Abstract
Iron-sulfur (Fe-S) cluster containing proteins are utilized in almost every biochemical pathway. The unique redox and coordination chemistry associated with the cofactor allows these proteins to participate in a diverse set of reactions, including electron transfer, enzyme catalysis, DNA synthesis and signaling within several pathways. Due to the high reactivity of the metal, it is not surprising that biological Fe-S cluster assembly is tightly regulated within cells. In yeast, the major assembly pathway for Fe-S clusters is the mitochondrial ISC pathway. Yeast Fe-S cluster assembly is accomplished using the scaffold protein (Isu1) as the molecular foundation, with assistance from the cysteine desulfurase (Nfs1) to provide sulfur, the accessory protein (Isd11) to regulate Nfs1 activity, the yeast frataxin homologue (Yfh1) to regulate Nfs1 activity and participate in Isu1 Fe loading possibly as a chaperone, and the ferredoxin (Yah1) to provide reducing equivalents for assembly. In this report, we utilize calorimetric and spectroscopic methods to provide molecular insight into how wt-Isu1 from S. cerevisiae becomes loaded with iron. Isothermal Titration Calorimetry (ITC) and an iron competition binding assay were developed to characterize the energetics of protein Fe(II) binding. Differential Scanning Calorimetry (DSC) was used to identify thermodynamic characteristics of the protein in the apo state or under iron loaded conditions. Finally, X-ray Absorption Spectroscopy (XAS) was used to characterize the electronic and structural properties of Fe(II) bound to Isu1. Current data are compared to our previous characterization of the D37A Isu1 mutant, and these suggest that when Isu1 binds Fe(II) in a manner not perturbed by the D37A substitution, and that metal binding occurs at a site distinct from the cysteine rich active site in the protein.
Keywords: Iron, Fe-S Cluster Biosynthesis, ISU Scaffold Protein, Iron Binding
Introduction
Iron-sulfur cluster containing proteins are utilized in all domains of life and found ubiquitously in nearly every cellular biochemical pathway (Johnson et al. 2005). Given the chemical diversity these clusters can impart to proteins, it is not surprising that Fe-S cluster containing proteins participate in a variety of biochemical reactions associated with catalysis, redox activity and even sensing of iron or oxidants (Stehling and Lill 2013). Elemental iron and sulfur, the two substrates required to form Fe-S clusters, are inherently reactive and toxic in high abundance, therefore cofactor assembly and utilization are tightly controlled within cells (Rouault 2014; Stehling et al. 2014). In eukaryotes, the mitochondrial Iron-Sulfur Cluster (ISC) pathway is the primary source of Fe-S clusters, and cluster biosynthesis is orchestrated directly on the ISU scaffold protein (Craig et al. 1999; Garland et al. 1999; Stehling and Lill 2013). In yeast, the Isu1 scaffold protein works in close association with other protein partners to control Fe-S cluster production and cofactor utilization (Lill et al. 2012), however the selective delivery of ferrous iron to Isu1 in order to accomplish cofactor assembly is a poorly understood process.
Molecular factors dictate the functionality of yeast Isu1 and its participation in cluster assembly and transfer. The active site of Isu1 contains 3 Cys residues that are completely conserved within Isu orthologs (Garland et al. 1999; Bridwell-Rabb et al. 2014). Mutation of Isu1 Asp 37 to Ala (numbered according to the position on the mature processed protein) precludes cluster release from the protein, imparting it with a greater stability for cluster retention than its wt- counterpart (Raulfs et al. 2008). ISU has been shown to exist in multiple conformational states with varying degrees of fold stability, associated with substrate binding and the cluster loaded/release status of the molecule (Kim et al. 2009; Cook et al. 2010). Finally, binding of yeast Isu1 to its protein partners at different stages in the assembly pathway drive the activity of the scaffold protein resulting in Fe-S cluster assembly or eventually in Fe-S cluster release (Lill 2009). Despite the large amount of information available related to the structure and function of Isu1, steps involved in iron delivery and loading onto Isu1 still need to be clarified.
In this report, we provide a comprehensive characterization of the binding affinity as well as a thermodynamic, structural and electronic characterization of Fe(II) bound to wt-Isu1 from S. cerevisiae. Isothermal titration calorimetry (ITC) was used to measure the affinity of Fe(II) binding to the Isu1 protein maintained in a homogeneous state. However, within a cell, protein metal binding is accomplished in the presence of additional metal-binding biomolecules that surely compete with the protein for metal acquisition. To simulate this environment, we developed a Fe(II) competition binding assay that allowed us to measure a protein’s metal binding affinity under more physiologically relevant competition conditions. Differential scanning calorimetry (DSC) was used to determine thermodynamic factors that dictate protein stability under apo and holo conditions. Finally, X-ray absorption spectroscopy (XAS) was used to characterize the structure and electronic properties of Fe(II) bound to Isu1. We have previously reported similar analysis of the D37A yeast Isu1 mutant (Cook et al. 2010), however this report highlights similarities with regards to Fe binding, serves as an initial identification of the yeast wt-Isu1 metal binding site and serves as a starting point for comparison of Fe binding by other ISU orthologs.
