Abstract
Improved understanding of the multilayer regulation of the human genome has led to a greater appreciation of environmental, nutritional, and epigenetic risk factors for human disease. Chromatin remodeling, histone tail modifications, and DNA methylation are dynamic epigenetic changes responsive to external stimuli. Careful interpretation can provide insights for actionable public health through collaboration between population and basic scientists and through integration of multiple data sources. We review key findings in environmental epigenetics both in human population studies and in animal models, and discuss the implications of these results for risk assessment and public health protection. To ultimately succeed in identifying epigenetic mechanisms leading to complex phenotypes and disease, researchers must integrate the various animal models, human clinical approaches, and human population approaches while paying attention to life-stage sensitivity, to generate effective prescriptions for human health evaluation and disease prevention.
Keywords: environmental epigenetics, life-stage exposures, integrated bioinformatics, risk assessment, molecular epidemiology
INTRODUCTION
The field of epigenetics is primed for translation of epigenetic knowledge to improve population health. Epigenetics is the study of DNA methylation and patterns of histone modifications (“histone marks”), which lead to changes in gene expression not accompanied by alterations in DNA sequence (see Appendix 1: Terminology for useful definitions). Unlike inherited genetic variation that is static through the life course, epigenetic changes are plastic, dynamic, and differ by tissue and disease state. This means that epigenetic changes may be useful as biomarkers of exposure and disease and as targets for modification through preventive and therapeutic interventions. Many challenges remain in measuring and interpreting epigenetic changes and in achieving actionable public health applications.
To succeed in identifying epigenetic mechanisms that lead to complex phenotypes and diseases, public health researchers must integrate animal models with human clinical and population approaches, paying close attention to windows of vulnerability, environmental and nutritional assessment, and cell type–specific epigenetic patterns (23). Animal models, in which exposures are well controlled and characterized in depth, continue to inform the evaluation of dose-response effects on the epigenome. Animal studies can also identify vulnerable time periods during gestation and development and test for multi- or transgenerational effects (65, 86). Human clinical and population approaches have identified epigenetic changes as important contributors to cancers and other diseases (100), and have revealed epigenetic drift with age (31). In addition, animal models and clinical samples are often useful for proteomic and chromatin structure evaluation. Finally, animal models and clinical studies can be used to evaluate cell type–specific effects and can validate assays of peripheral tissues (e.g., blood, urine, saliva) to serve as proxy measures of epigenetic change in epidemiology studies.
In this review, we introduce the reader to (a) the basic measurements of epigenetics, (b) the concept of using epigenetics as an environmental biosensor in public health research and surveillance, (c) observations about bioinformatic methods for epigenomics and epigenetics, and (d) applications of epigenetics and epigenomics to risk assessment for public health protection and for personalized medicine. We present animal, human, and in vitro approaches to epigenetic research. We conclude with a model of how a community of diverse researchers can come together in a National Institute of Environmental Health Sciences (NIEHS)–funded center to advance the application of epigenetics to population-based studies that address the contribution of early environmental exposures to the risks of adult disease.
THE BASIC MEASUREMENTS OF EPIGENETICS
The epigenome refers to all of the chemical modifications that are added to the genome to regulate gene expression and activity. “Epi-” is Greek for “above,” and thus we can think of the epigenome as the entirety of the modifications to the genome, from those modifications directly to DNA to modifications that attach to nucleosomes, the proteins around which DNA is wrapped. Although all the different cell types in the body share the same DNA sequence, epigenetic modifications and other regulatory mechanisms control whether cells become liver, lung, skin, or another organ.
The most commonly studied epigenetic modification is the addition of a methyl group to the cytosine of a cytosine-guanine pair (CpG), termed DNA methylation. If such methylation is located in close proximity to a gene, it often reduces or silences expression of that gene. DNA methylation is a component of the one-carbon metabolism pathway and is dependent upon several enzymes and dietary micronutrient cofactors, including folate, choline, and betaine. In mammals, the regulation of DNA methylation is more dynamic than previously believed (55). For example, in human embryonic stem cells, methylation of cytosine not part of CpG dinucleotide sites may be important to developmental homeostasis (51). As DNA methylation is a stable, covalently bound mark, it is consistently measurable in fresh and archived tissue and biofluid samples with various polymerase chain reaction, array, and sequencing methods (33). High-density regions of CpG dinucleotides are found in the regulatory regions of about 60% of the known genes (9). Molecular epidemiologic studies measure CpG methylation either near specific genes or as an average global measurement (where high levels of methylation correlate with increased genomic stability) (100).
Epigenetic manipulation of cellular phenotypes is also driven by alteration of chromatin packaging via covalent histone modifications and incorporation of histone variants into nucleosomes (81). Chromatin is a nucleoprotein complex that packages linear DNA, associated histones, and other proteins into structures called nucleosomes. Posttranslational modifications of histone proteins are numerous and include acetylation, methylation, ubiquitination, phosphorylation, and ADP-ribosylation (14, 20). Histone acetylation is usually associated with transcriptional activation because the affinity of histone proteins for DNA is reduced, leading to relaxation of chromatin packaging. Histone methylation typically occurs on lysine residues and results in various gene expression consequences depending on the specific location of the modification on the tail of the histone protein (46). One, two, or three methyl groups can be added to each lysine, adding enormous complexity to the histone code (41). Chromatin may be further modified by association with linker histones as well as by a myriad of coregulatory proteins. Informative specific patterns have emerged as indicators of gene regulation (64, 102). In addition, histone modifications can serve as targets for interventions with drugs that inhibit histone deacetylase or demethylase enzymes (39, 47, 105). Chromatin immunoprecipitation (ChIP), followed by gene-specific analyses or genome-wide analyses via next-generation chromatin immunoprecipitation sequencing (ChIP-seq), allows investigators to characterize protein-DNA complexes, including histone modifications (67). The requirements for protein-DNA isolation and storage are more stringent than those for DNA; thus, these modifications have not been utilized widely in epidemiologic studies.
EPIGENETICS AS A BIOMARKER OR SENSOR FOR ENVIRONMENTAL EXPOSURES IN MOLECULAR EPIDEMIOLOGIC STUDIES
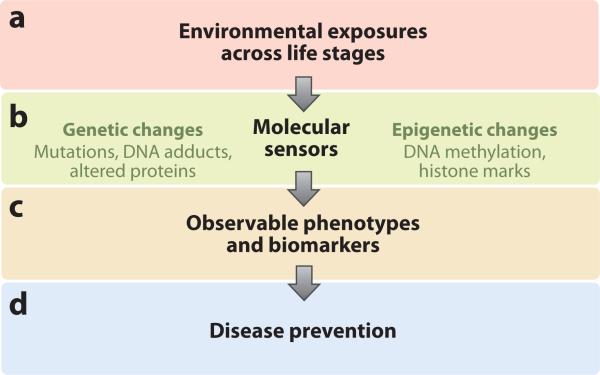
Humans are exposed to hazards throughout their life span, and the effects of these exposures are often not realized until decades later. This complicates traditional epidemiologic studies that rely on disease outcomes to identify cases and controls because exposure assessment is typically conducted after the likely relevant life stage has passed. However, what if researchers could identify epigenetic changes that are a consequence of potentially harmful exposures in populations and that could serve as an indicator (biomarker) of increased later-life disease risk (Figure 1)? Molecular epidemiology is a useful approach for linking exposures and disease in human populations. For example, carcinogenic DNA adducts in smokers’ blood cells provide a mechanistic link between tobacco smoke and the risk of lung cancer (79). Landmark studies by Perera et al. (74, 75) found an association between DNA adducts in cord blood, low birth weight, and decreased head circumference in children of pregnant mothers highly exposed to polycyclic aromatic hydrocarbons (PAHs) in Poland. Epigenetic epidemiology studies are complicated in that they must be attentive to the population distribution of epigenetic differences; multiple studies have identified divergent epigenetic profiles due to age, notably in monozygotic twins (31, 40), and with disease status, most commonly between cancerous and noncancerous tissues (see sidebar, Cancers: Epigenetic Diseases). Changes associated with modifiable environmental and lifestyle factors have been less clear and less consistent, confounded by differences in tissues examined, populations studied, and method of determining methylation status. These challenges are not insurmountable. Rapid advances in technology and in bioinformatics analyses (see below) facilitate the integration of data to identify relevant markers for translational studies of disease prediction and treatment in human populations.
