Abstract
Understanding the connection between protein structure and function requires a quantitative understanding of electrostatic effects. Structure-based electrostatics calculations are essential for this purpose, but their use have been limited by a long-standing discussion on which value to use for the dielectric constants (εeff and εp) required in Coulombic models and Poisson-Boltzmann models. The currently used values for εeff and εp are essentially empirical parameters calibrated against thermodynamic properties that are indirect measurements of protein electric fields. We determine optimal values for εeff and εp by measuring protein electric fields in solution using direct detection of NMR chemical shift perturbations (CSPs). We measured CSPs in fourteen proteins to get a broad and general characterization of electric fields. Coulomb's law reproduces the measured CSPs optimally with a protein dielectric constant (εeff) from 3 to 13, with an optimal value across all proteins of 6.5. However, when the water-protein interface is treated with finite difference Poisson-Boltzmann calculations, the optimal protein dielectric constant (εp) rangedsfrom 2-5 with an optimum of 3. It is striking how similar this value is to the dielectric constant of 2-4 measured for protein powders, and how different it is from the εp of 6-20 used in models based on the Poisson-Boltzmann equation when calculating thermodynamic parameters. Because the value of εp = 3 is obtained by analysis of NMR chemical shift perturbations instead of thermodynamic parameters such as pKa values, it is likely to describe only the electric field and thus represent a more general, intrinsic, and transferable εp common to most folded proteins.
Introduction
Some of the most fundamental biochemical reactions, such as enzymatic catalysis1, redox reactions2, H+ transfer3, and ion homeostasis4 are governed by electrostatic effects. To understand the structural and physical basis of such biological processes, it is necessary to know the magnitude and molecular determinants of electrostatic forces and energies in proteins. Owing to the difficulties inherent to the experimental measurement of electrostatic effects in proteins, we typically use structure-based calculations to estimate electrostatic energies in proteins. These theoretical calculations are an essential tool for dissecting structure-function relationships and properties of biomolecules, but they are notoriously sensitive to the input structure5,6 and to the parameters used such as the dielectric constants, and the charge-radius force field7. In particular, the value of the dielectric constants in these calculations remains highly contentious. What is clear is that the accuracy and utility of computational methods for structure-based electrostatics calculations is limited by our inability to describe dielectric effects quantitatively. Here we present data suggesting what the optimal value of the protein dielectric constant is when calculating electric fields with a Poisson-Boltzmann model framework and when using a simple Coulombic model.
Most structure-based calculations of electrostatic fields treat some part of the protein-water system as a dielectric continuum whose polarizability is described implicitly by a dielectric constant. To maximize the ability of a theoretical model to reproduce dielectric properties of proteins, the parameters that it employs (i.e. the charge-radius force field and its dielectric constant) are usually calibrated against benchmarks consisting of thermodynamic parameters for simpler systems, such as solvation free energies of ions in different polar solvents8, changes in stability induced by changes in pH, in ionic strength, or by mutations9, pKa values10, peptide acidity constants11 and redox potentials12. The problem is that although these thermodynamic parameters do reflect the magnitude of the electrostatic potential, they represent a convolution of many other factors as well. The dielectric constants obtained by calibration against thermodynamic data are therefore model dependent and experiment-dependent. They are empirical parameters calibrated to reproduce experimental benchmarks and to account implicitly for any physical factors that are not treated explicitly in the models13. In the present paper we focus on measuring protein electric fields via NMR spectroscopy and on using these experimental measurements to guide electrostatic field calculations. To this end, we analyze the measured electric field-dependent chemical shifts to extract the corresponding dielectric constants that reproduce them most accurately when using the Poisson-Boltzmann equation or Coulomb's law.
Spectroscopic observables such as Stark shifts and NMR chemical shifts offer a more direct measure of the magnitude and direction of electric fields in biomolecules than thermodynamic parameters such as pKa values. In proteins, chemical shifts of 1H, 15N and 13C nuclei measured with NMR spectroscopy represent a particularly rich source of information about electric fields. The relationship between the electric field, E, and the chemical shift reported by a nucleus, δef, was first identified by Pople14 and later formulated by Buckingham15 in what is known as Buckingham's equation:
| (1) |
Eγ in Buckingham's equation is the component of the uniform electric field at the nucleus along the coordinate γ, and ∂Eγ/∂rδ represents its gradient along the coordinate δ (See fig. S1b). Aγ, Bγ,γ and Aγδ are well-defined nucleus-dependent response properties that describe the sensitivity of the chemical shift δef to the applied uniform electric field and to its gradient. This sensitivity is particularly significant in 1HN and 15N nuclei involved in highly polarizable covalent bonds, such as the backbone N–H and C–N bonds. Therefore, chemical shifts reported by these nuclei can be used as high resolution probes of through-space electrostatic effects in proteins.
Stark shifts have already been shown to be useful as probes of electric fields in a few proteins and in an enzyme active site16-21. Chemical shift perturbations (CSP) measured with NMR spectroscopy hold promise of being equally useful to this end. CSPs can be induced by changing the charge state of a protein either by removing or inserting charged residues16, by binding charged ligands, by changing the ionic strength of the solution, by incorporation NMR sensitive nuclei22 or by changing pH17,19.
The CSP reported by a nucleus, Δδtot, can be affected primarily by three factors. It can be caused directly by changes in the electric field, Δδef, by changes in magnetic effects, Δδm, or by conformational changes, Δδs,:
| (2) |
To obtain as clean a measurement of the electric field effect as possible it is important to reduce the contribution of non-electrostatic effects to the measured Δδtot. For this reason, only proteins that do not experience significant pH-induced magnetic and conformational changes were studied16,17,19. In these proteins, Δδtot reflects predominantly the Δδef component, whose magnitude and direction is predictable (Eq. 1). The two other components, Δδm and Δδs, make minor contributions. The first of these, Δδm, is well-defined23,24 and often negligible except near aromatic rings that are structurally perturbed over the course of a titration. The second, Δδs, results from any titration-induced changes in structure, such as changes in torsion angles, bond lengths, bond angles or hydrogen bonding geometry close to a reporter nucleus25. The direction and magnitude of this component is expected to show a complex dependence on the structural environment of a given reporter nucleus25. Therefore, for the proteins studied here, Δδtot represents a measurement of the electric field, Δδef, with an added amount of noise proportional to (Δδs+Δδm), with a direction that is uncorrelated with the direction of Δδef. Any one measurement of the electric field at a single nucleus can include an error from conformational changes or magnetic effects. However, through simultaneous measurements performed for all nuclei in a single or several proteins, this error cancels out and hence reliable values of Δδef can be obtained.