Materials and Methods
Protein overexpression and isolation
A plasmid (pET21b-mIsu1) containing the coding sequence for the mature form of the wild type (WT) Sc Isu1 gene was transformed into BL21 codon plus RIL cells (Stratagene) for protein expression. Cells were grown at 37°C in Luria-Bertani broth containing Ampicillin (0.1 μg/μL) and Chloramphenicol (0.05 μg/μL) with shaking speed set at 250 rpm. At OD600 of 0.6, cells were induced for protein expression by the addition of 0.8 mM IPTG. Cells were induced at a temperature of 37°C, over a period of 3 hrs following which the cells were harvested by centrifugation and stored at -80°C until lysis. Steps for protein isolation and quality control follow exactly our published protocol for the D37A-Isu1 mutant (Cook et al. 2010).
Isothermal titration calorimetric (ITC) studies
The Fe(II) binding properties of wt-Isu1 were tested using ITC. Protein and aqueous ferrous ammonium sulfate solutions were prepared anaerobically using a Schlenk line degassed buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl and 5 mM β-ME. Both protein and metal solutions were prepared in identical buffer solutions to provide matched conditions. ITC data were collected using a VP-ITC microcalorimeter (MicroCal Inc.). The syringe contained a 1 mM aqueous ferrous iron solution was titrated into the adiabatic cell of 2.2 mL volume containing 50 μM of wt-Isu1. Data collection parameters followed a 2 μl initial injection of titrant solution with 29 additional injections of 10 μl titrant, performed using a 250 μl syringe. Injections were spaced at 10 minutes each and data were collected at 30°C with stirring speed set at 372 rpm. Data analysis was performed using the Origin 7.0 software provided by MicroCal and was fit to a two-site binding model, which calculates the stoichiometry of binding and dissociation constant parameters. All experiments were performed in duplicate on independent samples to ensure data reproducibility.
Fe(II) competition binding assay
The iron binding affinity of wt-Isu1 was determined by competition titrations in the presence of mag-fura-2 (Molecular Probes), a well-known metal chelator. Mag-fura-2 forms a 1:1 complex with most divalent cations and has an absorbance maximum at 366 nm in its apo state that shifts to a 325 nm signal upon metal binding. Changes in mag-fura-2 were monitored using a Shimadzu UV-1800 spectrophotometer located within an anaerobic box using a 1 cm pathlength quartz cuvette. Data were collected at room temperature. Mag-fura-2 was dissolved in high purity MiliQ water and stored at −80°C in 10 μL aliquots at a 2 mM stock concentration. Protein samples and ferrous ammonium sulfate solutions were prepared in buffer containing 20 mM HEPES (pH 7.5) and 150 mM NaCl. Prior to use, protein samples were reduced with 5 mM TCEP. Reagents and buffers were allowed to incubate at 4°C overnight within an anaerobic chamber before data collection. As a control, the initial starting solution contained between 2 to 5 μM mag-fura-2 in 1 ml of reaction buffer and aliquots of Fe(II), at progressive increments, were titrated until binding saturation was achieved. Each absorption spectrum was collected between 200–800 nm. For dilution correction, experiments were performed in the identical manner using apo buffer. For protein titrations, 3 to 15 μM protein aliquots were first mixed with mag-fura-2 before the addition of metal. Initial metal free mag-fura-2 concentrations were confirmed using the molecules’ molar absorptivity (ε) value of 29900 M−1cm−1 at 366 nm (Walkup and Imperiali 1997), while the protein concentration was determined using a Direct Detect IR system (BioRad). Binding data were fit using the program DYNAFIT (Kuzmic 1996) with both one and two-site binding models.
Differential scanning calorimetry (DSC) studies
Thermal stability of wt-Isu1 in the absence and presence of iron were evaluated using a differential scanning calorimeter (TA Instruments) located within a Coy anaerobic chamber. Protein and iron solutions were prepared anaerobically in 20 mM HEPES (pH 7.5), 150 mM NaCl and 5 mM β-Me. Buffer was extensively degassed prior to sample preparation and loading. Protein concentrations at ca. 1.5 mg/mL were used and iron solution was added to obtain samples of a 1:2 protein:Fe ratio, made up to a final volume of 600 μL. Scans were collected at a rate of 2°C per min over a temperature range of 10 to 90°C, while pressure was maintained at an excess of 2.95 atm throughout the scan period. Data obtained were analyzed using the Nano Analyze software provided by TA Instruments. Data was baseline subtracted using control runs of buffer alone or buffer with iron at concentrations that matched the amount within the protein samples. Peak deconvolution was achieved using the two state scaled mathematical model that incorporates the factor Aw, which accounts for inaccuracies in protein concentration upon denaturation. Data presented represents an average of two independent runs.