Figure 1.
Translating epigenetic and genetic markers of environmental exposure to public health interventions. (a) Environmental exposures throughout the life course induce genetic and epigenetic alterations, particularly in susceptible populations. (b) Genomic and epigenetic changes serve as molecular biosensors of environmental exposures’ toxic effects, and these effects can be quantified within populations. (c) Genetic and epigenetic changes could presage observable phenotypes, including both disease phenotypes and biomarkers indicative of disease. (d) At risk individuals and subpopulations identified by molecular sensors and biomarkers can be targeted for public health interventions.
CpG Methylation
Environmental epidemiology studies have measured both methylation of specific genes and global methylation over the whole genome. Tested genes are often limited to specific tumor suppressor genes identified as methylated in cancers, such as p16 (CDKN2A); silencing of a tumor suppressor releases cells to proliferate. Decreased global methylation, measured via bisulfite sequencing of LINE-1 and Alu elements, was identified in cohorts of healthy individuals exposed to benzene (10), persistent organic pollutants (80), air pollution (black carbon, PM2.5, SO2) (6), lead exposure (103), and arsenic (43). Genes found to be hypermethylated in response to environmental exposures include p15, MAGE-1, and H19 with benzene exposure (10), ACSL3 with PAH exposure (73), and p53 and p16 with arsenic exposure (18). One of the earliest studies of the epigenetic effects of exposure to an environmental toxicant examined the impact of benzene exposure on global as well as gene-specific promoter methylation (p15, MAGE-1, and H19) in blood from healthy individuals exposed to a wide range of airborne benzene concentrations (10). Benzene exposure was measured via a personal air monitor, and DNA methylation was determined via pyrosequencing. LINE-1, Alu, and MAGE-1 methylation were found to decrease with increasing airborne benzene exposure, whereas p15 methylation was increased with benzene exposure. The magnitudes of effect, however, were small, with a tenfold increase in benzene exposure associated with modest decreases in LINE-1, Alu, and MAGE-1 methylation, and increase in p15 methylation. Benzene exposure was also associated with an increase in methylation of the p15 and p16 promoter in a case-control study of benzene poisoning, with a corresponding decrease in p15 mRNA expression (55). The results from these early studies suggest that methylation at p15 and p16 is environmentally labile, although whether this region is directly modified by environmental exposures and how this methylation modifies disease risk are still to be determined.
In contrast to the benzene studies, where toxicant exposure was inversely associated with global methylation, PAH exposure was found to be positively associated with LINE-1 methylation. In a cohort of highly exposed male Polish coke-oven workers and matched controls, increased urinary levels of 1-pyrenol and benzo[a]pyrene diol-epoxide DNA adducts were found to be associated with an increase in both LINE-1 and Alu methylation (71). Another study reported a similar association between exposure to prenatal tobacco smoke, a potential source of PAHs, and increased global DNA methylation in blood as measured by the [3H]-methyl acceptance assay (92). Perera et al. (73) used methylation-sensitive restriction fingerprinting to identify specific genomic regions differentially methylated on the basis of PAH exposure in cord blood. They found that children who were exposed to higher concentrations of PAHs in utero had significantly higher methylation at the promoter of ACSL3. Hypermethylation of ACSL3 at birth was found to significantly predict future asthmatic status, suggesting that this gene may mediate the development of environmentally influenced asthma in children.
Perhaps most intriguing are studies that explore associations between social or behavioral factors and epigenetic regulation. The molecular basis underlying the response to social and environmental factors is not well understood. Epigenetics, early-life experiences, and stress-related outcomes in mice spurred an interest in the epigenetic basis of behavior in humans (17, 30). A study of the biological effects of shift work in a northern Italian cohort of male chemical plant workers found a significant increase in TNF-α promoter methylation in shift workers compared to day workers and an association between job seniority and Alu and IFN-γ hypomethylation (11). In a Scottish cohort, economically deprived individuals and manual laborers had significantly lower global DNA methylation in peripheral blood leukocytes (56). Also, increasing levels of plasma fibrinogen and IL-6 were associated with decreased global DNA methylation levels, suggesting a mechanistic link between systemic inflammation and epigenetic change in circulating cells. There is strong evidence for the association between antidepressant drugs and epigenetic modifications in mice (96, 97). Histone deacetylase inhibitors reversed epigenetic changes in schizophrenia with a concordant decrease in psychotic symptoms (95). These studies indicate that epigenetics may hold the key to a larger understanding of the social determinants of health, where early-life events shape later susceptibility to disease.
To date, molecular epidemiology studies that incorporate epigenetic measurements have rarely validated the biological effects of epigenetic changes via measurements of RNA or protein expression. Additionally, the epigenomic profiles of sorted-cell populations within a tissue could be characterized to interpret results from mixed-cell populations because cellular differentiation is an epigenetically controlled process. Functional validation paired with cell type–specific epigenomic profiles can elucidate whether small differences measured reflect simply a shift in cell population. These studies may identify subtypes of cells within a tissue that are more susceptible to epigenetic mechanisms of toxicity, which would not be reflected in the overall epigenetic profile of the mixed-cell population. The technology for cell type–specific epigenetic profiling is advancing rapidly, with multiple groups reporting epigenomic and transcriptomic profiles from single cells (42, 90). As high-throughput sequencing technologies become more widely available and more affordable, molecular epidemiologists will be able to incorporate selected cell type–specific epigenomic profiles into studies linking exposure and disease.
Specific Histone Marks
A few studies have investigated the influence of environmental exposures on global levels of specific histone modifications. These studies require availability of intact protein fractions, and quantification is currently not cost-effective for large studies. An investigation into the effects of metal-rich air particle exposure in healthy steelworkers found significantly increased levels of histone H3 with dimethylation of lysine-4 (H3K4me2) and histone H3 with acetylation of lysine-9 (H3K9ac) in peripheral blood (16). Specifically, individuals with higher inhalational exposure to iron, arsenic, and nickel, as quantified by personal air monitors, had significantly higher global levels of H3K4me2, a histone modification associated with transcriptional activation. Another study of individuals occupationally exposed to nickel found increased H3K4me3 and decreased H3K9me2 when compared to a reference group of age- and smoking-matched controls (4). The use of global histone measurements in environmental epidemiology has been limited by the difficulty in quantifying levels of modified histones in a large sample set as well as by the availability of appropriate samples. As technology becomes available to generate data from less optimally collected samples, histone modifications will be an informative addition to molecular epidemiologic studies.
Studies to date may be considered illustrative of the impact of toxicants on the epigenome; the next step is the incorporation and integration of diverse information. Bioinformaticists have developed tools to integrate multiple data sources to identify dysregulated pathways in diseases (83, 84). It is vital that collaborative groups design efficient prospective studies that maximize the use of samples to measure multiple toxicants, diet, and a range of health outcomes. These findings will result in a better understanding of the mechanisms of disease development and progression, leading to improvements in diagnosis, treatment, and potential interventions to reduce the burden of diseases in human populations. A logical path forward relies on interactions between basic scientists and molecular epidemiologists to identify and determine the risks associated with differential epigenetic regulation of labile genes. Basic scientists can take advantage of animal and in vitro systems to model life span exposures at a range of exposure levels and then take tissue samples at biologically relevant time points. Examples of such translational studies are early-life exposures in the Agouti mouse model (24, 25) and exposures in human primary breast epithelial cells (37, 101).
