Abstract
The highly virulent nature of Ebola virus, evident from the 2014 West African pandemic, highlights the need to develop vaccines or therapeutic agents that limit the pathogenesis and spread of this virus. While vaccines represent an obvious approach, targeting virus interactions with host proteins that critically regulate the virus lifecycle also represent important therapeutic strategies. Among Ebola virus proteins at this critical interface is its matrix protein, VP40, which is abundantly expressed during infection and plays a number of critical roles in the viral lifecycle. In addition to regulating viral transcription, VP40 coordinates virion assembly and budding from infected cells. Details of the molecular mechanisms underpinning these essential functions are currently being elucidated, with a particular emphasis on its interactions with host proteins that control virion assembly and egress. This review focuses on the strategies geared toward developing novel therapeutic agents that target VP40-specific control of host functions critical to virion transcription, assembly and egress.
Keywords: antiviral, CTD, Ebola, ESCRT, filovirus, L-domain, NTD, oligomerization, therapeutic agent, viral budding, virus-like particles, VLPs, VP40
The Filoviridae family is composed of the genera Ebola virus and Marburgvirus. The genus Marburgvirus includes one species, Marburg marburgvirus, populated by two viral strains: Marburg virus (MARV) and Ravn virus. There are five recognized species of the genus Ebola virus: Zaire Ebola virus (EBOV – previously known as ZEBOV), Sudan Ebola virus, Reston Ebola virus (RESTV), Taï Forest Ebola virus and Bundibugyo Ebola virus. A third genus, Cuevavirus, containing the species Lloviu cuevavirus and the virus Lloviu virus has been proposed and approved and awaits ratification by the International Committee on Taxonomy of Viruses [1].
With the exception of RESTV, human infection by the various Ebola virus and Marburgvirus species can lead to a syndrome with reported mortality rates as high as 90% [2]. This syndrome is characterized initially by malaise, body temperatures greater than 38.3°C (100.9°F), vomiting, and diarrhea. The virus can infect many organ systems and the adrenal gland, and the disease can progress to include respiratory distress and hypotension [3]. In many cases, viremia triggers an overwhelming inflammatory response that leads to profound hypovolemic shock, although death can sometimes be mitigated by intensive supportive therapy [4,5].
The ongoing West African EBOV pandemic, which has a reported case–fatality rate of 49% as of 19 October 2014 [6], highlights the global significance of this zoonotic pathogen. The current outbreak has devastated local healthcare communities and infrastructure [7] and the global community is now grappling with the consequences of living in an extensively interconnected world, where local outbreaks of disease may not remain scattered and isolated. Effective containment and control of the pandemic have been notably hindered in part by a lack of US FDA-approved vaccines and antifiloviral therapeutic agents. Given the many logistical constraints on the implementation of effective vaccination campaigns in underdeveloped countries, and the propensity for viruses to rapidly evade vaccine-induced immunity, it is critical that efforts also focus on developing therapeutic agents for postexposure and postinfection control of spread as a critical component of any global filoviral containment strategy [8]. As with any pathogen, our ability to devise vaccines or therapeutic agents hinges on first developing a fundamental understanding of the biology of the pathogen.
Indeed, we have learned much in recent years about the structure and biology of filoviruses. EBOV is an enveloped, negative-sense RNA virus which encodes seven structural proteins within its 19kb linear genome including nucleoprotein (NP), VP24, VP30, VP35, VP40, polymerase (L) and membrane-anchored glycoprotein (GP) as well as the nonstructural secreted glycoprotein (sGP) and small secreted glycoprotein (ssGP) [9]. Nucleoprotein, VP30, VP35 and L are constituents of the viral nucleocapsid and are essential for transcription and viral replication [10]. The VP24 protein has varied documented roles, including interferon antagonism, transcriptional and replication control during infection, and minor matrix protein function [11–16]. GP coordinates viral entry into host cells, in part, through the Niemann–Pick C1 internal receptor [17,18]. As the sole protein located on the viral membrane, GP is visible to the host immune system and has therefore been the target of many current vaccine efforts and antifiloviral therapeutic strategies. Viral proteins, in addition to GP, have been shown to enhance immune responses to EBOV, and thus may be important components of future vaccines [19,20]. Some of these efforts have shown promise in nonhuman primate models [21–32]; however, to date none have definitively demonstrated any preventative or therapeutic efficacy in humans.