X-ray absorption spectroscopy (XAS)
XAS was used to study the electronic properties and ligand coordination geometry of iron bound to wt-Isu1. Independent reproducible samples of iron loaded protein were prepared within an anaerobic chamber (Coy labs) at a metal:protein stoichiometry of 2:1. Samples were prepared at ~0.5 mM final protein concentrations in 20mM HEPES (pH: 7.5), 250 mM NaBr and 5 mM β-Me. Samples were diluted with 30% glycerol and loaded into Lucite cells pre-wrapped with Kapton tape, flash frozen and then stored in liquid nitrogen until data collection. XAS data were collected at the National Synchrotron Light Source on beamline X3B, equipped with a Si[110] crystal monochromator and Ni plated focusing/harmonic rejection mirror. Samples were maintained at 24 K using a helium Displex cryostat. Protein fluorescence excitation spectra were collected using 31-element Ge solid-state array detectors (Canberra). Spectral data analysis followed protocols previously outlined in detail (Cook et al. 2006).
Results and Discussion
Fe(II) binding characteristics
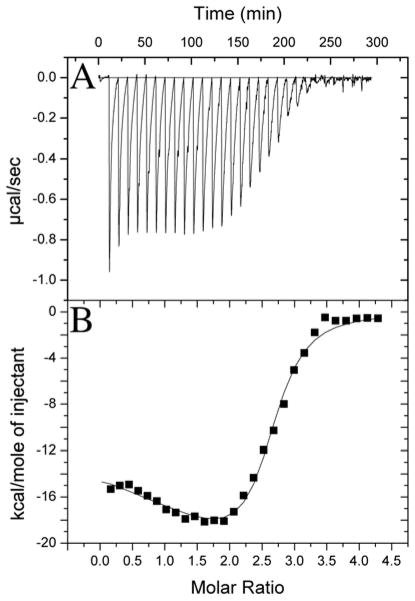
An early stage in yeast mitochondrial Fe-S cluster biosynthesis involves loading the substrate Fe(II) onto Isu1. To gain insight into the energetic factors driving this process, isothermal titration calorimetry was used to measure the metal:protein stoichiometry and metal binding affinity of iron onto S. cerevisiae wt-Isu1. Samples were prepared anaerobically and buffers extensively degassed prior to use. Å 1 mM solution of aqueous iron was titrated into a 50 μM Isu1 sample, and subsequent heat release profiles were converted to binding isotherm data, plotted for energy release relative to the molar ratio of protein to iron. These data produced a biphasic binding profile (Figure 1). ITC binding isotherms were best simulated using a 2 binding site model. Results showed binding affinities of Kd1 = 0.98 ± 0.75 μM and Kd2 = 18 ± 7 nM, and relative binding stoichiometries of 1.61 ± 0.16 and 0.60 ± 0.16, respectively. Following the assumption that iron binding drives these heat release events, these data suggest the S. cerevisiae wt-Isu1 monomer can bind up to two Fe(II) atoms within the micro- to sub-micromolar binding affinity range.
Figure 1. Isothermal titration calorimetric data for Isu1 Fe(II) loading.
(A) Raw isothermal titration calorimetry data are provided along with (B) the binding isotherm for protein ferrous iron binding in this figure. Data were collected anaerobically at 30°C in 20 mM HEPES (pH 7.5), 150 mM NaCl and 5 mM β-Me. Spacing between injections was 10 minutes. Syringe stirring speed was kept at 372 rpm.
The Fe(II) binding characteristics for wt-Isu1 are very similar to values published for the D37A Isu1 mutant from S. cerevisiae, as well as for other ISU orthologs. Ferrous iron binding isotherms, measured by ITC for the S. cerevisiae D37A Isu1 mutant, were best fit using a 2 binding site model with a Kd1 of 1.64 ± 1.68 μM and Kd2 of 6 ± 4 nM and Fe:protein binding stoichiometries of 1.19 ± 0.66 and 0.97 ± 0.31, respectively (Cook et al. 2010). Fe(ll) binding results for wt- and D37A-Isu1 are very similar and within error bars. Differences between wt- and D37A-Isu1 Fe(II) binding affinities may reflect subtle changes in the buffer conditions between experiments or may reflect subtle variations in metal binding between the two proteins. Published Fe(II) binding affinities for human and bacterial ISU orthologs were fit using only single site binding models with one iron atom at Kd’s of 2.0 ± 0.2 μM and 2.7 μM, respectively (Foster et al. 2000; Bertini et al. 2003). Collectively, these results suggest that ISU orthologs bind Fe(II) within the micro to sub-micromolar range. However, these values were measured collectively in the absence of protein partners and without competition from additional Fe(II) binding molecules, leaving open questions regarding the validity of the values with respect to in vivo systems.
Fe(II) competition binding assay
To test Isu1 iron binding under conditions that more closely represent those seen in vivo, we developed a Fe(II) binding competition assay for the protein in the presence of the small divalent cation chelator mag-fura-2 (Molecular Probes). This assay was performed within an anaerobic chamber to stabilize the ferrous iron solution. Spectroscopic characteristics regarding mag-fura-2 metal binding are well established (Simons 1993). Mag-fura-2 has a characteristic UV-Vis absorbance at 325 nm in its holo state and an absorbance maximum at 366 nm in the apo state. Progressive loading of Fe(II) onto mag-fura-2 showed an exponential decay profile at A366 vs [Fe(II)] (Figure 2A-Inset). The A366 decay profile could easily be fit using Equation 1 using the Dynafit program for enzyme kinetic data analysis (Kuzmic 1996). Equation 2 follows the known
Figure 2. (A) Spectra showing the Fe(II)-binding to mag-fura-2 and (B) Fe(II)-binding under conditions of Isu1/mag-fura-2 competition for metal are displayed.