Previous measurements of electric fields with Stark shifts or NMR chemical shifts have been limited to a few individual proteins16-19,26 or to a single location inside an enzyme active site20,21. To characterize the propagation of electric field in proteins in a more systematic manner, we used chemical shift perturbations measured with NMR spectroscopy in nine proteins, in four variants of these proteins, and in one enzyme-substrate intermediate. This large dataset allowed a self-consistent and systematic description of electric field propagation inside proteins, which in turn allowed an unprecedented determination of the value of the dielectric constant of proteins.
Materials and Methods
ACBP and hGRX
13C, 15N labeled bovine ACBP and hGRX were prepared as described previously27,28. A 1mM sample of the proteins was prepared in 100 mM KCl, 1 mM DSS, 0.02% NaN3 and 10% D2O. NMR spectra of ACBP (hGRX) were recorded in the pH interval from 3.3 (2.7) to 11.0 (8.9), in steps of approximately 0.25 by addition of NaOH. All spectra at each pH value were referenced to DSS. The pKa values for His residues in ACBP were determined from the 1H-13C HSQC, and for Asp/Glu from the 13CO and HN spectrum29. The pKa values for Cys residues in hGRX were determined from 2D 1H-13C HSQC of the aliphatic side-chain resonances.
Prediction of CSPs
The Δδef was calculated as a difference in chemical shielding of the nucleus upon the (de)protonation of an ionizable group using Buckingham's equation (Eq. 1). Nuclear shielding polarizabilities, Aγ, for 15N and 1HN nuclei were used from19. Although values for the second term in Buckingham's formula, Bγ,γ, have not been reported for 15N and 1HN in the literature, their negligible contribution to Δδef was assumed based on data reported for 19F and 13C nuclei22,30. The third term in Buckingham's formula, the gradient contribution, was calculated according to31. The electric field, Eγ, at the position of backbone 15N and 1HN nuclei was calculated with the PBE implemented in the APBS program (single Debye-Hückel boundary condition; final grid resolution of 0.25 Å/grid point)32 and with Coulomb's law. The values of εp and εeff were taken from the range [1, 30] with a step of 0.5. 20 ns MD simulations were generated as described in detail in SI.
MD simulations
MD simulations of 20 ns length were performed for each of the six protein systems using the GROMACS package with the Amber ff99SB force field33 and the TIP3P water model34. A real space cut-off of 10.0 Å on non-bonded interactions, periodic boundary conditions and particle-mesh Ewald electrostatics were used throughout the simulation. Prior to MD simulations, the systems were neutralized to a total charge of zero by adding Cl− and Na+ ions, energy minimized and equilibriated in the NVT ensemble over 200 ps. The temperature was controlled with a Berendsen coupling35 to a water bath at 300 K using a temperature-coupling constant of 0.1 ps. Snapshot structures were taken every 10 ps starting at 5 ns of the simulation time and each snapshot was used in CSP predictions (Eq. 1).
Results
Protein selection
Some of the pH-dependent 15N-1H HSQC spectra used in this analysis were recorded previously and were generously made available by other researchers. Included in this set were: (I) bovine beta-lactoglobulin (βLG)36, (II) B1 domain of protein G (GB1)37, (III) plastocyanin from A. variabilis (A.v. Pc)19, (IV) plastocyanin from Phormidium laminosum (P.l. Pc)38-40, (V) xylanase from Bacillus circulans (B.c. xylanase)41, (VI) the catalytic domain of β-(1,4)-glycosidase Cex from Cellulomonas fimi (CexCD)42, and (VII) a highly stable form of staphylococcal nuclease (SNase) known as Δ+PHS43. The pH-dependent 15N-1H HSQC spectra for two additional proteins were measured specifically for this study: (I) bovine acyl-coenzyme A binding protein (ACBP), and (II) human glutaredoxin 1 (hGRX). Spectra previously recorded for four variants of SNase with substitutions D21N, L38D, L38E and L38K, and for one long-lived enzyme-substrate intermediate of CexCD covalently modified by 2,4-dinitrophenyl 2-deoxy-2-fluoro-β-cellobioside (2FCb-CexCD)44 were also analyzed. The complete data set consisted of 1861 15N and 1861 1HN chemical shifts of backbone amides measured as a function of pH.
Identification of useful chemical shift perturbations (CSPs)
The titration of ionizable groups in a protein is reflected in the chemical shifts of backbone amides. To extract Δδtot values that originate from the titration of a single ionizable group, pH-dependent chemical shifts reported by a 15N or 1HN nucleus were fitted using an automated F-test based algorithm45,46. Only Δδtot values with a magnitude above the uncertainty level for the specific nucleus (0.1 ppm for 15N and 0.03 ppm for 1HN (ref. 47)) were considered. Each extracted Δδtot, denoted “observed CSP”, was assigned as arising from the (de)protonation of a particular ionizable group if its apparent pKa matched the pKa of the ionizable group (Figure S1 and Supporting Results in the Supporting Information). The spatial distribution of nuclei reporting Δδtot values associated with a specific ionization event provides information about the distribution of the electric field emanating from a single charge. Instances where it was clear that an ionization event did not induce a CSP in a reporter nucleus were also recorded. These are termed ‘absent CSPs’ and they are important, as these are cases where the contributions by a charge to the electric field dissipated before they could induce a CSP in the reporter nucleus.
The majority of the ionizable groups in the nine proteins that were investigated are solvent-exposed. Their pKa values are not perturbed significantly relative to the normal pKa values of isolated amino acids in water. Therefore, the ionizable groups of the same type of residues (Asp/Glu, His, Lys) tend to have similar pKa values in the proteins. This similarity of pKa values precludes unambiguous assignment of the majority of the Δδtot values in the experimental data to any one specific ionizable group. For this reason, these groups were not included in the analysis. The only groups used in the analysis of electric fields were the groups where the CSP could be associated with the change in charge of a specific residue. On this basis, 9 ionizable groups were identified whose apparent pKa values could be identified unambiguously in the experimental CSPs (details given in SI). These groups are: His14 (pKa=7.1) and His30 (pKa=6.7) in ACBP; His61 (pKa=7.1) and His92 (pKa=5.1) in A.v. Pc; His 92 (pKa=5.1) in P.l. Pc; Glu44 (pKa=4.0) and Glu94 (pKa=4.0) in hGRX, His80 (pKa=7.9) in CexCD and Asp21 (pKa=6.5) in SNase. The final dataset used to analyze electric fields included the effects of nine ionizable groups on a total of 786 15N and 786 1HN nuclei.