EPIGENETICS AS A BIOMARKER OR SENSOR FOR ENVIRONMENTAL EXPOSURES IN TOXICOLOGICAL STUDIES OF ANIMAL MODELS
Animal models are a valuable resource in environmental epigenetics. Epigenetic changes can be mediated through diet (25) and toxicant (2, 24) exposures. Rodent models are complementary to human studies in that epigenetic changes can be measured in relevant tissues throughout the life span with controlled exposure protocols. Although approaches to translate epigenetic discoveries from genetically identical animal strains to heterogeneous human populations are far from optimized, integrative computational approaches for joint analyses of human and animal data are promising.
We have utilized a multipronged approach with a mouse model, human clinical samples, and ongoing longitudinal epidemiological studies of bisphenol A (BPA) as a representative early environmental exposure (see sidebar, Bisphenol A as a Representative Developmental Epigenetic Toxicant). BPA has been associated with epigenetic alterations following developmental and adolescent exposures (2, 24, 26, 35, 91, 106). In a rat model, Ho and colleagues (35) observed multiple changes in gene-specific DNA methylation patterns in the adult male prostate, including hypomethylation of Pde4d4. Using the viable yellow agouti (Avy) mouse model, investigators have shown that maternal dietary exposure to moderate levels of BPA (50 mg BPA/kg diet) resulted in decreased DNA methylation at the Avy and CabpIAP loci (2, 24), whereas exposure to lower doses (50 ng and 50 μg BPA/kg diet) led to hypermethylating effects at these candidate loci (2). Recently, Bartolomei and colleagues (88) reported that fetal exposure to BPA alters expression and methylation status of imprinted genes in the mouse embryo and placenta. Imprinted genes and their associated regulatory components may be particularly sensitive to developmental environmental perturbations of the epigenome. In fact, individuals conceived during the Dutch hunger winter at the end of World War II were shown 60 years later to have altered DNA methylation at the imprinted IGF2 locus (33), a locus that plays an important role in growth.
These investigations of BPA's role in altering the developing epigenome have been limited to candidate gene-driven approaches, restricted in dose-response assessments, or confined to animal models. Dolinoy and coinvestigators (45) recently conducted a genome-wide environmental epidemiology study of BPA exposure and DNA methylation levels in preadolescent girls from Gharbiah, Egypt, showing that methylation profiles exhibit exposure-dependent trends. This work is significant because epigenetic epidemiology holds promise for the identification of biomarkers from previous exposures and for the development of epigenetic-based diagnostic strategies, including nutritional intervention approaches, which have been previously demonstrated in mouse models (8, 24).
NUTRITIONAL STATUS AND FOOD ADDITIVES
Epigenetics has been implicated as a mechanism linking early environmental exposures and nutritional status to disease risks later in life, yet studies claiming epigenetics as a mechanism often fail to follow individuals with well-characterized exposures beyond birth outcomes. Using physiologically relevant levels of dietary BPA exposure in an animal model, in 2012, Dolinoy and coinvestigators (2) showed that perinatal BPA exposure leads to dose-dependent, nonmonotonic effects on the fetal epigenome, with higher dietary exposure to BPA leading to hypomethylation at two candidate loci, but lower dietary levels were less effective at hypomethylating and led to the opposite—hypermethylating—effects. The same lab next demonstrated that perinatal exposure to BPA is associated with hyperactive and lean phenotypes in females, with improved hormone and free fatty acid profiles (3). These observations of hyperactivity and lean body mass following developmental BPA exposure are in contrast to cross-sectional epidemiological studies associating BPA with higher body weight and increased obesity (49, 94). The human studies may be confounded by food consumption practices and altered BPA metabolism associated with body composition. Ongoing studies are now closing the loop between perinatal exposure and phenotypes of the offspring by evaluating epigenome-wide methylation and chromatin profiles to understand the mechanism(s) linking early BPA exposure to later-in-life disease risks.
Folate supplementation has been a great success in the prevention of neural tube defects. However, in utero exposure to a maternal diet supplemented with methyl donors may also have unexpected effects, such as allergic airway disease. Mice born to mothers whose diets were supplemented with folic acid, vitamin B12, methionine, zinc, betaine, and choline experienced significantly higher rates of allergic airway disease caused by an alteration in T lymphocyte maturation (36). Additionally, DNA methylation differences, including hypermethylation of Runx3, a regulator of T lymphocyte differentiation, were observed in exposed pups (36). Epidemiologic studies have identified an association between perinatal folic acid supplementation and increased risk of wheezing at 18 months of age (32). These findings suggest that dietary interventions at key developmental time points can lead to unintended adverse consequences.
OBSERVATIONS ABOUT BIOINFORMATICS METHODS FOR EPIGENOMICS AND EPIGENETICS
The term epigenomics refers to genome-wide analyses of CpG methylation and/or histone modifications, whereas epigenetics refers to changes to individual genes or sets of genes. Epigenomics was revolutionized when microarray technology, originally used for genotyping and gene expression profiling, was adapted to measure epigenetic marks. Now massively parallel deep sequencing can measure genome-wide DNA methylation and histone marks after fragmentation and short-read DNA sequencing (48). These approaches can generate terabytes of data per experiment, leading to storage and processing challenges far beyond that of microarrays; research on how best to deal with and interpret such a deluge of data is critical.
Bioinformatics methods and tools are often developed with data arising from a specific technology, and then they are adopted by a different biomedical research community for a related use. Two examples are (a) gene set enrichment testing methods, developed for gene expression microarray data, now used for deep-sequencing applications, such as RNA-Seq and ChIP-Seq (89, 108), and (b) ChIP-Seq peak-finding software developed for identifying transcription factor binding sites, now adopted for detection of DNA methylation sites or regions with a certain histone modification (22, 104, 109). With many approaches currently available for assessing genome-wide epigenetic marks, multiple bioinformatics methods are required to fit the properties of the respective data types. For example, if using reduced-representation bisulfite sequencing to assess genome-wide DNA methylation, methods for preprocessing and identifying CpG site-specific differentially methylated regions are required (1).
As epigenomic deep-sequencing studies become mainstream, more population-based and context-specific studies are being performed, requiring novel bioinformatics algorithms and tools to properly and effectively analyze and interpret the data. For example, Sartor and coinvestigators developed PePr (meaning peak prioritization) (https://code.google.com/p/pepr-chip-seq/), which takes into account variability among biological replicates in the analysis of ChIP-Seq data, allowing the identification of consistent differences between exposed and unexposed individuals or animals. Another is ChIP-Enrich, which allows one to bridge from a list of genomic regions (for example, from a genome-wide histone modification study) to the targeted biological functions and pathways (http://chip-enrich.med.umich.edu). We applied one such tool, LRpath, to identify common pathways dysregulated via DNA methylation across cancer types (44). For a review of the available sequencing technologies and bioinformatics methods for performing, analyzing, and interpreting epigenomics studies, as well as integrative approaches for understanding the epigenetics in the context of the genome and transcriptome, see Sartor et al. (82).
SNYDEROME: INTEGRATED PERSONAL OMICS PROFILING: LONGITUDINAL ANALYSIS TO BRIDGE FROM OMICS TO PHENOTYPES
Multiple levels of omics data can be integrated with physiological, environmental, behavioral, and clinical information to traverse between genome and phenome (69). Longitudinal data gathering and analysis on selected individuals is highly complementary to population-based cross-sectional studies. Such repeated analyses have been performed and reported by Snyder, a pioneer in proteomics and functional genomics, and coworkers (19). They present integrated personalized omics profiling as a way to reveal dynamic molecular and clinical phenotypes, linking omics information with regular monitoring of physiological states and clinical symptoms. For example, because an unsuspected propensity to develop type 2 diabetes mellitus was recognized from genomic analysis and common laboratory tests, Snyder responded by modifying his diet, exercise, and monitoring practices to reduce his risk profile.