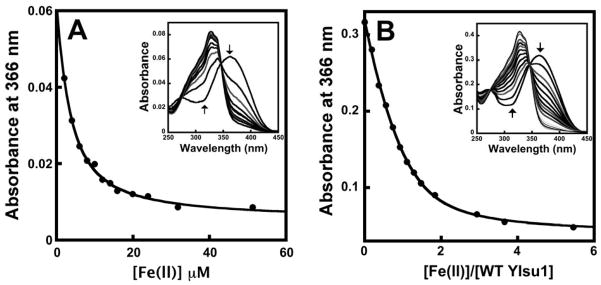
For (A), absorbance changes measured at 366 nm when increasing [Fe(II)] was added to 2 μM of apo mag-fura-2 are displayed. Inset shows absorbance spectral changes obtained during the titration. Arrows denote the direction of the absorbance changes upon increasing [Fe(II)]. For (B), absorbance changes measured at 366 nm when increasing [Fe(II)] was added to a mixture of apo mag-fura-2 and apo Isu1 are displayed. Inset shows absorbance spectral changes obtained during the titration of Fe(II) to mixture of 4.5 μM of mag-fura-2 and 3.7 μM of Isu1. Arrows denote the direction of the absorbance changes with increasing [Fe(II)].
| Eqn 1 |
1:1 metal:ligand stoichiometric profile for Fe binding to mag-fura-2, where M is ferrous metal, I is the mag-fura-2 small molecule indicator and Kd1 is the dissociation constant for the reaction. Simulations of the representative data in Figure 2A resulted in a calculated Kd1 of 2.05 (± 0.07) μM for iron binding to mag-fura-2. These data serve as a control for protein competition experiments below.
The mag-fura-2 Fe(II) binding constant was used to determine the Fe(II) binding characteristics of apo-Isu-1 under competition conditions. The mag-fura-2 to Isu1 ratio was varied between 2.5:1 to 1:1 to 1:2.5, and in each case the spectra showed a decay trend for the A366 apo mag-fura-2 feature (Figure 2B). This decay spectrum could be simulated using Dynafit by coupling Equation 1 for indicator iron binding with Equations 2 and 3 for the two metal binding site model for monomeric wt-Isu1. In Equations 2 and 3, M again refers to ferrous
| Eqn 2 |
| Eqn 3 |
metal, P refers to the protein, and Kd2 and Kd3 are dissociation constants for site 1 and the site 2 on the protein, respectively. Spectra were collected within an anaerobic chamber to stabilize ferrous iron. Although TCEP was added to the solution, there was no effect of reductant on the indicator Fe(II) binding spectrum. In addition, the presence of apo-Isu1 had no impact on the A366 spectral signal from apo-mag-fura-2. The affinity constants (Kd2 and Kd3) measured for Isu1-Fe(II) binding were 2.38 ± 0.85 μM and 4.55 ± 2.69 μM, in close agreement to the major binding constant measured by ITC. These data show wt-Isu1 can compete with mag-fura-2 for binding Fe(II) and that they have a similar affinity for metal. These data confirm that the micromolar Isu1-metal binding affinity measured by ITC, is also a relevant value for protein metal affinity under competition conditions. Given a Fe(II) concentration of 150 μM within the mitochondria(Holmes-Hampton et al. 2010), Isu1 binding affinities measured in this report are within the range to promote Fe(II) loading of the protein within the organelle. These affinities are also within a range where Isu1 could receive metal from an iron chaperone that binds Fe(II) at low μM affinity.
DSC characteristics of apo and holo wt-Isu1
Differential scanning calorimetry was used to characterize the thermal stability and molecular speciation of wt-Isu1 under apo and holo (metal preloaded) conditions. All DSC thermal denaturation curves were measured under anaerobic conditions. The apo wt-Isu1 DSC thermogram (Figure 3) was best simulated using a two state model. The approximation to unity of enthalpy values i.e., ΔHcal (kJ/mol), obtained upon peak integration, relative to the Van’t Hoff enthalpy ΔHvH (kJ/mol), obtained upon model fitting, is used as an indicator of validity of the model to the applied system (Grzesiak et al. 2000). Melting temps (TM) obtained for the two states in the apo protein were 47.34 ± 0.38°C and 51.36 ± 0.26°C (Table 1). The DSC thermogram for the Fe(II) loaded wt-Isu1 however shows a single melting transition, with a melting temp of 54.65 ± 0.06 °C (Figure 3, Table 1), a value slightly higher than that seen for the apo-protein.
Figure 3. Differential scanning calorimetric thermal denaturation curves for apo and holo S. cerevisiae wt-Isu1.