Statistics on the number of observed and absent CSPs associated with each ionizable group are presented in Table 1. The spatial distribution of CSPs is illustrated for ACBP in Figure 1 and in Figures S2-S7 for all remaining proteins. The number of observed CSPs originating from His80 in CexCD and Asp21 in SNase was further validated by requiring the absence of their observed CSPs in the spectra of the 2FCb-CexCD intermediate or in the D21N variant of SNase. The nuclei inside the protein core do not report CSPs originating from the solvent-exposed His14 in ACBP and Glu94 and Glu44 in hGRX. Indeed, only a handful of nuclei along the protein surface were sensitive to the electric field changes originating from the ionization of these groups. Only 9 15N (and 9 1HN) CSPs were associated with the titration of His14 in ACBP, whereas the corresponding numbers associated with the titration of Glu94 and Glu44 in hGRX were 10 15N (and 8 1HN) and 14 15N (and 11 1HN), respectively. In contrast, the CSPs associated with the ionization of partially or fully buried groups, such as His30 in ACBP, His92 and His61 in A.v. Pc, His92 in P.l. Pc, His80 in CexCD and Asp21 in SNase, are reported by nuclei positioned both deep in the core or on the protein surface. The number of CSPs originating from the titration of these groups were: His30 in ACBP (33 15N and 31 1HN), His92 (36 15N and 34 1HN) and His61 (30 15N and 26 1HN) in A.v. Pc, His92 (38 15N and 32 1HN) in P.l. Pc, His80 (57 15N and 46 1HN) in CexCD and Asp21 (33 15N and 31 1HN) in SNase.
Table 1.
Dataset used to measure the dielectric constant of proteins.
| Protein (pH range) | Titr.group | pKa | side chain SASA (Å2) | # of identified CSPs 15N (1HN) Δδef | # of absent CSPs 15N (1HN) Δδef |
|---|---|---|---|---|---|
| ACBP (pH 3.0-8.9) | His14 | 7.1 | 36 | 9 (9) | 70 (66) |
| His30 | 6.7 | 6 | 33 (31) | 46 (43) | |
| A.v. Pc (pH 4.0-8.4) | His92 | 5.1 | 7 | 36 (34) | 56 (58) |
| His61 | 7.1 | 14 | 30 (26) | 62 (66) | |
| P.l. Pc (pH 4.2-8.3) | His92 | 5.1 | 9 | 38 (32) | 58 (63) |
| hGRX (pH 2.8-11.1) | Glu44 | 4.0 | 19 | 10 (8) | 80 (80) |
| Glu94 | 4.0 | 24 | 14 (11) | 80 (80) | |
| CexCD (pH 4.1-9.3) | His80 | 7.9 | 2 | 57 (46) | 213 (232) |
| SNase (pH 1.4-11.1) | Asp21 | 6.5 | 2 | 33 (31) | 72 (87) |
| Total | 260 (228) | 737 (775) | |||
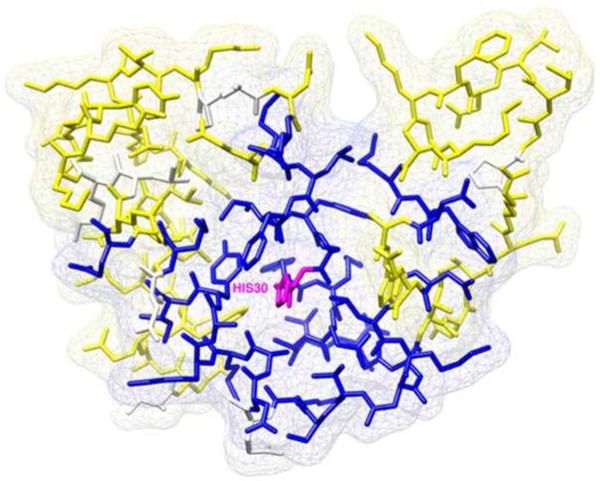
Figure 1.
Electric field propagation in ACBP caused by the protonation of His30 (magenta) and measured at backbone 15N: (blue) residues with identified CSPs above 0.1ppm; (yellow) residues with CSPs bellow 0.1ppm; (white) unassigned resonances and proline residues.
Differences in the number of the observed CSPs originating from the titration of solvent exposed and buried ionizable groups can be rationalized in terms of expected general properties of electric fields in proteins: electric fields originated from solvent exposed side chains are screened by bulk water, which has a high dielectric response. In contrast, electric fields inside the protein can propagate over larger distances because inside the protein the dielectric response is more muted than in water. Similar arguments can be applied to CSPs caused by titration-induced conformational changes (Δδs in Eq. 2): the titration of a buried ionizable group is more likely to induce a large conformational perturbation. Hence the ionization of a buried group can affect a much larger number of nuclei than the ionization of solvent exposed groups, which are less likely to induce conformational reorganization.
Corroboration that CSPs reflect through-space electric field effects
To analyze CSPs rigorously in terms of propagation of electric fields originating from the ionizable moieties, it is important to ascertain that the assigned Δδtot reflect primarily the propagation of through-space electrostatic effects. To test this we compared the direction of the observed Δδtot (downfield/upfield shift with increased pH) with the corresponding direction of the electric field-induced CSPs, Δδef, calculated from the crystal structures using Buckingham's equation and Coulomb's law. Whereas the local structural contribution to the CSP, Δδs, is expected to result in either upfield or downfield shifts depending on the structural environment of a given nucleus, the direction of the electric field-induced CSP, Δδef, is always determined by the position of the titrating site relative to the reporter nucleus (Eq. 1). Therefore, a high correlation between the directions predicted from Buckingham's equation and the direction of the observed Δδtot would indicate that electrostatic effects make the dominant contribution to the observed Δδtot.
A strong correlation was found between the directions of the observed Δδtot and the calculated Δδef values; 226 of 260 (87%) observed 15N Δδtot and 189 of 228 (83%) observed 1HN Δδtot displayed shifts in the direction predicted by Buckingham's equation. These results show convincingly that the electric field makes the dominant contribution to the observed Δδtot. Nevertheless, to further minimize artifacts of non-electrostatic effects in the electric field analysis, we excluded 34 of the 15N and 39 of the 1HN nuclei for which an agreement in CSPs direction was not obtained.
Dielectric constants obtained by analysis of electric fields with static structures
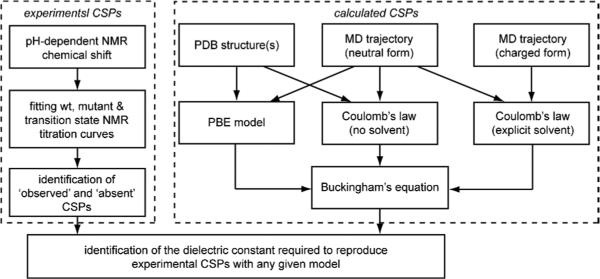
The ability of the Poisson-Boltzmann equation (PBE) and of Coulomb's law to reproduce experimental results was tested using both single static structures (see Figure 2 for an overview) as well as ensembles of structures. When a Coulomb interaction measured experimentally is analyzed with a primitive continuum model based on Coulomb's law, the dielectric parameter that reproduces the experimental quantity is referred to as the effective dielectric constant, εeff 13. More sophisticated models based on numerical solutions of the Poisson-Boltzmann equation describe dielectric effects of the protein with a parameter that we call εp. CSP values were used to identify values of εeff and εp that most accurately reproduce the experimental data.