The methods used in this initial paper (19) include whole-genome sequencing (using both the Complete Genomics and Illumina technology platforms), exome sequencing (with three different technologies), transcriptomics with microarrays and deep RNA sequencing, proteomics (with shotgun methods and with targeted analysis of cytokines), autoantibody profiling, and metabolomics. Snyder and coinvestigators are now performing whole-genome bisulfite sequencing, which will then be integrated and analyzed with the other data types. In addition, alternative splicing and differential expression of splice variants was documented; splicing will become important in the analysis of deep RNA-sequencing and proteomics data sets, as illustrated by Liu et al. (52) for our collaborative studies of chromosome 17 as part of the Human Proteome Project (54).
ENCYCLOPEDIA OF DNA ELEMENTS
A mammoth, highly coordinated project called the Encyclopedia of DNA Elements (ENCODE) has generated a tremendous array of data sets, including epigenomics data sets, comprehensively examining the organization and genome-wide control of expression of the genome (27, 107). Thirty-seven initial copublished ENCODE-related papers can be explored online via the Nature ENCODE explorer (http://www.nature.com/encode/#/threads), a specially designed visualization tool that allows users to access the linked papers and investigate topics that are discussed in multiple papers via interesting thematically organized threads. The wide spectrum of 125 cell and tissue types covered by this data greatly expands the horizons of cell-selective gene regulation analysis, enabling the recognition of systematic long-distance regulatory patterns and previously undescribed phenomena, such as mutation rate variation in normal versus immortal or pluripotent cells.
The histone information from ENCODE needs to be integrated with CpG methylation, the other main hallmark of epigenomics. This approach has been successfully used to interpret findings from genome-wide association studies that test the relationship between millions of single-nucleotide polymorphisms (SNPs) and human phenotypes. Integration of these population-level data with ENCODE has led to functional annotation of 80% of the SNPs reported to be associated with various phenotypes in humans (85). Mapping these SNPs to elements defined by ENCODE shows significant overall enrichment for regulatory function in SNP-disease-associated regions. These types of integrated approaches will be valuable in identifying regulatory regions of the genome for investigation in epidemiologic studies with carefully measured exposure data. Thus, it is crucial that population scientists collect biologic samples appropriate for the measurement of histone modifications for both validation and epigenetic discovery in informative populations.
APPLICATION OF EPIGENETICS AND EPIGENOMICS TO RISK ASSESSMENT FOR PUBLIC HEALTH PROTECTION AND FOR PERSONALIZED MEDICINE
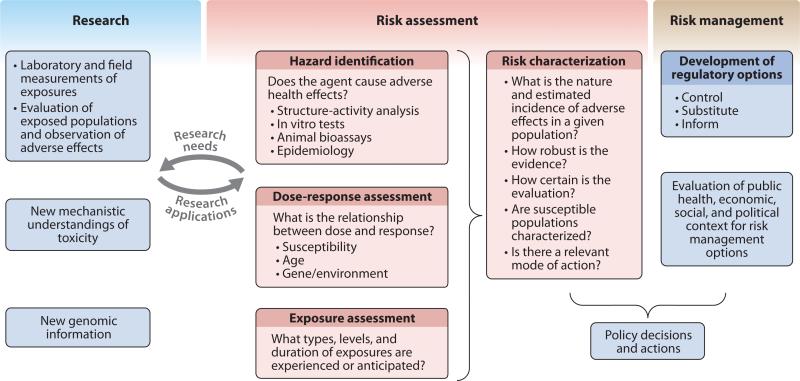
Molecular epidemiology has become a major input for risk assessment, addressing evidence of exposure, of individual variation in susceptibility to particular exposures, or of early effects of toxic agents. Reports from the National Research Council of the National Academies (61, 62) and the Presidential/Congressional Commission on Risk Assessment and Risk Management (77) have contributed to a useful scientific and regulatory framework for hazard identification, risk assessment, risk management, and risk communication. Risk communication is particularly important in public health practice. An ideal situation is when mechanistic research feeds directly into risk assessments and when critical data uncertainty for assessment, management, and communication of risks drive the agenda for research. A more encompassing term than risk assessment, which is often equated with quantitative risk assessment, is risk characterization, which also addresses the following qualitative questions: What is the nature of the adverse effects in a given population? Are they reversible? How robust is the evidence? How certain is the evaluation? Have susceptible populations been identified? Is there a relevant mode of action? What are the uncertainties? (See Figure 2.)
Figure 2.
Risk assessment/risk management framework. This framework shows, under the red highlighting, the four key steps of risk assessment: hazard identification, dose-response assessment, exposure assessment, and risk characterization. It shows an interactive, two-way process where research needs from the risk assessment process drive new research, and new research findings modify risk assessment outcomes. This figure is reprinted with permission from Faustman & Omenn (28).
As epigenomics and epigenetics reveal distinctive patterns of changes in DNA methylation and histone modification associated with particular toxic agent exposures, those findings will be highly useful in exposure assessment for qualitative and quantitative risk assessment. Such biomarkers will also enable comparison of exposures to and effects from structurally related chemicals, as well as enable the monitoring of responses to preventive and therapeutic interventions.
At present, experimental findings are often complex and inconsistent. For example, the nongenotoxic carcinogen arsenic produces both hypomethylation and hypermethylation, with overall greater hypomethylation genome wide, but results in hypermethylation of certain key promoter sites (such as for p53 and Ink4/Arf) (78). Deficiency of vitamins and substrates that generate S-adenosyl-methionine enhances the hypomethylation from arsenic. Arsenic also alters histone patterns (21). Similarly, Perera & Herbstman (72) reported combined general hypomethylation and specific hypermethylation of genes associated with in utero exposures to BPA, PAHs, arsenic, phthalates, and tobacco smoke; they highlighted transgenerational persistent changes and associated traits and diseases.
The validation and application of short-term in vitro assays of cultured cells can provide information about mechanisms of action of effects and typically are inexpensive and fast, especially compared with lifetime animal bioassays. The societal costs of relying on such in vitro tests for carcinogenicity, for example, with false positives (noncarcinogens classified as carcinogens) and false negatives (true carcinogens not detected), are the subject of the Lave-Omenn value-of-information model (50, 70). Meanwhile, lifetime rodent bioassays are no longer black-box studies to count tumors; numerous molecular assays, such as epigenomic assays, are also incorporated in these studies. In general, results in rodents can point to likely risk in humans; in several cases, however, the findings in rodents do not occur in humans, and the mechanisms for those differences are well characterized [see Faustman & Omenn (28), table 4-4]. Omics analyses also facilitate systems-based understanding of exposures, responses, and disease, moving risk assessment away from a linear single-event-based concept, and improve the biological plausibility of epidemiological associations (63).
THE UNIVERSITY OF MICHIGAN CENTER FOR LIFE-STAGE EXPOSURES AND ADULT DISEASE
The authors of this review are all members of the NIEHS Michigan Center for Life-Stage Exposures and Adult Disease at the University of Michigan. Epigenetics, life-stage environmental and nutritional evaluation, and bioinformatics are core elements of this center, organized by Dr. Howard Hu and now led by Dr. Rita Loch-Caruso. The goals are well matched to multiple features of the Annual Review of Public Health, Volume 34, symposium on “Developmental Origins of Adult Disease” (13, 34, 68, 76). The authors of these articles emphasized environmental, behavioral, socioeconomic, and dietary contributors to different risk profiles for later-in-life diseases. Center activities emphasize cross-disciplinary studies and activities that focus on exposures in vulnerable stages of life that represent critical windows of susceptibility. These activities span bioinformatics to epigenomics to bidirectional communication between center scientists and our regional community via a stakeholder advisory board.
We facilitate the integration of information through the development and use of new bioinformatics methods and tools for analyzing multidimensional data from these complex studies (44, 84). These approaches allow a broad range of research that spans studies of preterm birth to Alzheimer's disease. Early-life trauma, such as complications from preterm birth, are associated with later-life chronic health conditions (7). Projects led by the Michigan Center for Life-Stage Exposures and Adult Disease core center members John Meeker, Marie O’Neill, and Rita Loch-Caruso are linking specific environmental contaminants, including phthalates and air pollutants, with preterm birth (58, 66, 93). Other center members are gaining insights into how DNA methylation and other epigenetic mechanisms mediate environmentally driven risk for later development of certain neurological disorders, such as Alzheimer's disease, attention deficit-hyperactivity disorder, and amyotrophic lateral sclerosis.