VP40 (viral protein 40 kDa) is the most abundantly expressed filoviral protein and coordinates virion assembly at the plasma membrane (PM) through interactions with both viral and cellular components [10,33–34]. Indeed, exogenous expression of VP40 alone is sufficient to induce the formation of filamentous virus-like particles (VLPs) in mammalian cells (Figure 1) [35–38]. As EBOV is an NIAID Category A pathogen which must be studied under BSL-4 conditions, VP40-mediated VLP expression systems have become an essential tool for interrogating mechanisms of both EBOV and MARV virion formation under BSL-2 conditions. Indeed, by studying the mechanisms of VP40-mediated VLP formation, we have made important progress toward unraveling the molecular mechanisms of filovirus virion assembly and budding. While many similarities exist between EBOV VP40 and MARV VP40 [39–44], differences in amino acid sequence do exist [45,46] that could impact the approach to VP40-specific therapeutics. For example, EBOV VP40 has two overlapping L-domain motifs, 7PTAPPEY13, whereas MARV VP40 has one, 16PPPY19. Consequently, this review will focus on mechanisms by which EBOV VP40 controls important functions in the viral life cycle and current therapeutic strategies aimed at disrupting these functions.
Figure 1. Expression of Ebola virus VP40 results in VLP formation and egress.
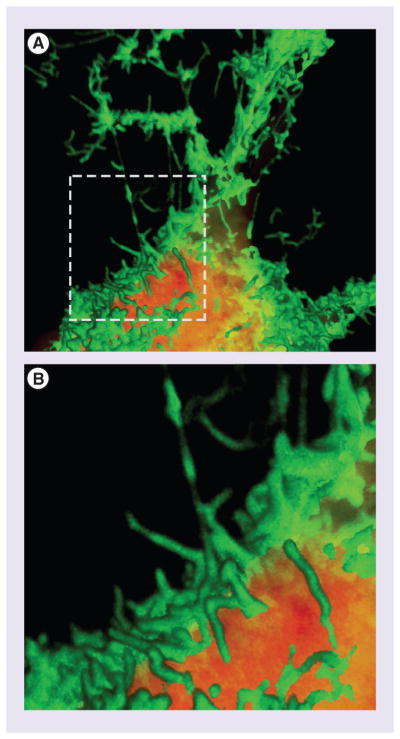
A GFP-VP40 fusion protein expressed inHEK293T cells shows robust virus-like particle assembly and egress at the plasma membrane (A). (B) corresponds to inset indicated in (A). Upper panel is approximately 21 μm wide. The cytoplasm is stained with HCS CellMask™ Deep Red. Images were rendered using Volocity software. This image was generated by the Freedman laboratory in the PennVet Imaging Core Facility.
VP40 structure & virion/VLP assembly
VP40 is a 326 amino acid protein (Figure 2A), and is the major matrix protein of EBOV that coordinates virion assembly and budding [33,35,37,47–50]. Crystallographic studies have identified two distinct domains within VP40, including an N-terminal domain (NTD) and a C-terminal domain (CTD) [48] (Figure 2B). NTD interactions are responsible for VP40 dimer formation in the cytoplasm, and CTD interactions are responsible for subsequent trafficking to and interaction with the PM [10,38]. Structural elements in VP40 relevant for therapeutic targeting are highlighted in Table 1. VP40 localization to the PM inner leaflet is important for VLP assembly and budding [10,51–52], and occurs at least in part by trafficking along actin filaments [53,54]. Deep penetration into the PM inner leaflet by a hydrophobic region of the VP40 CTD that includes residues L213, L295 and V298 is essential for appropriate localization, VLP assembly and VLP egress [10,38,55]. At the PM, VP40 forms butterfly-shaped dimers [10,34,38,52,56,57], which reorganize into linear hexamers [38]. These hexamers are essential building blocks of the viral matrix [38] and are critically important for virion assembly and budding.
Figure 2. Ebola virus VP40 with potential therapeutic agent targets highlighted.