Figure 2.
Overview of the approach for using CSPs for benchmarking computational methods for prediction of electric fields.
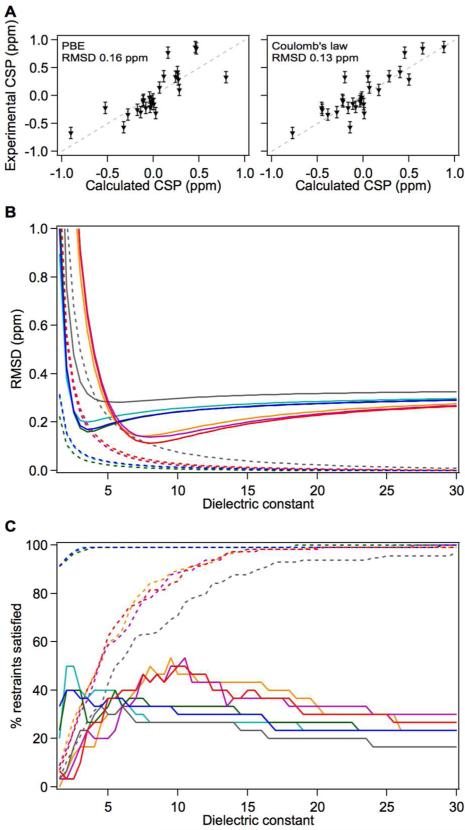
Analysis with Coulomb's law
Analysis of CSPs with Coulomb's law yields an optimal value of εeff of 6.5 for the 15N datasets, with an RMSD of 0.28 ppm between calculated and experimentally observed CSPs. For the 1HN datasets, the optimal εeff is 7.0 with an RMSD of 0.07 ppm; however εeff values from 6 to 15.0 (Table 2 and Table S1) yield RMSD values that are witin 0.1 ppm. This wide range of εeff values thus predicts the observed CSPs almost as accurately as the optimal εeff value. If we instead calculate the fraction of CSPs predicted correctly we find that εeff values between 6 and 15 do best for the observed CSPs, whereas the absent CSPs are predicted accurately only with εeff values above 15. This is illustrated for ACBP in Figure 3, and for all other proteins in Figures S3-S8. Figure 3B displays the RMSD between calculated and observed Δδef (solid line), and calculated and absent CSPs (dashed line). The fraction of reporter nuclei in which the observed Δδef (solid line) and the absent CSPs (dashed line) predicted within the experimental error are displayed in Figure 3C. In all proteins it was observed that optimal εeff in terms of RMSD, predicts the existence of CSPs in 20% of 15N and 1HN nuclei for which CSPs were not observed in the NMR spectra. This highlights the inability of Coulomb's law to correctly model the effect of solvent screening in a protein-water system.
Table 2.
Optimal values of the protein dielectric constant obtained with Poisson-Boltzmann electrostatics (εp) and with Coulomb's law (εeff) by analysis of CSPs reported by 15N or 1HN (given in brackets) nuclei using a single crystal structure, an ensemble of crystal structures, or an ensemble generated with MD simulations.
| Protein/group | εp from PBE |
εeff from Coulomb's law |
|||||
|---|---|---|---|---|---|---|---|
| static structure | ensemble of stat.struct. | MD simulations | static structure | ensemble of stat.struct. | MD without water | full MD | |
| ACBP | 3.5(3.5) | 3.5(3.0) | 3.5(4.0) | 7.5(8.0) | 8.0(9.0) | 8.0(9.0) | 5.5(8.5) |
| Hisl4 | 2.0(2.0) | 3.0(3.5) | 3.5(6.0) | 7.0(17.0) | 10.0(20) | 9.5(21.0) | 4.0(60) |
| His30 | 3.5(3.5) | 4.0(3.0) | 3.5(3.0) | 7.5(7.5) | 7.0(7.5) | 7.5(7.5) | 6.0 (8.0) |
| A.v. Pc | 3.0(3.0) | 2.5(3.0) | 2.0(2.5) | 5.5(4.5) | 5.5(5.5) | 5.5(7.5) | 11.0(31.5) |
| His6l | 5.0(5.0) | 4.0(9.5) | 2.5(7.5) | 8.0(16.5) | 8.0(18.0) | 9.5(20.0) | 60(41.5) |
| His92 | 2.0(2.5) | 2.0(2.0) | 2.0(2.5) | 4.0(4.0) | 4.0(4.0) | 4.0(5.0) | 5.5(17.5) |
| P.l. Pc | 2.0(2.5) | 2.0(2.5) | 2.5(3.5) | 4.0(4.5) | 4.0(4.5) | 4.0(5.0) | 60(60) |
| His92 | 2.0(2.5) | 2.0(2.5) | 2.5(3.5) | 4.0(4.5) | 4.0(4.5) | 4.0(5.0) | 60(60) |
| hGRX | 2.0(2.0) | 2.0(2.0) | 11.5(5.5) | 11.5(4.5) | 60(4.0) | ||
| Glu44 | 2.0(2.0) | 1.5(2.0) | 14.5(3.5) | 16.0(4.0) | 60(3.5) | ||
| Glu94 | 2.0(3.5) | 2.0(3.0) | 11.0(10.0) | 9.5(9.5) | 5.5(5.0) | ||
| CexCD | 5.0(5.0) | 5.0(5.0) | 4.5(10.0) | 7.5(7.0) | 7.5(7.0) | 8.0(9.0) | 4.5(60) |
| His80 | 5.0(5.0) | 5.0(5.0) | 4.5(10.0) | 7.5(7.0) | 7.5(7.0) | 8.0(9.0) | 4.5(60) |
| SNase | 2.5(3.5) | 2.5(3.5) | 2.0(2.5) | 6.0(10.0) | 6.0(9.0) | 7.0(9.0) | 46.5(51) |
| Asp21 | 2.5(3.5) | 2.5(3.5) | 2.0(2.5) | 6.0(10.0) | 6.0(9.0) | 7.0(9.0) | 46.5(51) |
| ALL | 3.0(3.0) | 3.0(3.5) | 3.0(4.0) | 7.0(6.5) | 6.0(7.0) | 7.0(7.0) | 20.0(28.5) |
Figure 3.
