural products from microbial strain collections, in particular from Actinomycetales.
Combining traditional natural product discovery methods with genomics-based technologies and bioinformatics will expedite the discovery process of novel enediynes. High-throughput methods to rapidly screen a large microbial strain collection for the conserved genes within the enediyne PKS cassettes are essential to allocate resources to strains that show biosynthetic potential for enediyne production.48,49 DNA sequencing of the conserved pksE genes or alternatively, the whole genome, allows further prioritization by identifying redundant or unique enediyne biosynthetic gene clusters through bioinformatics analysis (i.e., phylogenetic analysis, cluster annotation, genome neighborhood network).37,38 Once strains are prioritized, fermentation in multiple media and preparation of crude extracts allow chemical profiling of heptaene production, 46,47 and bioassays for DNA damage activity (i.e., by BIA)37,50 can then be utilized to determine which strains readily produce enediynes or enediyne-like metabolites. Fractionation of promising crude extracts guided by the BIA allows a simple detection method for isolating novel enediynes. With the conserved enediyne PKS gene cassettes known, ΔpksE deletion mutants can be easily constructed to correlate putative gene clusters with the production of heptaene or DNA-damaging natural products. Silent or cryptic gene clusters can be overcome by the genetic manipulation of positive or negative regulators found within the enediyne gene clusters or by extensive fermentation optimization.35,36 The expedient technologies and tools of recombinant DNA work in Streptomyces species and related actinomycetes that have been developed in the past two decades surely will facilitate these efforts.
It is evident from the rarity of known enediyne natural products that future enediyne discovery efforts must be approached using new and innovative means.37,38 The advancement of DNA sequencing technologies and genomics-based approaches in recent years has opened the door for a multi-disciplinary approach towards the discovery of natural products and enediynes in particular.52,53 The potential of the enediynes as relevant clinical drugs and the unique chemistry and enzymology involved in their biosynthesis should be of great interest to cancer biologists, natural product chemists, biochemists, enzymologists, structural biologists, and the community as a whole. We are optimistic that enediynes will continue to play an important role in natural product biosynthesis, engineering, and drug discovery.
Acknowledgments
Research on discovery, biosynthesis, engineering for structural diversity, and anticancer drug discovery of the enediyne natural products in the Shen Lab was supported in part by National Institutes of Health Grant CA78747 and the Natural Products Library Initiative at the Scripps Research Institute.
References and notes
- 1.Doyle TW, Borders DB. Enediyne Antibiotics as Antitumor Agents. Marcel-Dekker; New York: 1995. [Google Scholar]
- 2.Maeda H, Edo K, Ishida N. Neocarzinostatin: The Past, Present, and Future of an Anticancer Drug. Springer; New York: 1997. [Google Scholar]
- 3.Brukner I. Curr Opin Oncol Endocr Met Invest Drugs. 2000;2:344. [Google Scholar]
- 4.Xi Z, Goldberg IH. In: Comprehensive Natural Products Chemistry. Barton D, Nakanish K, Meth-Cohn O, editors. Vol. 7. Elsevier; New York: 1999. pp. 553–592. [Google Scholar]
- 5.Thorson JS, Sievers EL, Ahlert J, Shepard E, Whitwam RE, Onwueme KC, Ruppen M. Curr Pharm Des. 2000;6:1841. doi: 10.2174/1381612003398564. [DOI] [PubMed] [Google Scholar]
- 6.Galm U, Hager MH, Van Lanen SG, Ju H, Thorson JS, Shen B. Chem Rev. 2005;105:739. doi: 10.1021/cr030117g. [DOI] [PubMed] [Google Scholar]
- 7.Chari RVJ. Acc Chem Res. 2008;41:98. doi: 10.1021/ar700108g. [DOI] [PubMed] [Google Scholar]
- 8.Senter PD. Curr Opin Chem Biol. 2009;13:1. doi: 10.1016/j.cbpa.2009.03.023. [DOI] [PubMed] [Google Scholar]
- 9.Beck A, Senter P, Chari R. mAbs. 2011;3:331. doi: 10.4161/mabs.3.4.16612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Senter PD, Sievers EL. Annu Rev Med. 2013;64:15. doi: 10.1146/annurev-med-050311-201823. [DOI] [PubMed] [Google Scholar]
- 11.Thayer AM. C & E News. 2014. pp. 13–20. [Google Scholar]
- 12.Panowski S, Bhakta S, Raab H, Polakis P, Junutula JR. mAbs. 2011;6:34. doi: 10.4161/mabs.27022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shao RG, Zhen YS. Anticancer Agents Med Chem. 2008;8:123. doi: 10.2174/187152008783497055. [DOI] [PubMed] [Google Scholar]
- 14.Chowdari NS, Gangwar S, Sufi B. 2013/0209494 A1. US Patent Application Publication, Pub No US. Pub. Date, Aug. 15, 2013.