CONCLUDING REMARKS
For all types of toxicology and epidemiology studies and for public health actions, credible quantitative assessment of relevant exposures is critical. The use of multiple levels of omics analyses represents a rapidly emerging new generation of capabilities for exposure assessment or “the exposome.” Challenges remain in adapting emerging epigenetic technologies to epidemiology studies to identify mechanisms associated with exposure and disease. Well-characterized exposure data, especially for earlier-life exposures, are critical to identify important and functionally relevant case-control epigenetic differences. Especially with budgetary pressures on project costs, it is essential to carefully plan cohort studies that have the ability to biobank relevant samples and consider the types of samples that will be useful for exposure assessment in future studies. It will also be useful to develop and apply biomarkers of lifetime exposure to environmental toxicants, such as the measurement of heavy metals in primary teeth (5) or lead in bone (38), for an accurate modeling of exposure status throughout the life course in late-life diseases.
Another challenge is the recognition that epigenetic changes associated with environmental exposures or with disease outcomes have typically been small in magnitude. Although a 1–2% shift in global methylation as measured by LINE-1 may represent what appears to be a minor change, LINE-1 elements represent approximately 14% of the entire genome and may be functionally relevant. With an array of global methylation measurements to choose from, how to interpret results from a study that finds a statistically significant association between an exposure and Alu methylation, but not LINE-1, or vice versa, remains to be clarified, as with multiple measures in classical epidemiological studies.
Despite these challenges, the field of epigenetics will continue to make the crucial links between environmental and nutritional risk factors and human disease. The continued collaboration among basic scientists, bioinformaticists, and population researchers clarifies the biological relationship between exposures and epigenetics, facilitating risk assessment strategies in human populations. All of these factors lead to an exciting time in public health, as we realize our ability to link life-stage exposures and later disease to improve population health.
CANCERS: EPIGENETIC DISEASES.
Although numerous disease phenotypes have been associated with epigenetic etiology, including neurological dysfunction and metabolic syndrome, carcinogenesis remains the most actively studied disease category. Cancers are a heterogeneous set of diseases, displaying both genetic and epigenetic etiologies. Methylation profiles differ between cancer types; in general, however, the epigenome is widely hypomethylated compared to normal tissue, with the exception of hypermethylation in genic regions, notably tumor suppressor genes (29). Animal and cell line models of specific pathways of carcinogenesis serve as critical tools to understand the mechanisms of these diseases. The integration of laboratory and epidemiological approaches aims to translate findings to human clinical and population approaches to better prevent and treat cancers.
BISPHENOL A AS A REPRESENTATIVE DEVELOPMENTAL EPIGENETIC TOXICANT.
The epigenome is vulnerable to environmental factors during embryogenesis because the DNA synthetic rate is high, and the elaborate DNA methylation patterning required for normal tissue development is established at this time. An increasing number of animal and human studies now demonstrate that prenatal exposures to environmental conditions, including chemical and/or nutritional factors, increase the risk of developing adult-onset diseases. One of the routinely used chemical compounds in food and beverage containers, baby bottles, dental composites, and receipt paper is bisphenol A (BPA) (98). Accumulating work suggests that early exposure to endocrine-active compounds, such as BPA, increases susceptibility for adverse outcomes via epigenetic mechanisms.
BPA is a high-production-volume monomer used in the manufacture of polycarbonate plastic and epoxy resins. Several studies have reported detectable levels of total urinary BPA in a large proportion of populations around the world (15, 60, 110), and a recent study of human fetal liver samples indicated that there is considerable exposure to BPA during pregnancy (59). BPA exposure results in a variety of pathophysiological changes implicated in breast and prostate cancer, reproductive dysregulation, and behavioral abnormalities (53, 99). Epidemiology studies have associated increased BPA levels with cardiovascular disease risk, decreased semen quality, altered childhood behavior, and recurrent miscarriages (12, 49, 57, 87).
Owing to the uncertainty surrounding BPA's safety for humans, especially in young children, there was a sizeable consumer backlash against BPA in the United States and elsewhere. Companies such as Toys “R” Us and Walmart banned the sale of BPA in baby and water bottle products. In 2012, the US Food and Drug Administration banned BPA in baby bottles and children's drinking cups. Yet, BPA is still widely used in a number of other consumer goods, such as canned food and receipt paper. Thus, moving forward, the application of epigenetics and epigenomics to risk assessment for public health protection and for personalized medicine will be tremendously important for identifying potential developmental epigenetic toxicants.
ACKNOWLEDGMENTS
We recognize the role of Clyde Hertzman as director of the Human Early Learning Partnership at the University of British Columbia. Also as a member of the Annual Review of Public Health editorial committee, he organized and edited the symposium theme set of articles, mentioned above. Sadly, he died suddenly and unexpectedly in February 2013.
Support for work discussed in this article was provided by the University of Michigan NIEHS P30 ES017885, NIH ES017524, NIH/EPA P20 ES018171/RD 83480001, and R01CA158286-01 grants.
APPENDIX 1: TERMINOLOGY
Bisulfite conversion: a laboratory technique to analyze DNA methylation using the resistance of the conversion of a methylated cytosine to uracil compared with an unmethylated cytosine. Following PCR amplification, a methylated cytosine will appear as a cytosine (C), whereas a converted unmethylated cytosine will appear as a thymine (T).
Chromatin immunoprecipitation (ChIP): a procedure with protein-specific antibody to determine whether a given protein is localized to a particular DNA sequence. This technique helps characterize the interactions between histone marks and DNA.
Chromatin: a complex of genetic material and proteins (mostly histones) that condense to form chromosomes during cell division.
Cytosine–guanine pair: also known as a “CpG dinucleotide,” this is the primary site for DNA methylation.
DNA adduct: results when a cancer-causing chemical, such as a polycyclic aromatic hydrocarbon (PAH), chemically reacts with DNA. This affects the DNA replication machinery and leads to mutations.
DNA methylation: primarily a stable repressive mark found at cytosines in CpG dinucleotides, where the phosphate (P) indicates directionality (5′–CG–3′).
Epigenetics: literally means “on top of or in addition to genetics”; the study of DNA methylation and patterns of histone modifications (“histone marks”) which lead to changes in gene expression not accompanied by alterations in DNA sequence.
Epigenome: the global pattern of epigenetic marks that may distinguish or be variable among cell types.
Exome sequencing: DNA sequencing restricted to the sequences coding for proteins.
HDAC inhibitor (histone deacetylase): a drug that targets the epigenome through inhibition of enzymes that alter the histone proteins.
Histones: family of proteins that act like spools around which DNA is wrapped to assist in condensing DNA into structures called nucleosomes.
Pluripotent cell: a cell that can differentiate, or become, most cell types in the body, induced by specific transcription factors.
Proteomics: the analysis of many, potentially all, the proteins in a specimen.
Reduced representation bisulfite sequencing (RRBS): a procedure that analyzes DNA methylation in a large subset of the genome.
Regulatory regions: regions in genes that are important to the initiation of transcription. These are usually in the 5′ areas of genes and are called the “promoters” but can also be in the early exon regions of the gene.
Transcriptomics: analysis of all (not including rRNA) of the messenger RNA in the cell, or the message from the DNA to guide production of proteins.