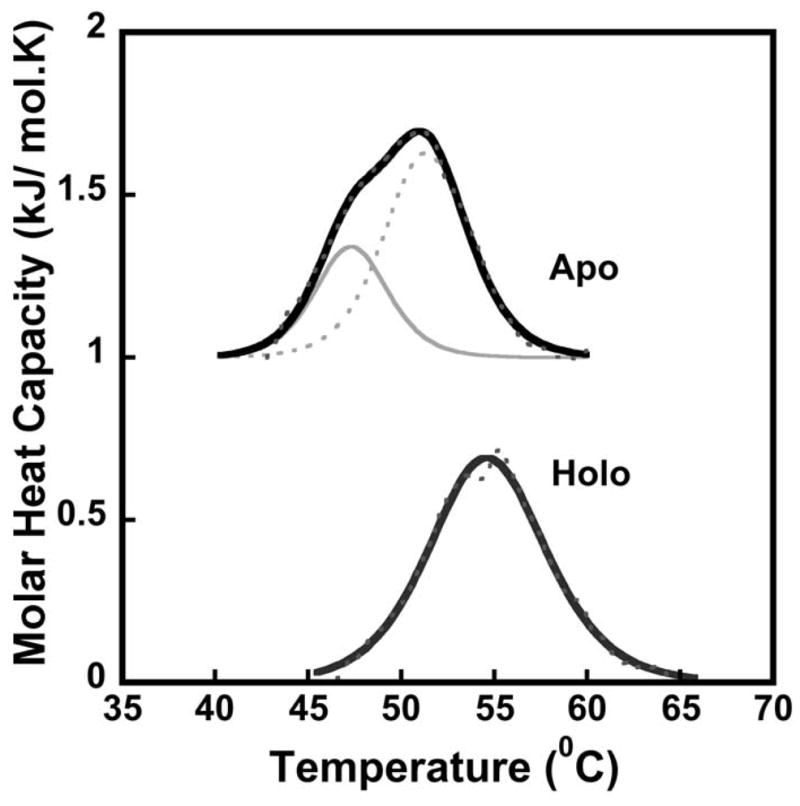
Data were collected between 10–90°C at a scan rate of 2°C with excess pressure of 2.95 atm. Holo protein was pre-loaded with 2 equivalents of Fe(ll) and apo protein curves were offset for clarity. Black solid lines represent the fit to raw data, black dashed lines represent raw data, and grey solid & dashed lines represent the deconvolution of raw data using the two-state scaled model.
Table 1.
Thermodynamic parameters for melting temp determination of apo & holo WT Sc Isu1.
| State | Tm (1) (°C) | Tm(2) (°C) | Tmax (°C) | ΔHvH (kJ/mol) | ΔHcal (kJ/mol) | ΔHcal/ ΔHvH | ΔScal (kJ/mol•K) |
|---|---|---|---|---|---|---|---|
| Apo | 47.3 ± 0.4 | 51.4 ± 0.3 | 51.3 | 8.40 | 7.94 | 0.95 | 0.025 |
| Holo | 54.7 ± 0.1 | --- | 55.3 | 5.76 | 5.10 | 0.90 | 0.016 |
Tm(1 & 2) represent temperature maximum for each transition fit to the raw data. ΔHvH represents Van't Hoff enthalpy (total enthalpy under each fit obtained upon correction with scaling factor Aw). ΔHcal, ΔScal and Tmax represent enthalpy, entropy and maximum temperatures respectively obtained from peak integration after baseline correction of the raw data. Fitting parameters obtained from simulations of at least 2 independent data sets.
The two transitions seen for the apo protein may indicate that the protein exists in two states, while the single transition suggests that the holo protein is in a single state. Two species (unstructured and structured forms) were characterized in solution for the bacterial protein using NMR spectroscopy by the Markley lab (Kim et al. 2009) and using fluorimetry by our laboratory (Cook et al. 2010). The subtle thermal stabilization due to Fe(II) binding suggests metal may bind in a manner that drives the protein fold to a more structured state. A conformational shift of this nature may drive Fe-S cluster formation of Fe-S cluster release following successful bioassembly.
X-ray absorption near edge spectroscopy (XANES) analysis
X-ray absorption spectroscopy was used to characterize the electronic and structural characteristics of Fe(II) bound to wt-Isu1. The XANES region of an XAS spectrum provides direct insight into the electronic and spin state of iron within the sample, as well as the symmetry of the metal-ligand coordination environment. The XANES spectrum for iron bound to wt-Isu1 is consistent with metal bound in the Fe(II) redox state (first inflection edge energy of 7123.0 eV), as compared to Fe(II) and Fe(III) controls (Figure 4). Spectral features in the wt-Isu1 XANES closely resemble those observed for D37A Isu1 (Cook et al. 2010). Fitting analysis for the 1s→3d feature in the XANES pre-edge region shows transitions at 7111.5 and 7114.0 eV, and a transition area of 15.4 × 10−2 eV, consistent with Fe(II) bound to the protein being high spin and contained within a pseudo-octahedral coordination environment, again in close agreement with the D37A Isu1 mutant data (Cook et al. 2010). Based on our XANES analysis, we know that iron bound to wt-Isu1 in a high spin ferrous state is in a pseudosymmetric octahedral coordination geometry. Fe(II) bound to both wt- and D37A Isu1 share common electronic and structural properties, indicating the Asp 37 mutation has minimal impact on the metal binding to Isu1.
Figure 4. XANES spectra comparison of Fe(ll) loaded wt-Isu1 from S. cerevisiae with Fe(II) and Fe(III) model compounds.