- 15.Ravandi F. J Clin Oncol. 2011;29:349. doi: 10.1200/JCO.2010.32.2693. [DOI] [PubMed] [Google Scholar]
- 16.Edo K, Mizugaki M, Koide Y, Seto H, Furihata K, Otake N, Ishida N. Tetrahedron Lett. 1985;26:331. [Google Scholar]
- 17.Van Lanen SG, Shen B. Curr Top Med Chem. 2008;8:448. doi: 10.2174/156802608783955656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang ZX. Nat Prod Rep. 2010;27:499. doi: 10.1039/b908165h. [DOI] [PubMed] [Google Scholar]
- 19.Otani T, Yoshida K, Sasaki T, Minami Y. J Antibiot. 1999;52:415. doi: 10.7164/antibiotics.52.415. [DOI] [PubMed] [Google Scholar]
- 20.Ren F, Hogan PC, Anderson AJ, Myers AG. J Am Chem Soc. 2007;129:5381. doi: 10.1021/ja071205b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komano K, Shimamura S, Norizuki Y, Zhao D, Kabuto C, Sato I, Hirama M. J Am Chem Soc. 2009;131:12072. doi: 10.1021/ja905397p. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi S, Ashizawa S, Takahashi Y, Sugiura Y, Nagaoka M, Lear MJ, Hirama M. J Am Chem Soc. 2001;12:11294. doi: 10.1021/ja011779v. [DOI] [PubMed] [Google Scholar]
- 23.Buchanan GO, Williams PG, Feling RH, Kauffman CA, Jensen PR, Fenical W. Org Lett. 2005;7:2731. doi: 10.1021/ol050901i. [DOI] [PubMed] [Google Scholar]
- 24.Oh DC, Williams PG, Feling RH, Kauffman CA, Jensen PR, Fenical W. Org Lett. 2006;8:1021. doi: 10.1021/ol052686b. [DOI] [PubMed] [Google Scholar]
- 25.Lane AL, Nam SJ, Fukuda T, Yamanaka K, Kauffman CA, Jensen PR, Fenical W, Moore BS. J Am Chem Soc. 2013;135:4171. doi: 10.1021/ja311065v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nam SJ, Gaudencio SP, Kauffman CA, Jensen PR, Kondratyuk TP, Marler LE, Pezzuto JM, Fenical W. J Nat Prod. 2010;73:1080. doi: 10.1021/np100087c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee MD, Dunne TS, Chang CC, Ellestad GA, Siegel MM, Morton GO, McGahren WJ, Borders DB. J Am Chem Soc. 1987;109:3466. [Google Scholar]
- 28.Myers AG, Fraley ME, Tom NJ, Cohen SB, Madar DJ. Chem Biol. 1995;2:33. doi: 10.1016/1074-5521(95)90078-0. [DOI] [PubMed] [Google Scholar]
- 29.Golik J, Dubay G, Groenewold G, Kawaguchi H, Konishi M, Krishnan B, Ohkuma H, Saitoh K, Doyle TW. J Am Chem Soc. 1987;109:3462. doi: 10.7164/antibiotics.38.1605. [DOI] [PubMed] [Google Scholar]
- 30.Kennedy DR, Gawron LS, Ju J, Liu W, Shen B, Beerman TA. Cancer Res. 2007;67:773. doi: 10.1158/0008-5472.CAN-06-2893. [DOI] [PubMed] [Google Scholar]
- 31.Kennedy DR, Ju J, Shen B, Beerman TA. Proc Natl Acad Sci USA. 2007;104:17632. doi: 10.1073/pnas.0708274104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beerman TA, Gawron LS, Shin S, Shen B, McHugh MM. Cancer Res. 2009;69:593. doi: 10.1158/0008-5472.CAN-08-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beerman TA, Gawron LS, Shen B, Kennedy DR. DNA Repair. 2014;21:165. doi: 10.1016/j.dnarep.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen B, Liu W, Nonaka K. Curr Med Chem. 2003;10:2317. doi: 10.2174/0929867033456701. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y, Yin M, Horsman GP, Huang SX, Shen B. J Antibiot. 2010;63:482. doi: 10.1038/ja.2010.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y, Yin M, Horsman GP, Shen B. J Nat Prod. 2010;74:420. doi: 10.1021/np100825y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zazopoulos E, Huang K, Staffa A, Liu W, Bachmann BO, Nonaka K, Ahlert J, Thorson JS, Shen B, Farnet CM. Nat Biotechnol. 2003;21:187. doi: 10.1038/nbt784. [DOI] [PubMed] [Google Scholar]