(A) Schematic diagram of Ebola virus VP40 showing established or potential therapeutic targets. (B) The asymmetric butterfly-shaped VP40 dimer shown (PDB ID: 4LDB [38]) is adapted from [8] with critical interaction domains highlighted with colors that correspond to the matching sequences shown in (A). The N-terminal domain dimerization interface is comprised of residues 52–65 and 108–117 and is highlighted in pink. The C-terminal domain basic domain, including residues K221, K224, K225, K270, K274 and K275, is highlighted in red. Residues L295, and V298 of the C-terminal domain hydrophobic domain are highlighted in blue. Residues of the recently identified plasma membrane-interacting N-terminal domain loop (residues K127, T129, and N130) are highlighted in yellow. A residue critical for RNA binding of the octameric conformation of VP40 (R134) is highlighted in purple. The crystal structure of the VP40 dimer does not include residues 1–44 and 322–326, therefore L-domain motifs are not represented in (B).
Table 1.
Functional regions of Ebola virus VP40 as potential targets for inhibition.
| VP40 sequence | Function | Effect of inhibition | Ref. |
|---|---|---|---|
| 7PTAP10 | Interaction with ESCRT Tsg101 | Reduce VLP assembly and egress | [40,58,59] |
| 10PPEY13 | Interaction with ESCRT Nedd4 | Reduce VLP assembly and egress | [38,40,60,61] |
| Residues 52–65, 108–117 | NTD dimerization interface | Block VP40 dimer formation | [38] |
| 127KATN130 | NTD loop for PM interaction | Block VP40 PM localization, VLP assembly and egress | [62] |
| R134 | RNA binding of VP40 octamer | Block Ebola virus viral replication | [38,57] |
| K221, K224, K225, K270, K274, K275 | CTD Basic Patch for electrostatic interactions with the PM | Block PM interaction, VLP assembly and egress | [38] |
| 295LDPV298 | CTD Hydrophobic loop for PM insertion | Block VP40 PM localization, VLP assembly and egress | [10,38,56] |
CTD:C-terminal domain; PM: Plasma membrane; NTD: N-terminal domain; VLP: Virus-like particle.
In addition to the hydrophobic CTD region, a recently identified basic CTD region composed of lysine residues (K221, K224, K225, K270, K274 and K275) has also been implicated in VP40-PM association. Some of these positively charged lysines are essential for electrostatic VP40–PM interactions and subsequent matrix assembly and VLP egress [38]. In fact, mutations of K224 and K225 abolish matrix assembly and subsequent VLP egress without disrupting VP40 localization to the PM [38], highlighting the nuanced role of these distinct CTD motifs. Surprisingly, an NTD loop adjacent to the PM-penetrating CTD hydrophobic loop, comprising residues K127, T129 and N130, has also recently been shown to interact with the PM, demonstrating a previously unrecognized role for NTD–PM interactions in efficient VP40-PM localization, oligomerization, matrix assembly and egress [62]. This intriguing development further establishes the critical nature of VP40-PM interactions and underscores how a wide array of functionality can be encoded within this single viral gene product.
The critical role for CTD-dependent VP40 localization and PM interactions in VLP assembly and egress [38,55] establishes these interactions as attractive therapeutic targets that could be targeted to disrupt virus assembly and budding. Indeed, as mutations in the VP40 CTD that inhibit its insertion into the PM significantly reduces VLP formation [55], small molecules that prevent membrane insertion would also be expected to block budding. Indeed, small molecule inhibitors that generally interfere with membrane binding of proteins via lipid-binding domains have been identified [63] and these may represent useful therapeutics [8]. For example, Soni and Stahelin [64] have recently reported that plasma membrane phosphatidylserine (PS) may be an important component of EBOV budding, and that VP40 selectively induces vesiculation of membranes rich in PS such as the inner leaflet of the PM. In addition to targeted inhibition of the hydrophobic VP40–PM interactions, targeting the electrostatic interactions between the CTD and the PM [38] would also likely lead to significant reductions in EBOV matrix assembly and subsequent VLP formation and egress. Finally, small molecules that disrupt functions of the recently identified NTD interacting loop [62] may also inhibit EBOV assembly and egress and warrant additional attention. Taken together, strategies that target hydrophobic and electrostatic CTD (and NTD) VP40 interactions, both between VP40 and the PM and within VP40 oligomers, represent an important avenue for antifiloviral therapeutic research.
Role of VP40 oligomerization in viral transcription via ring formation
One important aspect of VP40 that has profound biological implications is its oligomeric state, as distinct protein conformations control different essential functions during the viral life cycle [38,47,65]. Bornholdt et al. [38] elegantly demonstrated that VP40 exists in a wide range of oligomeric conformations at distinct intracellular locations and, for example, that in addition to plasma membrane-localized VP40 hexamers, there are perinuclear octameric ring conformations [38,41,50,57]. Recent work from a number of groups has established that this octameric form controls virion replication [41,50,57]. NTD interactions alone appear to be sufficient to orchestrate the formation of perinuclear octameric rings as their formation is not affected by deletion of the CTD-domain of VP40 [38,66].