Whole-genome bisulfite sequencing (WGBS): DNA sequencing that can identify the CpGs that are methylated in the genome.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, et al. MethylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson OS, Nahar MS, Faulk C, Jones TR, Liao C, et al. Epigenetic responses following maternal dietary exposure to physiologically relevant levels of bisphenol A. Environ. Mol. Mutagen. 2012;53:334–42. doi: 10.1002/em.21692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson OS, Peterson KE, Sanchez BN, Zhang Z, Mancuso P, Dolinoy DC. Perinatal bisphenol A exposure promotes hyperactivity, lean body composition, and hormonal responses across the murine life course. FASEB J. 2013;27:1784–92. doi: 10.1096/fj.12-223545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arita A, Niu J, Qu Q, Zhao N, Ruan Y, et al. Global levels of histone modifications in peripheral blood mononuclear cells of subjects with exposure to nickel. Environ. Health Perspect. 2012;120:198–203. doi: 10.1289/ehp.1104140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arora M, Kennedy BJ, Elhlou S, Pearson NJ, Walker DM, et al. Spatial distribution of lead in human primary teeth as a biomarker of pre- and neonatal lead exposure. Sci. Total Environ. 2006;371:55–62. doi: 10.1016/j.scitotenv.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 6.Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, et al. Rapid DNA methylation changes after exposure to traffic particles. Am. J. Respir. Crit. Care Med. 2009;179:572–78. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Behrman RE, Butler AS. Preterm Birth: Causes, Consequences, and Prevention. Natl. Acad.; Washington, DC: 2006. [PubMed] [Google Scholar]
- 8.Bernal AJ, Dolinoy DC, Huang D, Skaar DA, Weinhouse C, Jirtle RL. Adaptive radiation-induced epigenetic alterations mitigated by antioxidants. FASEB J. 2013;27:665–71. doi: 10.1096/fj.12-220350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 10.Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–80. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 11.Bollati V, Baccarelli A, Sartori S, Tarantini L, Motta V, et al. Epigenetic effects of shiftwork on blood DNA methylation. Chronobiol. Int. 2010;27:1093–104. doi: 10.3109/07420528.2010.490065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braun JM, Kalkbrenner AE, Calafat AM, Yolton K, Ye X, et al. Impact of early-life bisphenol A exposure on behavior and executive function in children. Pediatrics. 2011;128:873–82. doi: 10.1542/peds.2011-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bygren LO. Intergenerational health responses to adverse and enriched environments. Annu. Rev. Public Health. 2013;34:49–60. doi: 10.1146/annurev-publhealth-031912-114419. [DOI] [PubMed] [Google Scholar]
- 14.Caiafa P, Zampieri M. DNA methylation and chromatin structure: the puzzling CpG islands. J. Cell Biochem. 2005;94:257–65. doi: 10.1002/jcb.20325. [DOI] [PubMed] [Google Scholar]
- 15.Calafat A, Ye X, Wong L, Reidy J, Needham L. Exposure of the U.S. population to bisphenol A and 4-tertiary-octylphenol: 2003–2004. Environ. Health Perspect. 2008;116:39–44. doi: 10.1289/ehp.10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cantone L, Nordio F, Hou L, Apostoli P, Bonzini M, et al. Inhalable metal-rich air particles and histone H3K4 dimethylation and H3K9 acetylation in a cross-sectional study of steel workers. Environ. Health Perspect. 2011;119:964–69. doi: 10.1289/ehp.1002955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Champagne FA, Weaver IC, Diorio J, Dymov S, Szyf M, Meaney MJ. Maternal care associated with methylation of the estrogen receptor-α1b promoter and estrogen receptor-α expression in the medial preoptic area of female offspring. Endocrinology. 2006;147:2909–15. doi: 10.1210/en.2005-1119. [DOI] [PubMed] [Google Scholar]
- 18.Chanda S, Dasgupta UB, Guhamazumder D, Gupta M, Chaudhuri U, et al. DNA hypermethylation of promoter of gene p53 and p16 in arsenic-exposed people with and without malignancy. Toxicol. Sci. 2006;89:431–37. doi: 10.1093/toxsci/kfj030. [DOI] [PubMed] [Google Scholar]
- 19.Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HY, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheung P, Lau P. Epigenetic regulation by histone methylation and histone variants. Mol. Endocrinol. 2005;19:563–73. doi: 10.1210/me.2004-0496. [DOI] [PubMed] [Google Scholar]
- 21.Chu F, Ren X, Chasse A, Hickman T, Zhang L, et al. Quantitative mass spectrometry reveals the epigenome as a target of arsenic. Chem. Biol. Interact. 2011;192:113–17. doi: 10.1016/j.cbi.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colaneri A, Staffa N, Fargo DC, Gao Y, Wang T, et al. Expanded methyl-sensitive cut counting reveals hypomethylation as an epigenetic state that highlights functional sequences of the genome. Proc. Natl. Acad. Sci. USA. 2011;108:9715–20. doi: 10.1073/pnas.1105713108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dolinoy DC, Faulk C. The use of animal models to advance epigenetic science. ILAR J. 2012;53:227–31. doi: 10.1093/ilar.53.3-4.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A–induced DNA hypomethylation in early development. Proc. Natl. Acad. Sci. USA. 2007;104:13056–61. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ. Health Perspect. 2006;114:567–72. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doshi T, Mehta SS, Dighe V, Balasinor N, Vanage G. Hypermethylation of estrogen receptor promoter region in adult testis of rats exposed neonatally to bisphenol A. Toxicology. 2011;289:74–82. doi: 10.1016/j.tox.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 27.Dunham I, Birney E, Lajoie BR, Sanyal A, Dong X, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faustman EM, Omenn GS. Risk assessment. In: Klaassen CD, editor. Casarett and Doull's Toxicology: The Basic Science of Poisons. 8th ed. McGraw-Hill; New York: 2013. pp. 123–49. [Google Scholar]
- 29.Feinberg AP. The epigenetics of cancer etiology. Semin. Cancer Biol. 2004;14:427–32. doi: 10.1016/j.semcancer.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 30.Fish EW, Shahrokh D, Bagot R, Caldji C, Bredy T, et al. Epigenetic programming of stress responses through variations in maternal care. Ann. NY Acad. Sci. 2004;1036:167–80. doi: 10.1196/annals.1330.011. [DOI] [PubMed] [Google Scholar]
- 31.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl. Acad. Sci. USA. 2005;102:10604–9. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Håberg SE, London SJ, Stigum H, Nafstad P, Nystad W. Folic acid supplements in pregnancy and early childhood respiratory health. Arch. Dis. Child. 2009;94:180–84. doi: 10.1136/adc.2008.142448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl. Acad. Sci. USA. 2008;105:17046–49. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hertzman C. Commentary on the symposium: biological embedding, life course development, and the emergence of a new science. Annu. Rev. Public Health. 2013;34:1–5. doi: 10.1146/annurev-publhealth-031912-114500. [DOI] [PubMed] [Google Scholar]