- 38.Liu W, Ahlert J, Gao Q, Wendt-Pienkowski E, Shen B, Thorson JS. Proc Natl Acad Sci USA. 2003;100:11959. doi: 10.1073/pnas.2034291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu W, Christenson SD, Standage S, Shen B. Science. 2002;297:1170. doi: 10.1126/science.1072110. [DOI] [PubMed] [Google Scholar]
- 40.Liu W, Nonaka K, Nie P, Zhang J, Christenson SD, Bae J, Van Lanen SG, Zazopoulos E, Farnet CM, Yang CF, Shen B. Chem Biol. 2005;12:293. doi: 10.1016/j.chembiol.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 41.Van Lanen SG, Oh TJ, Liu W, Wendt-Pienkowski E, Shen B. J Am Chem Soc. 2007;129:13082. doi: 10.1021/ja073275o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lohman JR, Huang SX, Horsman GP, Dilfer P, Huang T, Chen Y, Wendt-Pienkowski E, Shen B. Mol BioSyst. 2013;9:478. doi: 10.1039/c3mb25523a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGlinchey RP, Nett M, Moore BS. J Am Chem Soc. 2008;130:2406. doi: 10.1021/ja710488m. [DOI] [PubMed] [Google Scholar]
- 44.Ahlert J, Shepard E, Lomovskaya N, Zazopoulos E, Staffa A, Bachmann BO, Huang K, Fonstein L, Czisny A, Whitwam RE, Farnet CM, Thorson JS. Science. 2002;297:1173. doi: 10.1126/science.1072105. [DOI] [PubMed] [Google Scholar]
- 45.Gao Q, Thorson JS. FEMS Microbiol Lett. 2008;282:105. doi: 10.1111/j.1574-6968.2008.01112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Van Lanen SG, Ju J, Liu W, Dorrestein PC, Li W, Kelleher NL, Shen B. Proc Natl Acad Sci USA. 2008;105:1460. doi: 10.1073/pnas.0711625105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horsman GP, Chen Y, Thorson JS, Shen B. Proc Natl Acad Sci USA. 2010;107:11331. doi: 10.1073/pnas.1003442107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie P, Ma M, Rateb ME, Shaaban KA, Yu Z, Huang SX, Zhao LX, Zhu X, Yan Y, Peterson RM, Lohman JR, Yang D, Yin M, Rudolf JD, Jiang Y, Duan Y, Shen B. J Nat Prod. 2014;72:377. doi: 10.1021/np401063s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hindra, Huang T, Yang D, Rudolf JD, Xie P, Xie G, Teng Q, Lohman JR, Zhu X, Huang Y, Zhao L-X, Jiang Y, Duan Y, Shen B. J Nat Prod. 2014;77:2296. doi: 10.1021/np5006168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elespuru RK, Yarmolinsky MB. Environ Mutagen. 1979;1:65. doi: 10.1002/em.2860010113. [DOI] [PubMed] [Google Scholar]
- 51.Walsh CT, Fischbach MA. J Am Chem Soc. 2010;132:2469. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cox R, Piel J, Moore BS, Weissman KJ. Nat Prod Rep. 2009;26:1361. [Google Scholar]