Optimal values of εp and εeff using 15N CSPs measured in ACBP. (A) Correlation between experimentally measured 15N Δδef emanating from the His30 charge and calculated 15N Δδef obtained from the static structure PDB ID: 1HB6 and with optimal dielectric constant values that minimize RMSD (εp=3.5; εeff=7.5). (B) Average RMSD between measured and predicted 15N Δδef in ACBP as a function of dielectric constant. Solid lines depict the RMSD of observed Δδef and dashed lines depict the RMSD of absent Δδef. Color scheme: calculations with static structure with PBE (cyan) or with Coulomb's law (orange), calculations with an ensemble of static structures using PBE (blue) or Coulomb's law (red), calculations with an ensemble generated with MD simulations and PBE (green) or Coulomb's law (magenta), microscopic approach (grey). (C) Fraction of 15N reporter nuclei whose predicted Δδef are within the nucleus-specific experimental errors of the observed Δδef (solid line) and absent Δδef (dashed line). Color scheme is identical to (B).
Analysis with a finite difference Poisson-Boltzmann model
The optimal value of εp obtained by analysis of both 15N and 1HN datasets in all proteins is 3.0. (Tables 2 and S1). With this value the RMSD between the observed and calculated Δδefs is 0.25 ppm for 15N data and 0.06 ppm for 1HN data. The analysis with the PBE methodology yielded the following observations: (1) The magnitudes of the observed Δδef are predicted in the PBE model most successfully when a low value of εp is used regardless of whether performance was evaluated with RMSD or with fractional statistics. (2) The optimal value of εp = 3 predicted both the presence and the absence of the CSPs in the reporter nuclei. (3) The optimal values of εp obtained by analysis of both observed and absent CSPs confirmed the ability of the PBE method to account for the screening of electric fields with the two-dielectric system (protein, εp; solvent, εs=80). (4) The range of εp values obtained by analysis of nine different ionizable groups in six different proteins is unexpected low, and falls in the range 2 to 5, which is much narrower than the range obtained in calculations with Coulomb's law (Table 2). (5) The value of εp = 3 contrasts sharply with the values of εp in the range 6 to 20 commonly used in structure-based pKa calculations with PB models to maximize agreement between experimental and calculated thermodynamic parameters such as pKa values and redox potentials48. The value of value of εp = 3 is strikingly similar to the value of the protein dielectric constant measured with dry proteins in experiments with parallel plate capacitors.
The difference between the magnitude of the protein dielectric constant resolved by analysis with the one dielectric model in Coulomb's law and with the two-dielectric calculations with PB electrostatics suggest that screening by bulk water plays an important role in attenuating the propagation of electric fields inside proteins. This effect cannot be accounted for correctly with Coulomb's law because the role of bulk water is subsumed into the single dielectric constant used in the analysis, but the effect is reproduced quite well in finite difference Poisson-Boltzmann calculations. This shows that screening by bulk solvent cannot be neglected in theoretical models for the calculation of electrostatic effects inside proteins.
Analysis of electric fields with conformational ensembles
NMR chemical shifts reflect ensemble-averaged chemical properties of a nucleus. For this reason, it is necessary to move beyond analysis based on calculations with single static structures and to examine the extent to which the conformational diversity present in the ensemble in solution could reproduce the structural diversity probed by the NMR spectroscopy experiment in solution. To this end ensembles were generated from crystal structures and from molecular dynamics (MD) trajectories.
Ensemble of static crystal structures
An ensemble of static structures was generated using structures available in the Protein Data Bank (Table S2). These different structures were obtained from different crystallization conditions, with variants, or in the presence of ligands. When the number of existing crystal structures for any one protein of interest was less than 5, models solved by NMR spectroscopy were included in the ensemble. The total number of structures used to create the ensemble of each protein (average Cα-RMSD to the representative static structure shown in parenthesis) was: 5 (1.5 Å) for ACBP, 4 (0.8 Å) for A.v. Pc, 8 (0.5 Å) for P.l. Pc, 14 (0.3 Å) for CexCD and 36 (0.9 Å) for SNase.
Use of these ensembles of static structures improved quantitative agreement between the experimentally observed and calculated CSPs (Table S1, Figures 3 and S3-S8). Improvement in RMSD between the observed and calculated Δδef was on average 0.02 ppm (0.01 ppm) for the PBE model and the 15N (1HN) chemical shift dataset. The corresponding improvement for calculations with Coulomb's law was 0.04 ppm (0.02 ppm). The largest improvement was for ACBP, 0.03 ppm for both 15N and 1HN datasets, whose structures show the largest diversity, with an average RMSD of 1.46 Å. These results, similar to those reported by Best et al.49, highlight the importance of accurate accounting of protein dynamics in making comparisons with ensemble-averaged experimental data. Although the improvement is quite modest in our case, the results suggest that even a modest number of structures determined under different conditions can improve the agreement with NMR chemical shift experiments although the improvement is quite modest in our case.
Ensemble from MD trajectory
Ensembles of MD structures were generated from 20 ns trajectories in which the ionizable groups of interest were kept in the neutral state. The average RMSD between the generated conformations and the initial static structures were: 2.18 Å for ACBP, 2.43 Å for A.v. Pc, 3.61 Å for P.l. Pc, 1.74 Å for hGRX, 1.52 Å for CexCD, and 1.46 Å for SNase.
Use of ensembles of MD-generated proteins did not improve quantitative agreement between experimentally measured and calculated secondary titrations with the PBE model or with Coulomb's law (Table S1, Figures 3 and S2-S8). The RMSD between observed and calculated CSPs increased on average by 0.01 ppm (0.01 ppm) using the PBE model and 15N (or 1HN) chemical shift dataset, and 0.01 ppm (0.01 ppm) using Coulomb's model. The inability of the MD-generated conformational ensembles to reproduce ensemble-averaged experimental data may originate with differences in time scales between the NMR experiment and the relatively short MD simulations, or with the inherent inability of MD simulations to sample relevant conformations far from the crystal structure. Whereas chemical shifts are averaged by dynamic processes on the microsecond time-scale, the MD trajectories were much shorter and consequently not able to account for the true ensemble of the protein in solution, as reflected in the NMR spectroscopy data.
Microscopic modeling
The response of permanent and induced dipoles to the gain or loss of charge is not homogenous throughout a protein. In continuum electrostatic models this reorganization is supposed to be accounted for implicitly with the protein dielectric constant. The agreement between measured and calculated data can be greatly affected by how this response is handled in the calculations. To examine this question, the reorganization of permanent dipoles was modeled explicitly by generating two separate ensembles of MD conformations. In one of the ensembles the ionizable group of interest was treated as being charged and in the other one it was assigned the neutral state. Using the ensembles in both states, Δδef values were calculated using Coulomb's law and Buckingham's formula and then compared against the experimental results. This is a crude approach to microscopic modeling of structural reorganization in response to ionization events.