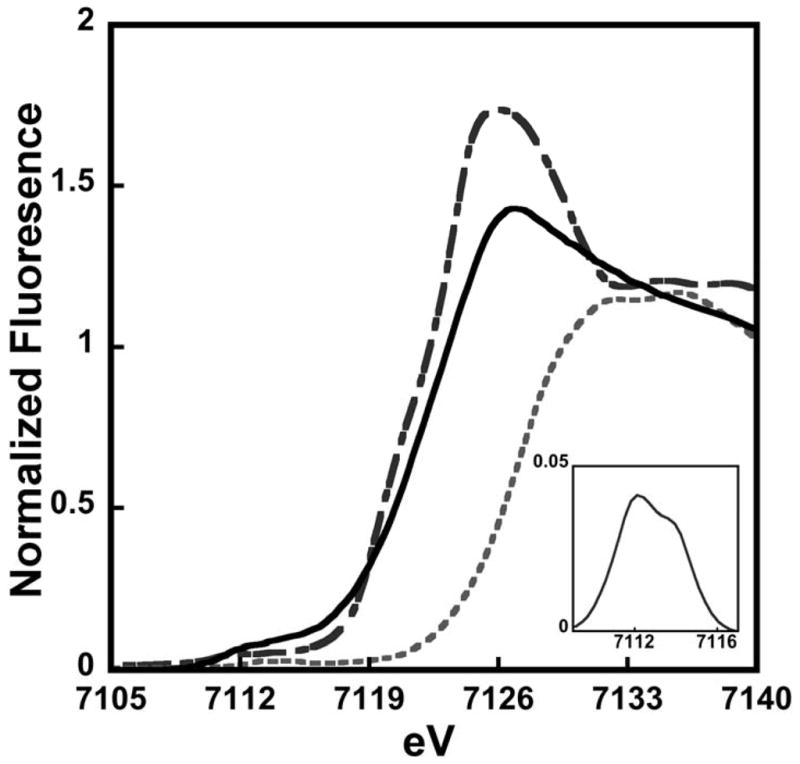
Spectrum for wt-Isu1 shown in black solid line is compared with ferrous ammonium sulfate (long dash) and ferric ammonium sulfate (short dash). Inset: Baseline subtracted 1s→3d feature for Fe(II) bound to wt-Isu1.
Extended X-ray absorption fine structure (EXAFS) spectroscopic analysis
The EXAFS region of a XAS spectrum provides high resolution metrical details of the ligand coordination environment for metals bound to a protein (Bencze et al. 2007). EXAFS was used to characterize the Fe(II) bound to wt-Isu1 to clarify details of the protein’s metal binding site. Fourier transforms of the EXAFS provide a radial distribution function of the ligand environment surrounding the metal. Both the Fe(II) EXAFS and the Fourier transform for iron bound wt-Isu1 are shown in Figure 5. Following simulations of the data, fitting parameters that provided the best fit simulation are listed in Table 2. The nearest neighbor Fe(II) ligand environment showed two distinct scattering environments consisting of 6 Oxygen/Nitrogen based ligands centered at 1.97 Å and 2.12 Å. There is no evidence for any sulfur scattering in this data. Long-range scattering, observed in this data, were constructed with only carbon atoms at average bond lengths of 2.85 Å, 3.39 Å and 4.02 Å.
Figure 5. EXAFS (A) and Fourier transform of the EXAFS (B) for Fe(II) bound to wt-Isu1.
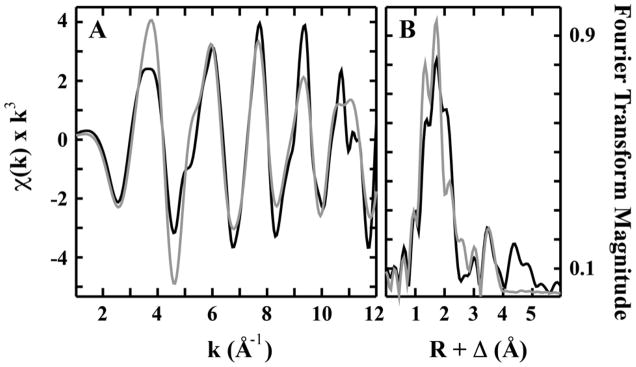
Raw data is shown in back while simulated spectra are in grey.
Table 2.
Summary of the best fit EXAFS simulation analysis for 2 iron atoms bound to wt-Isu1 from S. cerevisiae.