In contrast to the role established for the NTD, it is not clear that CTD plays any role in VP40-mediated control of viral transcription, as a point mutation in the CTD domain (R134A) abrogating VP40 RNA binding does not prevent VP40 dimerization, PM localization or VLP formation and egress [38,57]. Moreover, a distinct mutation (I307R) buried in the CTD interface abrogates PM localization and promotes the formation of perinuclear octameric rings [38]. Thus, while these studies demonstrate that the formation of VP40 octameric ring structures is not absolutely dependent on RNA binding [38], the formation of VP40 octamers is essential for production of replication competent virus in vitro [57]. These findings highlight the need for further efforts focused on how the octameric VP40 controls viral replication.
Importantly, the involvement or requirement for VP40 ring octamers in transcriptional control is distinct from the role of hexamers in matrix assembly and VLP budding functions [38,57]. This relationship between oligomeric structure and function prompts additional questions. For example, although mutations of the RNA-binding site do not abrogate VP40 ring formation, the RNA-binding site in the VP40 ring structure may nonetheless represent an attractive drug target for inhibition of EBOV replication. Indeed, small molecules targeting RNA-binding by HIV-1 Gag matrix protein have been shown to inhibit viral replication [67]. Further exploration of the mechanism that controls formation of the octameric VP40 ring structure will help us better understand how VP40 regulates EBOV replication and these may illuminate the best direction to pursue in our efforts to develop antifiloviral therapeutics that target steps of VP40 oligomerization that critically control virus transcription.
Given that VP40 oligomerization is controlled by complex intermolecular interactions and interactions with cellular membranes, and the fact that different oligomeric forms participate in distinct steps in the EBOV life cycle, small molecules designed to inhibit or modify the formation of oligomeric forms may be useful therapeutics. For example, as the VP40 NTDs coordinate formation of dimers, which are precursors of higher order oligomers, inhibiting NTD-mediated dimerization might effectively halt formation of higher order oligomers. Another strategy, that has not yet been explored, would be to limit VP40 oligomerization by preferentially destabilizing higher order conformations, or conversely stabilizing the dimeric or monomeric form to prevent formation of higher order structures [38]. Although it has been suggested that EBOV may be able to circumvent these strategies by simply increasing VP40 protein production [8], they nonetheless represent an important direction for antifiloviral agent research and may be useful in concert with other therapeutic modalities that target sequential or overlapping functions of VP40.
Role of VP40 in ESCRT protein recruitment for viral assembly & egress
Broadly, RNA viruses are challenging pathogens to control as their error-prone polymerases generate mutations that enable them to evade virus-targeted agents. The development of ‘host-oriented’ therapeutics that target, in part, conserved host as opposed to specific viral functions may therefore represent a more effective antiviral strategy. Among the host mechanisms used by EBOV and other enveloped RNA viruses is the host Endosomal Sorting Complex Required for Transport (ESCRT) pathway [68–70]. The ESCRT pathway is normally involved in protein trafficking and recycling through endosomes, as well as in regulating cytokinesis. However, VP40 hijacks these ESCRT functions, primarily via interactions involving two overlapping N-terminal tetra-peptide motifs known as ‘L-domain’ motifs to efficiently assemble and bud at the PM [35,40]. Specifically, a PTAP motif mapping to VP40 amino acids 7–10 interacts with the host protein Tsg101 to recruit the ESCRT-1 complex, and an overlapping PPEY motif mapping to amino acids 10–13 of VP40 interacts with WW domains of host Nedd4 ubiquitin ligase [40,58,60]. These well-characterized interactions between VP40 and ESCRT proteins are important for efficient VLP and virion formation as each EBOV VP40 L-domain functions independently in virion assembly and egress. Indeed, nonfunctional mutations of both the EBOV VP40 L-domain motifs reduces budding of VSV pseudotypes with EBOV L-domains and live recombinant EBOV by approximately tenfold [59,71].