- 35.Ho SM, Tang WY, Belmonte de Frausto J, Prins GS. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res. 2006;66:5624–32. doi: 10.1158/0008-5472.CAN-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hollingsworth JW, Maruoka S, Boon K, Garantziotis S, Li Z, et al. In utero supplementation with methyl donors enhances allergic airway disease in mice. J. Clin. Investig. 2008;118:3462–69. doi: 10.1172/JCI34378. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Hsu PY, Deatherage DE, Rodriguez BA, Liyanarachchi S, Weng YI, et al. Xenoestrogen-induced epigenetic repression of microRNA-9-3 in breast epithelial cells. Cancer Res. 2009;69:5936–45. doi: 10.1158/0008-5472.CAN-08-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu H, Rabinowitz M, Smith D. Bone lead as a biological marker in epidemiologic studies of chronic toxicity: conceptual paradigms. Environ. Health Perspect. 1998;106:1–8. doi: 10.1289/ehp.981061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huertas D, Soler M, Moreto J, Villanueva A, Martinez A, et al. Antitumor activity of a small-molecule inhibitor of the histone kinase Haspin. Oncogene. 2011;31:1408–18. doi: 10.1038/onc.2011.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20:170–79. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 42.Kantlehner M, Kirchner R, Hartmann P, Ellwart JW, Alunni-Fabbroni M, Schumacher A. A high-throughput DNA methylation analysis of a single cell. Nucleic Acids Res. 2011;39:e44. doi: 10.1093/nar/gkq1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kile ML, Baccarelli A, Hoffman E, Tarantini L, Quamruzzaman Q, et al. Prenatal arsenic exposure and DNA methylation in maternal and umbilical cord blood leukocytes. Environ. Health Perspect. 2012;120:1061–66. doi: 10.1289/ehp.1104173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J, Karnovsky A, Mahavisno V, Weymouth T, Pande M, et al. LRpath analysis reveals common pathways dysregulated via DNA methylation across cancer types. BMC Genomics. 2012;13:526. doi: 10.1186/1471-2164-13-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim JH, Rozek LS, Soliman AS, Sartor MA, Hablas A, et al. Bisphenol A-associated epigenomic changes in prepubescent girls: a cross-sectional study in Gharbiah. Egypt. Environ. Health. 2013;12:33. doi: 10.1186/1476-069X-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 47.Lai C-J, Bao R, Tao X, Wang J, Atoyan R, et al. CUDC-101, a multitargeted inhibitor of histone deacetylase, epidermal growth factor receptor, and human epidermal growth factor receptor 2, exerts potent anticancer activity. Cancer Res. 2010;70:3647–56. doi: 10.1158/0008-5472.CAN-09-3360. [DOI] [PubMed] [Google Scholar]
- 48.Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat. Rev. Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 49.Lang IA, Galloway TS, Scarlett A, Henley WE, Depledge M, et al. Association of urinary bisphenol A concentration with medical disorders and laboratory abnormalities in adults. JAMA. 2008;300:1303–10. doi: 10.1001/jama.300.11.1303. [DOI] [PubMed] [Google Scholar]
- 50.Lave LB, Omenn GS. Cost-effectiveness of short-term tests for carcinogenicity. Nature. 1985;324:29–34. doi: 10.1038/324029a0. [DOI] [PubMed] [Google Scholar]
- 51.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu S, Im H, Bairoch A, Cristofanilli M, Chen R, et al. A chromosome-centric human proteome project (C-HPP) to characterize the sets of proteins encoded in chromosome 17. J. Proteome Res. 2012;12:45–57. doi: 10.1021/pr300985j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maffini MV, Rubin BS, Sonnenschein C, Soto AM. Endocrine disruptors and reproductive health: the case of bisphenol-A. Mol. Cell Endocrinol. 2006;254–255:179–86. doi: 10.1016/j.mce.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 54.Marko-Varga G, Omenn GS, Paik Y-K, Hancock WS. A first step toward completion of a genome-wide characterization of the human proteome. J. Proteome Res. 2012;12:1–5. doi: 10.1021/pr301183a. [DOI] [PubMed] [Google Scholar]
- 55.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–57. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McGuinness D, McGlynn LM, Johnson PCD, MacIntyre A, Batty DG, et al. Socio-economic status is associated with epigenetic differences in the pSoBid cohort. Int. J. Epidemiol. 2012;41:151–60. doi: 10.1093/ije/dyr215. [DOI] [PubMed] [Google Scholar]
- 57.Meeker JD, Ehrlich S, Toth TL, Wright DL, Calafat AM, et al. Semen quality and sperm DNA damage in relation to urinary bisphenol A among men from an infertility clinic. Reprod. Toxicol. 2010;30:532–39. doi: 10.1016/j.reprotox.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meeker JD, Hu H, Cantonwine DE, Lamadrid-Figueroa H, Calafat AM, et al. Urinary phthalate metabolites in relation to preterm birth in Mexico City. Environ. Health Perspect. 2009;117:1587–92. doi: 10.1289/ehp.0800522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nahar MS, Liao C, Kannan K, Dolinoy DC. Fetal liver bisphenol A concentrations and bio-transformation gene expression reveal variable exposure and altered capacity for metabolism in humans. J. Biochem. Mol. Toxicol. 2013;27:116–23. doi: 10.1002/jbt.21459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nahar MS, Soliman AS, Colacino JA, Calafat AM, Battige K, et al. Urinary bisphenol A concentrations in girls from rural and urban Egypt: a pilot study. Environ. Health. 2012;11:20. doi: 10.1186/1476-069X-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Natl. Res. Counc. Risk Assessment in the Federal Government: Managing the Process. Natl. Acad.; Washington, DC: 1983. [PubMed] [Google Scholar]
- 62.Natl. Res. Counc. Biological Markers in Pulmonary Toxicology. Natl. Acad.; Washington, DC: 1989. [Google Scholar]
- 63.Natl. Res. Counc. Toxicity Pathway-Based Risk Assessment: Preparing for Paradigm Change: A Symposium Summary. Natl. Acad.; Washington, DC: 2010. [PubMed] [Google Scholar]
- 64.Nie Y, Liu H, Sun X. The patterns of histone modifications in the vicinity of transcription factor binding sites in human lymphoblastoid cell lines. PLoS ONE. 2013;8:e60002. doi: 10.1371/journal.pone.0060002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nilsson E, Larsen G, Manikkam M, Guerrero-Bosagna C, Savenkova MI, Skinner MK. Environmentally induced epigenetic transgenerational inheritance of ovarian disease. PLoS ONE. 2012;7:e36129. doi: 10.1371/journal.pone.0036129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O'Neill MS, Osornio-Vargas A, Buxton MA, Sáanchez BN, Rojas-Bracho L, et al. Air pollution, inflammation and preterm birth in Mexico City: study design and methods. Sci. Total Environ. 2012;448:79–83. doi: 10.1016/j.scitotenv.2012.10.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.O'Geen H, Echipare L, Farnham PJ. Using ChIP-seq technology to generate high-resolution profiles of histone modifications. In: Tollefsbol TO, editor. Methods in Molecular Biology, Vol. 791: Epigenetics Protocols. Humana; New York: 2011. pp. 265–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Odgers CL, Jaffee SR. Routine versus catastrophic influences on the developing child. Annu. Rev. Public Health. 2013;34:29–48. doi: 10.1146/annurev-publhealth-031912-114447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Omenn GS. Gene-environment interactions: eco-genetics and toxicogenomics. In: Ginsburg GS, Willard HF, editors. Genomic and Personalized Medicine. 2nd ed. Academic; London: 2012. pp. 50–59. [Google Scholar]
- 70.Omenn GS, Stuebbe S, Lave LB. Predictions of rodent carcinogenicity testing results: interpretation in light of the Lave-Omenn value-of-information model. Mol. Carcinog. 1995;14:37–45. doi: 10.1002/mc.2940140108. [DOI] [PubMed] [Google Scholar]
- 71.Pavanello S, Bollati V, Pesatori AC, Kapka L, Bolognesi C, et al. Global and gene-specific promoter methylation changes are related to anti-B[a]PDE-DNA adduct levels and influence micronuclei levels in polycyclic aromatic hydrocarbon-exposed individuals. Int. J. Cancer. 2009;125:1692–97. doi: 10.1002/ijc.24492. [DOI] [PubMed] [Google Scholar]
- 72.Perera F, Herbstman J. Prenatal environmental exposures, epigenetics, and disease. Reprod. Toxicol. 2011;31:363–73. doi: 10.1016/j.reprotox.2010.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Perera F, Tang WY, Herbstman J, Tang D, Levin L, et al. Relation of DNA methylation of 5′-CpG island of ACSL3 to transplacental exposure to airborne polycyclic aromatic hydrocarbons and childhood asthma. PLoS ONE. 2009;4:e4488. doi: 10.1371/journal.pone.0004488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perera FP, Jedrychowski W, Rauh V, Whyatt RM. Molecular epidemiologic research on the effects of environmental pollutants on the fetus. Environ. Health Perspect. 1999;107(Suppl. 3):451–60. doi: 10.1289/ehp.99107s3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Perera FP, Whyatt RM, Jedrychowski W, Rauh V, Manchester D, et al. Recent developments in molecular epidemiology: a study of the effects of environmental polycyclic aromatic hydrocarbons on birth outcomes in Poland. Am. J. Epidemiol. 1998;147:309–14. doi: 10.1093/oxfordjournals.aje.a009451. [DOI] [PubMed] [Google Scholar]