With this more microscopic approach it was more difficult to reproduce the experimental data with structure-based calculations. The RMSD for 15N or 1HN data increased by 0.12 ppm or 0.02 ppm, respectively, relative to the calculations with single static structures. A more detailed microscopic approach was not pursued, as this is an open-ended problem in sampling that probably reflects differences in the conformational reorganization sampled in solution and in silico. The calculations that were performed were detailed enough to reveal how challenging it will be to reproduce the propagation of electric fields inside the anisotropic and heterogeneous interior of proteins using detailed atomistic methods devoid of any implicit parameters, such as dielectric constants, to account for attenuation of electric fields by polarization of the protein.
Discussion
Structure-based computation of electrostatic effects in proteins is challenging because quantitative treatment of the dielectric response of proteins is not straightforward. Most models treat at least some parts of the protein-water system as a dielectric continuum and use dielectric constants to capture implicitly the effects of polarization. The problem is that the magnitude of the dielectric constants used in the different models is not transferable because these are simply ad hoc empirical parameters without any clear physical meaning.
The dielectric constant measured in parallel plate capacitor experiments using dry protein powders (εpowder) is 2 in the high-frequency limit, and closer to 4 in the equilibrium regime and in the presence of a few water molecules per protein50. In contrast, εp is usually assigned values of 6 to 20 in PBE calculations to maximize agreement between experimental and calculated thermodynamic data (e.g. pKa values, redox potentials, pH dependence of ΔG)10,13,48,51-58. The difference between εpowder = 2 to 4 and εp = 6 to 20 is due mainly to two factors: 1) in dehydrated proteins all dynamic relaxation processes are suppressed and we therefore have a muted dielectric response, and 2) εp is found by maximizing the agreement with experimental thermodynamic properties that are influenced by many factors in addition to the electric field. The εp = 6 to 20 value are high because they account implicitly for all forms of dynamic, structural reorganization that are usually not treated explicitly in a computational model. In the case of thermodynamic data describing ionization events, the local structural reorganization is likely significant as indicated by the high dielectric constants needed to reproduce the experimental values. The correct explicit modeling of these reorganization events present a formidable challenge for quantitative structure-based energy calculations.
The experimental value of εp = 3 as determined here by analysis of CSPs, is significant because this dielectric constant likely represents a value closer to a general εp (i.e. the dielectric constant within a PBE framework that describes only the electric field) of proteins in solution. This value was obtained by analysis of the behavior of several proteins, with different folds, and with different structural and functional properties. This suggests that εp = 3 represent a general, intrinsic and probably transferable property of proteins with a size and dynamic characteristics similar to the set of proteins studied here. The similarity with the values of εp = 2 to 4 measured with dry protein powders suggests that it reflects primarily the polarization of electrons and contributions from fluctuations of permanent dipoles although this agreement could be coincidental.
The difference between the value of εp measured in this study using the propagation of electric fields and the value of εp obtained through reverse pKa calculations, reflects differences in the goals and the physical processes monitored in these two different types of experiments. The goal of a reverse pKa calculation is to identify the value εp that reproduces an experimental pKa value. In such an analysis, anything that affects the pKa and is not treated explicitly in the model becomes absorbed in the dielectric constant. Because ionizable side chains (the reporters of pKa values) are located in solvent-exposed regions that can undergo considerable structural reorganization, the εp values they report reflect the substantial structural heterogeneity and reorganization experienced by ionizable side chains when they titrate. εp values obtained by analysis of pKa values therefore reflect dynamic properties of proteins and implicitly describe non-electrostatic effects that cause significant perturbation of pKa values. In contrast, the sensors of the charge-dipole interactions examined in this study are the non-titratable backbone amides. Unlike side chains, the mobility of backbone C-N and N-H bonds is much lower and therefore less sensitive to ionization-induced structural changes. The backbone amides themselves do not titrate and it is therefore possible to measure an εp value in the absence of nearby structural reorganization - such a measurement is never possible in a reverse pKa calculation. In fact, the data used in this analysis purposely excluded cases where conformational reorganization could contribute to the CSP, thus the value of εp = 3 obtained by analysis of electric fields reflected in CSPs is likely to reflect more directly an intrinsic εp of proteins, devoid of contributions from structural reorganization, water penetration, or any other factor that can affect the magnitude of electrostatic effects in proteins. These results suggest that the most accurate method for calculation of electrostatic effects in proteins (with PBE solvers) should use εp = 3 to reproduce implicitly the contributions from electronic polarization and high frequency dipolar fluctuations of the protein matter, with all other dynamic contributions treated explicitly. The latter if of course very challenging because it is difficult to sample correctly all the different microstates that contribute to thermodynamic properties of proteins in solution. It is important to note that the value of εp found here is dependent on the scaling factors used in Buckingham's equation, and although all evidence points to the values used here being quite accurate (see supplementary material), it is nevertheless possible that future findings could change this and thus affect our conclusions on the value of εp.
We used the exquisite sensitivity of 15N and 1HN chemical shifts to the magnitude and direction of electric fields along the highly polarizable C-N and N-H bonds to assess the magnitude of electric fields in 14 proteins. The magnitudes of the measured electric field emanating from a single charge ranged from 1.2 MV/cm to 20.0 MV/cm, with a resolution of 1.2 MV/cm. The lower limit, and hence the sensitivity of the measurements, was determined by the uncertainty of the extracted Δδef from the CSPs (0.1 ppm for 15N and 0.03 ppm for 1HN nuclei47); the upper limit was detected for Leu36 in A.v. Pc (−1.66 ppm). To illustrate the significance of such large variations in the measured electric fields emanating from a single charge, consider how a single charge could affect the rate of a catalytic reaction. If a charge were able to produce electric fields in this range, parallel to a dipolar transition state separating unit charges over a distance of 1Å, the resulting increase on the reaction rate would range from 1.5 (1.2 MV/cm) to more than 2000 fold (20.0 MV/cm).
Conclusions
To reproduce electric fields inside proteins accurately with continuum electrostatics models it is necessary to treat the dielectric properties of proteins accurately. The CSP has exquisite information about the protein dielectric constant that has not been tapped before. Our goal is to determine the range of the dielectric constants, εp in Poisson-Boltzmann electrostatics and εeff in Coulomb's law, that best reproduce the experimental CSPs. The optimal value of εp in the PBE was of particular interest because the value of this quantity and its physical meaning has been the most contentious topic in theoretical calculations of electrostatic interactions in the last three decades18,59.
The results obtained in this study have two important conclusions. (1) A low value of εp = 3 and a narrow range of values of εp from 2 to 5 is optimal for reproducing electric fields in the proteins studied. Because these εp values were obtained by analysis of NMR chemical shift perturbations and not by analysis of thermodynamic variables such as pKa values, which are contaminated by non-electrostatic factors, they likely represent the best estimates of the most general and intrinsic value of the εp of proteins. (2) The differences between results from analysis with PB electrostatics and with a simple Coulomb formalism underscore the importance of screening effects of bulk water, which attenuates the propagation of the electric field inside proteins. This screening effect should always be accounted for in the theoretical models. It is reproduced well with a simple PBE-based model that treats the dielectric response of water and of protein with separate dielectric constants, but it is underestimated by a simple Coulomb expression as shown by the significant over-prediction of CSPs.