While abrogation of these virus–host interactions does not fully disrupt virion assembly and egress, small molecules have been identified that inhibit VP40 PTAP-dependent interactions with host ESCRT proteins that are effective blockers of EBOV VP40 VLP egress in vitro [72]. Interestingly, as PPxY domains are more ubiquitously distributed among enveloped RNA virus matrix proteins, small molecules that disrupt PPxY interaction appear to be effective broad-spectrum antiviral agents that block both MARV and EBOV VP40-mediated VLP formation, but also broadly inhibit formation of arenavirus VLPs and budding of live rhabdoviruses in vitro [73]. Most importantly, both the PTAP and PPxY series of budding inhibitors specifically interfere with EBOV VP40–host interactions and exhibit little if any cytotoxicity [72,73]. Together, studies of L-domain mutants and small molecule inhibitors of L-domain interactions validate the idea of targeting L-domain-mediated VP40-ESCRT protein interactions to block EBOV budding and spread.
Given these important roles established for VP40–Tsg101 and VP40–Nedd4 interactions in efficient viral egress, studies with inhibitors and mutants outlined above also demonstrate that other mechanisms play some role in productive viral replication in vitro [71]. Consistent with this idea, a PTAP L-domain VP40 mutant recruits the ESCRT proteins VPS4, VPS28 and VPS37B in vitro [61]. In fact, a distinct L-domain motif (YPx(n)L) identified in other enveloped virus matrix proteins, such as HIV-1 Gag [68,74–81], plays a key role in their assembly and budding. Thus, additional VP40–ESCRT interactions may regulate this residual budding observed for EBOV double L-domain mutants, and these may be important additional therapeutic targets. Ultimately, we may discover that cocktails containing multiple L-domain blockers provide a synergistic clinical benefit, even if individual inhibitors do not fully abrogate EBOV budding alone.
In summary, the development of host-oriented therapeutics has the potential benefit of inhibiting essential pathways required by viruses, as these pathways also control critical cellular functions that viruses have been unable to circumvent. While care must be taken to address issues of cytotoxicity with this class of inhibitors as they may also have some impact on host functions, preliminary efforts suggest that these are effective therapeutic strategies. Nevertheless, ongoing efforts to further characterize filoviral VP40-ESCRT pathway interactions should provide additional insights that will facilitate the development of more potent, selective and broad-spectrum antifiloviral therapeutic compounds. Moreover, given the conserved requirement by EBOV, MARV and a range of other enveloped RNA viruses for viral matrix proteins – ESCRT protein interactions, small molecules that target these interactions may prove effective against both current strains of filoviruses and strains that may emerge as these viruses evolve.
Conclusion & future perspective
The current EBOV outbreak in West Africa and emergence of EBOV in the USA and European countries has highlighted the need for effective vaccines and antifiloviral therapeutic agents. Wide ranging strategies for developing therapeutics are currently being pursued, and some have proven efficacious in nonhuman primate models [21–32]. Among these strategies, those outlined in this review that target VP40 are extremely promising as they interrupt multiple sequential steps of the filoviral life cycle [8]. Antiviral therapeutics directed against these VP40 domains and the functions they orchestrate are proceeding at a rapid pace and ongoing efforts will undoubtedly further inform these strategies.
Recent studies have identified several additional mechanisms of budding that may ultimately prove to be useful therapeutic targets. For example, phosphorylation at VP40 amino acid 13 was recently shown to be regulated by the c-Abl-1 tyrosine kinase, and inhibition of this phosphorylation by the small molecule IkT-001 reduces EBOV production in culture [82]. Approaches that reduce or prevent VP40 phosphorylation represent yet another new therapeutic strategy but, ultimately, combination therapies that target multiple VP40 functions and interactions may prove more effective.
Also, EBOV has demonstrated a remarkable ability to encode a variety of functions within individual gene products [11,13,83–85]. For example, a recent study identified host IQGAP1 as an EBOV VP40 interacting partner that plays a role in efficient egress of VP40 VLPs [86]. As IQGAP1 links EBOV budding to the actin cytoskeleton of the cell, molecules that disrupt viral particle transport may represent an additional family of targets for antifiloviral therapeutics. Our understanding of the mechanisms by which VP40 interacts with host pathways to facilitate EBOV virion assembly and budding have been of tremendous importance to efforts geared toward developing effective antifiloviral therapeutics. Given the extensive conformational plasticity of the EBOV VP40 protein, it would not be surprising to discover additional mechanisms by which VP40 co-opts normal host pathways to facilitate viral assembly and egress and these might be revealed by unbiased high-throughput efforts to define the range of host genes that modulate VLP budding or production of live virus.