- 76.Power C, Kuh D, Morton S. From developmental origins of adult disease to life course research on adult disease and aging: insights from birth cohort studies. Annu. Rev. Public Health. 2013;34:7–28. doi: 10.1146/annurev-publhealth-031912-114423. [DOI] [PubMed] [Google Scholar]
- 77.Pres./Congr. Comm. Risk Assess. Risk Manag . Risk Assessment and Risk Management in Regulatory Decision-Making. US Gov. Print. Off; Washington, DC: 1997. [Google Scholar]
- 78.Reichard JF, Puga A. Effects of arsenic exposure on DNA methylation and epigenetic gene regulation. Epigenomics. 2010;2:87–104. doi: 10.2217/epi.09.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rudiger HW, Nowak D, Hartmann K, Cerutti P. Enhanced formation of benzo(a)pyrene:DNA adducts in monocytes of patients with a presumed predisposition to lung cancer. Cancer Res. 1985;45:5890–94. [PubMed] [Google Scholar]
- 80.Rusiecki JA, Baccarelli A, Bollati V, Tarantini L, Moore LE, Bonefeld-Jorgensen EC. Global DNA hypomethylation is associated with high serum-persistent organic pollutants in Greenlandic Inuit. Environ. Health Perspect. 2008;116:1547–52. doi: 10.1289/ehp.11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 2006;7:437–47. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- 82.Sartor MA, Dolinoy DC, Rozek LS, Omenn GS. Bioinformatics for high-throughput toxicoepigenomic studies. In: Sahu SC, editor. Toxicology and Epigenetics. Wiley; New York: 2012. pp. 569–88. [Google Scholar]
- 83.Sartor MA, Leikauf GD, Medvedovic M. LRpath: a logistic regression approach for identifying enriched biological groups in gene expression data. Bioinformatics. 2009;25:211–17. doi: 10.1093/bioinformatics/btn592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sartor MA, Mahavisno V, Keshamouni VG, Cavalcoli J, Wright Z, et al. ConceptGen: a gene set enrichment and gene set relation mapping tool. Bioinformatics. 2010;26:456–63. doi: 10.1093/bioinformatics/btp683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–59. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Skinner MK, Manikkam M, Guerrero-Bosagna C. Epigenetic transgenerational actions of endocrine disruptors. Reprod. Toxicol. 2011;31:337–43. doi: 10.1016/j.reprotox.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sugiura-Ogasawara M, Ozaki Y, Sonta S, Makino T, Suzumori K. Exposure to bisphenol A is associated with recurrent miscarriage. Hum. Reprod. 2005;20:2325–29. doi: 10.1093/humrep/deh888. [DOI] [PubMed] [Google Scholar]
- 88.Susiarjo M, Sasson I, Mesaros C, Bartolomei MS. Bisphenol A exposure disrupts genomic imprinting in the mouse. PLoS Genet. 2013;9:e1003401. doi: 10.1371/journal.pgen.1003401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Taher L, Ovcharenko I. Variable locus length in the human genome leads to ascertainment bias in functional inference for non-coding elements. Bioinformatics. 2009;25:578–84. doi: 10.1093/bioinformatics/btp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–82. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 91.Tang WY, Morey LM, Cheung YY, Birch L, Prins GS, Ho SM. Neonatal exposure to estradiol/bisphenol A alters promoter methylation and expression of Nsbp1 and Hpcal1 genes and transcriptional programs of Dnmt3a/b and Mbd2/4 in the rat prostate gland throughout life. Endocrinology. 2012;153:42–55. doi: 10.1210/en.2011-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Terry MB, Ferris JS, Pilsner R, Flom JD, Tehranifar P, et al. Genomic DNA methylation among women in a multiethnic New York City birth cohort. Cancer Epidemiol. Biomark. Prev. 2008;17:2306–10. doi: 10.1158/1055-9965.EPI-08-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tetz LM, Cheng A, Korte C, Giese R, Wang P, et al. Mono-2-ethylhexyl phthalate induces oxidative stress responses in human placental cells in vitro. Toxicol. Appl. Pharmacol. 2013;268:47–54. doi: 10.1016/j.taap.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Trasande L, Attina TM, Blustein J. Association between urinary bisphenol A concentration and obesity prevalence in children and adolescents. JAMA. 2012;308:1113–21. doi: 10.1001/2012.jama.11461. [DOI] [PubMed] [Google Scholar]
- 95.Tremolizzo L, Doueiri M-S, Dong E, Grayson DR, Davis J, et al. Valproate corrects the schizophrenia-like epigenetic behavioral modifications induced by methionine in mice. Biol. Psychiatry. 2005;57:500–9. doi: 10.1016/j.biopsych.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 96.Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat. Neurosci. 2006;9:519–25. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 97.Tsankova NM, Kumar A, Nestler EJ. Histone modifications at gene promoter regions in rat hippocampus after acute and chronic electroconvulsive seizures. J. Neurosci. 2004;24:5603–10. doi: 10.1523/JNEUROSCI.0589-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vandenberg LN, Chahoud I, Heindel JJ, Padmanabhan V, Paumgartten FJ, Schoenfelder G. Urinary, circulating, and tissue biomonitoring studies indicate widespread exposure to bisphenol A. Environ. Health Perspect. 2010;118:1055–70. doi: 10.1289/ehp.0901716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vandenberg LN, Colborn T, Hayes TB, Heindel JJ, Jacobs DR, Jr, et al. Hormones and endocrine-disrupting chemicals: low-dose effects and nonmonotonic dose responses. Endocr. Rev. 2012;33:378–455. doi: 10.1210/er.2011-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Virani S, Colacino JA, Kim JH, Rozek LS. Cancer epigenetics: a brief review. ILAR J. 2012;53:359–69. doi: 10.1093/ilar.53.3-4.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Weng YI, Hsu PY, Liyanarachchi S, Liu J, Deatherage DE, et al. Epigenetic influences of low-dose bisphenol A in primary human breast epithelial cells. Toxicol. Appl. Pharmacol. 2010;248:111–21. doi: 10.1016/j.taap.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wiencke J, Zheng S, Morrison Z, Yeh R. Differentially expressed genes are marked by histone 3 lysine 9 trimethylation in human cancer cells. Oncogene. 2007;27:2412–21. doi: 10.1038/sj.onc.1210895. [DOI] [PubMed] [Google Scholar]
- 103.Wright RO, Schwartz J, Wright RJ, Bollati V, Tarantini L, et al. Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ. Health Perspect. 2010;118:790–95. doi: 10.1289/ehp.0901429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wu G, Yi N, Absher D, Zhi D. Statistical quantification of methylation levels by next-generation sequencing. PLoS ONE. 2011;6:e21034. doi: 10.1371/journal.pone.0021034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yacqub-Usman K, Duong CV, Clayton RN, Farrell WE. Epigenomic silencing of the BMP-4 gene in pituitary adenomas: a potential target for epidrug-induced re-expression. Endocrinology. 2012;153:3603–12. doi: 10.1210/en.2012-1231. [DOI] [PubMed] [Google Scholar]
- 106.Yaoi T, Itoh K, Nakamura K, Ogi H, Fujiwara Y, Fushiki S. Genome-wide analysis of epigenomic alterations in fetal mouse forebrain after exposure to low doses of bisphenol A. Biochem. Biophys. Res. Commun. 2008;376:563–67. doi: 10.1016/j.bbrc.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 107.Yavartanoo M, Choi JK. ENCODE: a sourcebook of epigenomes and chromatin language. Genomics Inform. 2013;11:2–6. doi: 10.5808/GI.2013.11.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–58. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang Z, Alomirah H, Cho HS, Li YF, Liao C, et al. Urinary bisphenol A concentrations and their implications for human exposure in several Asian countries. Environ. Sci. Technol. 2011;45:7044–50. doi: 10.1021/es200976k. [DOI] [PubMed] [Google Scholar]