Of the observables in NMR spectroscopy, only the chemical shift is sensitive to long-range electric field effects60. Therefore, the optimal values of εp and εeff obtained from the analysis of CSPs cannot be corroborated independently with data from other high-resolution spectroscopic observable, such as J-coupling, residual dipolar coupling, order parameters or nuclear Overhauser effect restraints. The analysis presented here, however, was performed separately for different proteins and using experimental data reported by different nuclei types, 15N and 1HN nuclei, known to report different sensitivities to the different factors that determine pH-dependent CSPs (e.g. Δδm, Δδs and Δδef in Eq. 2)37. In all protein systems studied the results obtained for optimal εp are correlated and therefore validate its narrow range in proteins in general. The almost identical, self-consistent results for optimal values of εp and εeff obtained by analysis of data reported by two nuclei types, 15N and 1HN, further support the validity of this analysis and its conclusions. These results represent yet another application of the NMR chemical shift; the most readily and accurately measured NMR observable that should lead to improved electrostatic calculation methodology and understanding of protein properties and functions in general. They constitute the first general, systematic description of general properties of electric field propagation probed by chemical shifts in proteins.
The observables obtained from NMR spectroscopy experiments are rich in information useful to validate the ability of computational models to account for the ensemble properties of proteins. We therefore probed the ability of ensembles, defined in two different ways, to reproduce the diversity probed by the NMR chemical shift in solution. Ensembles consisting of multiple crystal structures of the same protein (or similar proteins), gave a slightly improved prediction of the measured CSPs. In contrast, conformational averaging generated from MD trajectories was unable to capture the dynamics reflected in experimental data acquired on the NMR time scale. These results suggest that standard MD simulations should be used with caution for this purpose.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by an SFI PIYR award (04/YI1/M537) to J.E.N., an SFI Research Frontiers Programme award (08/RFP/BIC1140) to J.E.N., a PhD scholarship from The Irish Research Council for Science Engineering and Technology (IRCSET) Graduate Research Education Programme (GREP) to P.K, a grant from the Danish Natural Science Research Council (272-060251) to K.T and grant GM073838 from the National Institutes of Health USA to B.G-M. E. NMR instrument support (L.P.M.) was provided by the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, the British Columbia Knowledge Development Fund, the UBC Blusson Fund, and the Michael Smith Foundation for Health Research. The authors are grateful for experimental data provided by Peter B. Crowley, Jens Jørgen Led, Kazumasa Sakurai and Yuji Goto.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Supporting Materials and Mehods; Supporting Results; Figure S1. Example of pH-dependent CSP on an amide group; Figure S2-S7. Structural distribution of CSPs in six proteins; Figure S8. Optimal values of εp and εeff using cumulative 15N and 1HN CSPs measured in all proteins; Table S1. RMSD between experimental and calculated Δδef; Table S2. Sets of crystal structures used to ensembles of static structures. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Warshel A, Florian J. Proc. Natl. Acad. Sci. USA. 1998;95:5950–5955. doi: 10.1073/pnas.95.11.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varadarajan R, Zewert TE, Gray HB, Boxer SG. Science. 1989;243:69–72. doi: 10.1126/science.2563171. [DOI] [PubMed] [Google Scholar]
- 3.Luecke H, Richter HT, Lanyi JK. Science. 1998;280:1934–1937. doi: 10.1126/science.280.5371.1934. [DOI] [PubMed] [Google Scholar]
- 4.Doyle D, Cabral JM, Pfuetzner R, Kuo A, Gulbis J, Cohen S, Chait B, MacKinnon R. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 5.Nielsen JE, Andersen KV, Honig B, Hooft RW, Klebe G, Vriend G, Wade RC. Protein Eng. 1999;12:657–662. doi: 10.1093/protein/12.8.657. [DOI] [PubMed] [Google Scholar]
- 6.Nielsen JE, McCammon JA. Protein Sci. 2003;12:313–326. doi: 10.1110/ps.0229903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carstensen T, Farrell D, Huang Y, Baker NA, Nielsen JE. Proteins. 2011;79:3287–3298. doi: 10.1002/prot.23091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warshel A, Russell SQ. Rev. Biophys. 1984;17:283–422. doi: 10.1017/s0033583500005333. [DOI] [PubMed] [Google Scholar]
- 9.Isom D, Castaneda CA, Cannon BR, Velu P, Garcia-Moreno B. Proc. Natl. Acad. Sci. USA. 2010;107:16096–16100. doi: 10.1073/pnas.1004213107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tynan-Connolly BM, Nielsen JE. Protein Sci. 2007;16:239–249. doi: 10.1110/ps.062538707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernandez G, Anderson JS, LeMaster DM. Biochemistry. 2009;48:6482–6494. doi: 10.1021/bi900526z. [DOI] [PubMed] [Google Scholar]
- 12.Churg A, Warshel A. Biochemistry. 1986;25:1675–1681. doi: 10.1021/bi00355a035. [DOI] [PubMed] [Google Scholar]
- 13.Schutz CN, Warshel A. Proteins. 2001;44:400–417. doi: 10.1002/prot.1106. [DOI] [PubMed] [Google Scholar]
- 14.Schneider W, Bernstein H, Pople J. J. Chem. Phys. 1958;28:601–607. [Google Scholar]
- 15.Buckingham AD. Can. J. Chem. 1960;38:300–307. [Google Scholar]
- 16.LeMaster DM, Minnich M, Parsons PJ, Anderson JS, Hernández G. J. Inorg. Biochem. 2006;100:1410–1412. doi: 10.1016/j.jinorgbio.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 17.Boyd J, Domene C, Redfield C, Ferraro MB, Lazzeretti P. J. Am. Chem. Soc. 2003;125:9556–9557. doi: 10.1021/ja034855y. [DOI] [PubMed] [Google Scholar]