Additionally, pivotal questions about the functional implications of VP40’s 3D structure remain unanswered. For example, though VP40 octamers are essential for productive EBOV replication in culture [57], we know little about the mechanisms that control their formation. Also, while RNA binding seems dispensable for the formation of these octameric structures, RNA binding is critical for regulating viral transcription [38]. Efforts to better understand the mechanisms of VP40 octamer formation and their role in productive EBOV replication may reveal additional targets or strategies for regulating this virus.
Ultimately, while results from initial in vitro experiments with PTAP and PPxY inhibitors have been extremely promising [72,73], animal studies utilizing these inhibitors have not yet been performed, so we do not know yet whether these compounds will prevent budding and spread of virus in vivo. However, the potential application of L-domain interaction inhibitors for more ‘broad-spectrum’ control of enveloped RNA virus infections is tantalizing. Further research efforts to refine and optimize these agents for clinical use will be timely, particularly in light of the ongoing, and potential future, EBOV pandemics.
Executive summary.
VP40 is a critical target for novel antifiloviral therapeutic agent development
VP40 is the most abundantly expressed viral protein during filoviral infection.
Exogenous expression of VP40 alone is sufficient for the formation and budding of virus-like particles (VLPs) in vitro under BSL-2 conditions.
VP40 performs a number of essential functions during EBOV infection, including regulation of viral transcription and coordination of virion assembly and budding.
VP40 exists in multiple conformational states during EBOV infection
-
Role of VP40 oligomerization in viral transcription via ring formation:
VP40 exists primarily in a dimeric conformation in the cytoplasm;
N-terminal domain (NTD) interactions are responsible for coordinating dimer formation and perinuclear ring formation;
C-terminal domain (CTD) interactions are responsible for coordinating higher order oligomerization of VP40 dimers;
Higher order oligomers have different functions in the viral life cycle: linear hexamers are responsible for subsequent matrix filament assembly and virion formation at the plasma membrane, while octamers form perinuclear ring structures and regulate viral transcription;
Inhibiting VP40 oligomerization represents a viable mechanism for antifiloviral therapeutic agent development.
VP40 interacts directly with the plasma membrane to promote virion assembly
VP40 NTD and CTD hydrophobic loops insert into the plasma membrane, positioning VP40 for subsequent VLP assembly.
VP40 CTD basic residues interact electrostatically with the plasma membrane to coordinate matrix filament assembly.
Inhibiting VP40–PM interactions represents a promising approach for antifiloviral therapeutics.
VP40 recruits host Endosomal Sorting Complex Required for Transport family members to mediate efficient virion assembly & egress
-
Role of VP40 in Endosomal Sorting Complex Required for Transport protein recruitment for viral assembly and egress:
VP40 has two N-terminal overlapping late domain (‘L-domain’) motifs which function to recruit host Endosomal Sorting Complex Required for Transport proteins to mediate efficient virion assembly and egress;
The VP40 PTAP L-domain motif (amino acids 7–10) recruits host Tsg101;
The VP40 PPxY L-domain motif (amino acids 10–13) recruits host Nedd4;
Small molecules that inhibit L-domain-mediated virus–host interactions reduce virus assembly and egress.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest;
•• of considerable interest
- 1.Bukreyev AA, Chandran K, Dolnik O, et al. Discussions and decisions of the 2012–2014 International Committee on Taxonomy of Viruses (ICTV) Filoviridae Study Group, January 2012–June 2013. Arch Virol. 2014;159(4):821–830. doi: 10.1007/s00705-013-1846-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Team WER. Ebola virus disease in West Africa – the first 9 months of the epidemic and forward projections. N Engl J Med. 2014;371(16):1481–1495. doi: 10.1056/NEJMoa1411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoenen T, Groseth A, Falzarano D, Feldmann H. Ebola virus: unravelling pathogenesis to combat a deadly disease. Trends Mol Med. 2006;12(5):206–215. doi: 10.1016/j.molmed.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Takada A, Kawaoka Y. The pathogenesis of Ebola hemorrhagic fever. Trends Microbiol. 2001;9(10):506–511. doi: 10.1016/s0966-842x(01)02201-6. [DOI] [PubMed] [Google Scholar]
- 5.Kreuels B, Wichmann D, Emmerich P, et al. A case of severe Ebola virus infection complicated by Gram-negative septicemia. N Engl J Med. 2014;371(25):2394–2401. doi: 10.1056/NEJMoa1411677. [DOI] [PubMed] [Google Scholar]
- 6.ProMed Mail. PRO/AH/EDR> Ebola virus disease – West Africa (196): WHO, Mali conf case ex Guinea, Liberia (Promed Mail Post, 2014–2010–24 00:49:04) http://promedmail.org.