- 53.Bachmann BO, Van Lanen SG, Baltz RH. J Ind Microbiol Biotechnol. 2014;41:175. doi: 10.1007/s10295-013-1389-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmitt EK, Moore CM, Krastel P, Petersen F. Curr Opin Chem Biol. 2011;15:497. doi: 10.1016/j.cbpa.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 55.Galm U, Shen B. Exp Opin Drug Dis. 2006;1:409. doi: 10.1517/17460441.1.5.409. [DOI] [PubMed] [Google Scholar]
- 56.Berdy J. J Antibiot. 2012;65:385. doi: 10.1038/ja.2012.27. [DOI] [PubMed] [Google Scholar]
- 57.The new enediyne producers with GenBank accession numbers (in the parentheses) for their respective genome, gene cluster, or enediyne PKS cassette. Actinomadura flavalba DSM 45200 (ARFO00000000), Actinomadura madurae (AY271660), Actinomadura verrucosospora (AY267372), Actinosynnema mirum DSM 43827 (CP001630), Amycolatopsis alba DSM 44262 (ARAF00000000), Amycolatopsis azurea DSM 43854 (ANMG00000000), Amycolatopsis balhimycina FH 1894 (ARBH00000000), Amycolatopsis decaplanina DSM 44594 (AOHO00000000), Amycolatopsis mediterranei U32 (CP002000), Amycolatopsis nigrescens CSC17Ta-90 (ARVW00000000), Amycolatopsis orientalis DSM 40040 (ASJB00000000), Bacillus sp. 2_A_57_CT2 (ACWD00000000), Catenulispora acidiphila DSM 44928 (CP001700), Cyanothece sp. PCC 7822 (CP002198), Haliangium ochraceum DSM 14365 (CP001804), Herpetosiphon aurantiacus DSM 785 (CP000875), Kitasatospora sp. CECT 4991 (AF546142), Methylomicrobium alcaliphilum (NC_016112), Microbispora sp. PTA-5024 (AWEV00000000), Micromonospora aurantiaca ATCC 27029 (CP002162), Micromonospora chersina (EF552206), Micromonospora echinospora (AF497482), Micromonospora lupini str. Lupac 08 (CAIE00000000), Micromonospora sp. 046/Eco11 (AF546153), Micromonospora sp. CNB394 (ARGW00000000), Micromonospora sp. L5 (CP002399), Nocardia brasiliensis ATCC 700358 (CP003876), Nostoc sp. PCC 7524 (CP003552), Saccharomonospora halophila 8 (AICX00000000), Saccharomonospora marina XMU15 (CM001439), Saccharothrix espanaensis (HE804045), Salinispora pacifica CNS055 (ARGH00000000), Salinispora arenicola CNS205 (CP000850), Salinispora pacifica CNS-143 (KC863955), Salinispora tropica CNB-440 (CP000667), Scytonema hofmanni PCC 7110 (ANNX00000000), Streptoalloteichus sp. ATCC 53650 (JX679499), Streptomyces carzinostaticus subsp. neocarzinostaticus (AY117439), Streptomyces citricolor (AH012470), Streptomyces clavuligerus plasmid (CM000914), Streptomyces ghanaensis (ABYA00000000), Streptomyces globisporus (AY048670), Streptomyces griseus NBRC 13350 (AP009493), Streptomyces kaniharaensis (AH012469), Streptomyces macromomyceticus (AF546155), Streptomyces sp. 100/Eco52 (AF546152), Streptomyces sp. 171/Eco105 (AF546154), Streptomyces sp. AA4 (ACEV00000000), Streptomyces sp. ATexAB-D23 (AREH00000000), Streptomyces sp. CNS615 (AQPE00000000), Streptomyces sp. CNT-179 (KC863954), Streptomyces sp. CNT302 (ARIM00000000), Streptomyces sp. Tü6071 (CM001165), Streptomyces sp. W007 (AGSW00000000), Streptomyces viridosporus T7A (AJFD00000000), Streptosporangium roseum DSM 43021 (CP001814), and Verrucosispora maris AB-18-032 (CP002638).