- 18.Kukic P, Farrell D, Søndergaard CR, Bjarndóttir U, Bradley J, Pollastri G, Nielsen JE. Proteins. 2010;78:971–984. doi: 10.1002/prot.22621. [DOI] [PubMed] [Google Scholar]
- 19.Hass MA, Jensen MR, Led JJ. Proteins. 2008;72:333–343. doi: 10.1002/prot.21929. [DOI] [PubMed] [Google Scholar]
- 20.Webb LJ, Boxer SG. Biochemistry. 2008;47:1588–1598. doi: 10.1021/bi701708u. [DOI] [PubMed] [Google Scholar]
- 21.Suydam IT, Snow CD, Pande VS, Boxer SG. Science. 2006;313:200–204. doi: 10.1126/science.1127159. [DOI] [PubMed] [Google Scholar]
- 22.Pearson J, Oldfield E, Lee F, Warshel A. J. Am. Chem. Soc. 1993;115:6851–6861. [Google Scholar]
- 23.Perkins SJ, Dower SK, Gettins P, Wain-Hobson S, Dwek RA. Biochem. J. 1977;165:223–225. doi: 10.1042/bj1650223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williamson MP, Asakura T. J. Magn. Reson. B. 1993;101:63–71. [Google Scholar]
- 25.de Dios A, Laws D, Oldfield E. J. Am. Chem. Soc. 1994;116:7784–7786. [Google Scholar]
- 26.Anderson JS, Hernandez G, LeMaster DM. Biophys. Chem. 2009;141:124–130. doi: 10.1016/j.bpc.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 27.Thomsen J, Kragelund B, Teilum K, Knudsen J, Poulsen F. J. Mol. Biol. 2002;318:805–814. doi: 10.1016/S0022-2836(02)00159-6. [DOI] [PubMed] [Google Scholar]
- 28.Jensen K, Winther J, Teilum K. J. Am. Chem. Soc. 2011;133:3034–3042. doi: 10.1021/ja1096539. [DOI] [PubMed] [Google Scholar]
- 29.Tollinger M, Forman-Kay J, Kay L. J. Am. Chem. Soc. 2002;124:5714–5717. doi: 10.1021/ja020066p. [DOI] [PubMed] [Google Scholar]
- 30.Augspurger J, Pearson J, Oldfield E, Dykstra C, Park KD, Schwartz D. J. Magn. Reson. 1992;100:342–357. [Google Scholar]
- 31.Batchelor J. J. Am. Chem. Soc. 1975;97:3410–3415. [Google Scholar]
- 32.Baker NA, Sept D, Holst MJ, McCammon JA. IBM J. Res. Dev. 2001;45:427–438. [Google Scholar]
- 33.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahoney M, Jorgensen W. J. Chem. Phys. 2000;112:8910–8922. [Google Scholar]
- 35.Berendsen H, Postma J, van Gunsteren W, Di Nola A, Haak J. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 36.Sakurai K, Goto Y. Proc. Natl. Acad. Sci. USA. 2007;104:15346–15351. doi: 10.1073/pnas.0702112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomlinson JH, Green VL, Baker PJ, Willamson MP. Proteins. 2010;78:3000–3016. doi: 10.1002/prot.22825. [DOI] [PubMed] [Google Scholar]
- 38.Crowley PB, Otting G, Schlarb-Ridley, Canters BG, Ubbink M. J. Am. Chem. Soc. 2001;123:10444–10453. doi: 10.1021/ja0112700. [DOI] [PubMed] [Google Scholar]
- 39.Crowley P, Matias P, Khan A, Roessle M, Svergun D. Chem. Eur. J. 2009;15:12672–12680. doi: 10.1002/chem.200901410. [DOI] [PubMed] [Google Scholar]
- 40.Crowley P, Matias P, Mi H, Firbank S, Banfield M, Dennison C. Biochemistry. 2008;47:6583–6589. doi: 10.1021/bi800125h. [DOI] [PubMed] [Google Scholar]
- 41.McIntosh LP, Naito D, Baturin SJ, Okon M, Joshi M, Nielsen JE. J. Biomol. NMR. 2011;51:5–19. doi: 10.1007/s10858-011-9537-x. [DOI] [PubMed] [Google Scholar]
- 42.Schubert M, Poon D, Wicki J, Tarling C, Kwan E, Nielsen J, Withers S, McIntosh L. Biochemistry. 2007;46:7383–7395. doi: 10.1021/bi700249m. [DOI] [PubMed] [Google Scholar]
- 43.Castañeda C, Fitch C, Majumdar A, Khangulov V, Schlessman J, García-Moreno B. Proteins. 2009;77:570–588. doi: 10.1002/prot.22470. [DOI] [PubMed] [Google Scholar]
- 44.White A, Tull D, Johns K, Withers S, Rose D. Nat. Struct. Biol. 1996;3:149–154. doi: 10.1038/nsb0296-149. [DOI] [PubMed] [Google Scholar]
- 45.Farrell D, Sa-Miranda E, Webb H, Georgi N, Nielsen JE. Proteins. 2010;78:843–857. doi: 10.1002/prot.22611. [DOI] [PubMed] [Google Scholar]
- 46.Webb H, Tynan-Connolly BM, Lee GM, Farrell D, O'Meara F, Søndergaard CR, Teilum K, Hewage C, McIntosh LP, Nielsen JE. Proteins. 2011;79:685–702. doi: 10.1002/prot.22886. [DOI] [PubMed] [Google Scholar]
- 47.Bartik K, Redfield C, Dobson CM. Biophys. J. 1994;66:1180–1184. doi: 10.1016/S0006-3495(94)80900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Antosiewicz J, McCammon J, Gilson M. J. Mol. Biol. 1994;238:415–436. doi: 10.1006/jmbi.1994.1301. [DOI] [PubMed] [Google Scholar]
- 49.Best R, Lindorff-Larsen K, DePristo M, Vendruscolo M. Proc. Natl. Acad. Sci. USA. 2006;103:10901–10906. doi: 10.1073/pnas.0511156103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bone S, Pethig R. J. Mol. Biol. 1985;181:323–326. doi: 10.1016/0022-2836(85)90096-8. [DOI] [PubMed] [Google Scholar]
- 51.Nielsen JE, Vriend G. Proteins. 2001;43:403–412. doi: 10.1002/prot.1053. [DOI] [PubMed] [Google Scholar]
- 52.Demchuk E, Wade RC. J. Phys. Chem. 1996;100:17373–17387. [Google Scholar]
- 53.Karshikoff A. Protein Eng. 1995;8:243–248. doi: 10.1093/protein/8.3.243. [DOI] [PubMed] [Google Scholar]
- 54.Yang AS, Gunner MR, Sampogna R, Sharp K, Honig B. Proteins. 1993;15:252–265. doi: 10.1002/prot.340150304. [DOI] [PubMed] [Google Scholar]
- 55.Alexov EG, Gunner MR. Biophys. J. 1997;72:2075–2093. doi: 10.1016/S0006-3495(97)78851-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bashford D, Karplus M. Biochemistry. 1990;29:10219–10225. doi: 10.1021/bi00496a010. [DOI] [PubMed] [Google Scholar]
- 57.Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A. Nucleic Acids Res. 2005;33:W368–371. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Warwicker J. Protein Sci. 1999;8:418–425. doi: 10.1110/ps.8.2.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Warshel A, Sharma P, Kato M, Parson W. Biochim. Biophys. Acta. 2006;1764:1647–1676. doi: 10.1016/j.bbapap.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 60.Osapay K, Case D. J. Am. Chem. Soc. 1991;113:9436–9444. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.