- 7.Wolz A. Face to face with Ebola – an emergency care center in Sierra Leone. N Engl J Med. 2014;371(12):1081–1083. doi: 10.1056/NEJMp1410179. [DOI] [PubMed] [Google Scholar]
- 8••.Stahelin RV. Could the Ebola virus matrix protein VP40 be a drug target? Expert Opin Ther Targets. 2014;18(2):115–120. doi: 10.1517/14728222.2014.863877. Review of potential therapeutic targets within Ebola virus (EBOV) VP40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehedi M, Falzarano D, Seebach J, et al. A new Ebola virus nonstructural glycoprotein expressed through RNA editing. J Virol. 2011;85(11):5406–5414. doi: 10.1128/JVI.02190-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10•.Adu-Gyamfi E, Soni SP, Xue Y, Digman MA, Gratton E, Stahelin RV. The Ebola virus matrix protein penetrates into the plasma membrane: a key step in viral protein 40 (VP40) oligomerization and viral egress. J Biol Chem. 2013;288(8):5779–5789. doi: 10.1074/jbc.M112.443960. Demonstrates importance of EBOV VP40 CTD insertion into plasma membrane. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han Z, Boshra H, Sunyer JO, Zwiers SH, Paragas J, Harty RN. Biochemical and functional characterization of the Ebola virus VP24 protein: implications for a role in virus assembly and budding. J Virol. 2003;77(3):1793–1800. doi: 10.1128/JVI.77.3.1793-1800.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noda T, Halfmann P, Sagara H, Kawaoka Y. Regions in Ebola virus VP24 that are important for nucleocapsid formation. J Infect Dis. 2007;196(Suppl 2):S247–S250. doi: 10.1086/520596. [DOI] [PubMed] [Google Scholar]
- 13.Watanabe S, Noda T, Halfmann P, Jasenosky L, Kawaoka Y. Ebola virus (EBOV) VP24 inhibits transcription and replication of the EBOV genome. J Infect Dis. 2007;196(Suppl 2):S284–S290. doi: 10.1086/520582. [DOI] [PubMed] [Google Scholar]
- 14.Mateo M, Carbonnelle C, Martinez MJ, et al. Knockdown of Ebola virus VP24 impairs viral nucleocapsid assembly and prevents virus replication. J Infect Dis. 2011;204(Suppl 3):S892–S896. doi: 10.1093/infdis/jir311. [DOI] [PubMed] [Google Scholar]
- 15.Zhang AP, Bornholdt ZA, Liu T, et al. The Ebola virus interferon antagonist VP24 directly binds STAT1 and has a novel, pyramidal fold. PLoS Pathog. 2012;8(2):e1002550. doi: 10.1371/journal.ppat.1002550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watt A, Moukambi F, Banadyga L, et al. A novel life cycle modeling system for Ebola virus shows a genome length-dependent role of VP24 in virus infectivity. J Virol. 2014;88(18):10511–10524. doi: 10.1128/JVI.01272-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carette JE, Raaben M, Wong AC, et al. Ebola virus entry requires the cholesterol transporter Niemann–Pick C1. Nature. 2011;477(7364):340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Côté M, Misasi J, Ren T, et al. Small molecule inhibitors reveal Niemann–Pick C1 is essential for Ebola virus infection. Nature. 2011;477(7364):344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson JA, Bray M, Bakken R, Hart MK. Vaccine potential of Ebola virus VP24, VP30, VP35, and VP40 proteins. Virology. 2001;286(2):384–390. doi: 10.1006/viro.2001.1012. [DOI] [PubMed] [Google Scholar]
- 20.Warfield KL, Swenson DL, Olinger GG, Kalina WV, Aman MJ, Bavari S. Ebola virus-like particle-based vaccine protects nonhuman primates against lethal Ebola virus challenge. J Infect Dis. 2007;196(Suppl 2):S430–S437. doi: 10.1086/520583. [DOI] [PubMed] [Google Scholar]
- 21.Warren TK, Warfield KL, Wells J, et al. Advanced antisense therapies for postexposure protection against lethal filovirus infections. Nat Med. 2010;16(9):991–994. doi: 10.1038/nm.2202. [DOI] [PubMed] [Google Scholar]