| Fe-Nearest Neighbor Ligands a | Fe-Long Range Ligands a | |||||||
|---|---|---|---|---|---|---|---|---|
| Atom b | R(Å) c | C.N. d | σ2 e | Atom b | R(Å) c | C.N. d | σ2 e | F’ f |
| O/N | 1.97 | 2 | 1.80 | C | 2.85 | 2 | 3.90 | 0.62 |
| O/N | 2.12 | 4 | 1.09 | C | 3.39 | 2 | 5.26 | |
| C | 4.02 | 4 | 5.13 | |||||
Independent metal-ligand scattering environment
Scattering atoms: O (Oxygen), N (Nitrogen), C (Carbon)
Metal-ligand bond length
Metal-ligand coordination number
Debye-Waller factor given in A2 × 103
Number of degrees of freedom weighted mean square deviation between data and fit
The coordination information for iron bound to wt-Isu1 closely matches the structure determined for Fe(II) bound to the D37A Isu1 mutant (Cook et al. 2010). Similar to the current data, Fe(II) bound to the D37A mutant showed a nearest neighbor ligand environment consisting only of 6 Oxygen/Nitrogen based ligands. In both samples, there is no indication of any sulfur scattering, a fact that is surprising since one model for iron delivery to Isu suggests metal is delivered directly to the Cysteine rich protein active site. These data indicate that this is not the case and instead there must exist an initial metal binding site on Isu1 distinct from the Cys rich active site that is used during Fe-S cluster assembly. The Fe(II)-nearest neighbor ligand bond lengths for wt-Isu1 (O/N ligand environments at 1.97 Å and 2.12 Å) are within error bars of the technique relative to values obtained for the D37A mutant (at 2.01 Å and 2.14 Å). In both cases, only long range Fe••• Carbon scattering was observed. Combined, these structural studies indicate that wt- and D37A-Isu1 from S. cerevisiae bind iron at a 6 coordinate metal binding site that has pseudo-octahedral coordination geometry with only O and N based ligands. These results suggest that metal, bound as high-spin Fe(II), must be coordinated at a site distinct from the Cys rich Fe-S cluster assembly site on the protein.
Summary
Mitochondrial Fe-S cluster assembly pathways are universally conserved in Eukaryotes and the obvious importance of bioassembly is highlighted by the fact that Fe-S clusters are utilized in nearly every other cellular pathway. In yeast, assembly is tightly controlled through the highly coordinated activity of a set of protein partners that form a complex made up of the Isu1 scaffold protein, the Nfs1 cysteine desulfurase, the Isd11 accessory protein, the Yah1 ferredoxin and the Yfh1 yeast frataxin homologue (Stehling et al. 2014). Although intricacies of the mechanism for coordinated Fe-S cluster assembly are still not clear, delivery of iron to Isu1 is an absolute requirement in order for the process to proceed. Iron delivery most likely occurs in a chaperone assisted manner supported in some way by the protein frataxin (Bencze et al. 2006), but to understand the metal delivery process it is useful to have a biophysical characterization of the Isu1 iron binding process. This report promotes that objective.
In this report, we provide a complete characterization of the yeast wt-Isu1 Fe(II) binding under homo and heterogeneous environmental conditions, with an identification of the thermodynamic stability of the protein and clarification of the bound metal structure for the protein. Comparison of the wt-Isu1 iron binding characteristics with the D37A mutant suggest that while this mutation has a dramatic impact on Isu1 Fe-S cluster release, there is no real perturbation in the initial metal binding activity of the protein. DSC data indicate that while apo-wt-Isu1 exists in two thermally stable species, the holoprotein is more homogeneous in its nature and has a higher molecular thermodynamic stability. Structural details of the protein’s Fe(II) binding site indicate that wt-Isu1 binds iron in a six coordinate O/N ligand binding environment, possibly in a location rich in acidic/basic surface exposed residues. The lack of any cysteine ligation indicates the initial metal binding site on the protein is distinct from the protein’s cluster assembly site. In the bacterial IscU/IscS co-crystal structure, the Isu1 ortholog binds to the cysteine desulfurase in a manner that orients the Cys rich active site to be in close proximity to the persulfide delivery loop of the cysteine desulfurase (Shi et al. 2010). Given the potential for toxicity of free sulfide within a cell, tight structural coupling of the two proteins seems reasonable to preserve the innocuousness and efficiency of the pathway (Bridwell-Rabb et al. 2014). In this binding orientation, the highly conserved ISU C-terminus is solution exposed and available for possible metal delivery by a protein-binding partner. The ISU C-terminal helix is therefore a possible candidate for the initial Fe(II) binding site on the protein (Shi et al. 2010). Given the large number of conserved surface exposed acidic and basic residues in this region, it is easy to envision that the Isu1 Fe(II) initial binding site could be located on this secondary structural region.
Acknowledgments
This work was supported by funds for A.V.R. from the American Heart Association and the Friedreich’s Ataxia Research Alliance (12PRE11720005), and funds from the National Institutes of Health for A.D. (DK53953) and T.L.S. (DK068139). Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource (SSRL). SSRL is a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the NIH, National Center for Research Resources, Biomedical Technology Program.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- Bencze KZ, Kondapalli KC, Cook JD, McMahon S, Millan-Pacheco C, Pastor N, Stemmler TL. The structure and function of frataxin. Crit Rev Biochem Mol Biol. 2006;41(5):269–291. doi: 10.1080/10409230600846058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencze KZ, Kondapalli KC, Stemmler TL. In: X-Ray Absorption Spectroscopy. Applications of Physical Methods in Inorganic and Bioinorganic Chemistry: Handbook, Encyclopedia of Inorganic Chemistry. 2. Scott RA, Lukehart CM, editors. Chichester, UK: John Wiley & Sons,LTD; 2007. pp. 513–528. [Google Scholar]
- Bertini I, Cowan JA, Del Bianco C, Luchinat C, Mansy SS. Thermotoga maritima IscU. Structural characterization and dynamics of a new class of metallochaperone. J Mol Biol. 2003;331(4):907–924. doi: 10.1016/S0022-2836(03)00768-X. [DOI] [PubMed] [Google Scholar]
- Bridwell-Rabb J, Fox NG, Tsai CL, Winn AM, Barondeau DP. Human frataxin activates Fe-S cluster biosynthesis by facilitating sulfur transfer chemistry. Biochemistry. 2014;53(30):4904–4913. doi: 10.1021/bi500532e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook JD, Bencze KZ, Jankovic AD, Crater AK, Busch CN, Bradley PB, Stemmler AJ, Spaller MR, Stemmler TL. Monomeric yeast frataxin is an iron-binding protein. Biochemistry. 2006;45(25):7767–7777. doi: 10.1021/bi060424r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook JD, Kondapalli KC, Rawat S, Childs WC, Murugesan Y, Dancis A, Stemmler TL. Molecular details of the yeast frataxin-Isu1 interaction during mitochondrial Fe-S cluster assembly. Biochemistry. 2010;49(40):8756–8765. doi: 10.1021/bi1008613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig EA, Voisine C, Schilke B. Mitochondrial iron metabolism in the yeast Saccharomyces cerevisiae. Biol Chem. 1999;380(10):1167–1173. doi: 10.1515/BC.1999.148. [DOI] [PubMed] [Google Scholar]
- Foster MW, Mansy SS, Hwang J, Penner-Hahn JE, Surerus KK, Cowan JA. A Mutant Human IscU Protein Contains a Stable [2Fe-2S]2+ Center of Possible Functional Significance. J Am Chem Soc. 2000;122:6805–6806. doi: 10.1021/ja000800+. [DOI] [Google Scholar]
- Garland SA, Hoff K, Vickery LE, Culotta VC. Saccharomyces cerevisiae ISU1 and ISU2: members of a well-conserved gene family for iron-sulfur cluster assembly. J Mol Biol. 1999;294(4):897–907. doi: 10.1006/jmbi.1999.3294. [DOI] [PubMed] [Google Scholar]
- Grzesiak A, Helland R, Smalas AO, Krowarsch D, Dadlez M, Otlewski J. Substitutions at the P(1) position in BPTI strongly affect the association energy with serine proteinases. J Mol Biol. 2000;301(1):205–217. doi: 10.1006/jmbi.2000.3935. [DOI] [PubMed] [Google Scholar]
- Holmes-Hampton GP, Miao R, Garber Morales J, Guo Y, Munck E, Lindahl PA. A nonheme high-spin ferrous pool in mitochondria isolated from fermenting Saccharomyces cerevisiae. Biochemistry. 2010;49(19):4227–4234. doi: 10.1021/bi1001823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- Kim JH, Fuzery AK, Tonelli M, Ta DT, Westler WM, Vickery LE, Markley JL. Structure and dynamics of the iron-sulfur cluster assembly scaffold protein IscU and its interaction with the cochaperone HscB. Biochemistry. 2009;48(26):6062–6071. doi: 10.1021/bi9002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: Application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460(7257):831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- Lill R, Hoffmann B, Molik S, Pierik AJ, Rietzschel N, Stehling O, Uzarska MA, Webert H, Wilbrecht C, Muhlenhoff U. The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism. Biochim Biophys Acta. 2012;1823(9):1491–1508. doi: 10.1016/j.bbamcr.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Raulfs EC, O'Carroll IP, Dos Santos PC, Unciuleac MC, Dean DR. In vivo iron-sulfur cluster formation. Proc Natl Acad Sci U S A. 2008;105(25):8591–8596. doi: 10.1073/pnas.0803173105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault TA. Mammalian iron-sulphur proteins: novel insights into biogenesis and function. Nat Rev Mol Cell Biol. 2014 doi: 10.1038/nrm3909. E-pub ahead of print. [DOI] [PubMed] [Google Scholar]
- Shi R, Proteau A, Villarroya M, Moukadiri I, Zhang L, Trempe JF, Matte A, Armengod ME, Cygler M. Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions. PLoS Biol. 2010;8(4):1–18. doi: 10.1371/journal.pbio.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons TJ. Measurement of Zn(II) ion concentration with fluorescence probe magfura-2. J Biochem Biophys Methods. 1993;27:25–37. doi: 10.1016/0165-022X(93)90065-V. [DOI] [PubMed] [Google Scholar]
- Stehling O, Lill R. The role of mitochondria in cellular iron-sulfur protein biogenesis: mechanisms, connected processes, and diseases. Cold Spring Harb Perspect Biol. 2013;5(8):1–17. doi: 10.1101/cshperspect.a011312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehling O, Wilbrecht C, Lill R. Mitochondrial iron-sulfur protein biogenesis and human disease. Biochimie. 2014;100:61–77. doi: 10.1016/j.biochi.2014.01.010. [DOI] [PubMed] [Google Scholar]
- Walkup GK, Imperiali B. Fluorescent chemosensors for divalent zinc based on zinc finger domains. J Am Chem Soc. 1997;119:3443–3450. doi: 10.1021/ja9642121. [DOI] [Google Scholar]