- 22.Warren TK, Warfield KL, Wells J, et al. Antiviral activity of a small-molecule inhibitor of filovirus infection. Antimicrob Agents Chemother. 2010;54(5):2152–2159. doi: 10.1128/AAC.01315-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Basu A, Li B, Mills DM, et al. Identification of a small-molecule entry inhibitor for filoviruses. J Virol. 2011;85(7):3106–3119. doi: 10.1128/JVI.01456-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iversen PL, Warren TK, Wells JB, et al. Discovery and early development of AVI-7537 and AVI-7288 for the treatment of Ebola virus and Marburg virus infections. Viruses. 2012;4(11):2806–2830. doi: 10.3390/v4112806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madrid PB, Chopra S, Manger ID, et al. A systematic screen of FDA-approved drugs for inhibitors of biological threat agents. PLoS ONE. 2013;8(4):e60579. doi: 10.1371/journal.pone.0060579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warren TK, Wells J, Panchal RG, et al. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature. 2014;508(7496):402–405. doi: 10.1038/nature13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ansari AA. Clinical features and pathobiology of Ebola virus infection. J Autoimmun. 2014;55:1–9. doi: 10.1016/j.jaut.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 28.Marzi A, Feldmann H, Geisbert TW, Falzarano D. Vesicular stomatitis virus-based vaccines for prophylaxis and treatment of filovirus infections. J Bioterror Biodef. 2011;S1(4) doi: 10.4172/2157-2526.S1-004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marzi A, Engelmann F, Feldmann F, et al. Antibodies are necessary for rVSV/ZEBOV-GP-mediated protection against lethal Ebola virus challenge in nonhuman primates. Proc Natl Acad Sci USA. 2013;110(5):1893–1898. doi: 10.1073/pnas.1209591110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marzi A, Feldmann H. Ebola virus vaccines: an overview of current approaches. Expert Rev Vaccines. 2014;13(4):521–531. doi: 10.1586/14760584.2014.885841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu X, Wong G, Audet J, et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature. 2014;514(7520):47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi JH, Croyle MA. Emerging targets and novel approaches to Ebola virus prophylaxis and treatment. Bio Drugs. 2013;27(6):565–583. doi: 10.1007/s40259-013-0046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33•.Timmins J, Scianimanico S, Schoehn G, Weissenhorn W. Vesicular release of ebola virus matrix protein VP40. Virology. 2001;283(1):1–6. doi: 10.1006/viro.2001.0860. Demonstrates importance of VP40 for virus-like particle (VLP) formation. [DOI] [PubMed] [Google Scholar]
- 34.Adu-Gyamfi E, Digman MA, Gratton E, Stahelin RV. Investigation of Ebola VP40 assembly and oligomerization in live cells using number and brightness analysis. Biophys J. 2012;102(11):2517–2525. doi: 10.1016/j.bpj.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35••.Harty RN, Brown ME, Wang G, Huibregtse J, Hayes FP. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc Natl Acad Sci USA. 2000;97(25):13871–13876. doi: 10.1073/pnas.250277297. Demonstrates role for VP40 PPxY motif during EBOV VLP assembly and budding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geisbert TW, Jahrling PB. Differentiation of filoviruses by electron microscopy. Virus Res. 1995;39(2–3):129–150. doi: 10.1016/0168-1702(95)00080-1. [DOI] [PubMed] [Google Scholar]
- 37•.Noda T, Sagara H, Suzuki E, Takada A, Kida H, Kawaoka Y. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J Virol. 2002;76(10):4855–4865. doi: 10.1128/JVI.76.10.4855-4865.2002. Demonstrates importance of VP40 for VLP formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38••.Bornholdt ZA, Noda T, Abelson DM, et al. Structural rearrangement of Ebola virus VP40 begets multiple functions in the virus life cycle. Cell. 2013;154(4):763–774. doi: 10.1016/j.cell.2013.07.015. Demonstrates roles for various VP40 conformations at different cellular locations based on elegant X-ray crystallography. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bukreyev AA, Volchkov VE, Blinov VM, Netesov SV. The VP35 and VP40 proteins of filoviruses. Homology between Marburg and Ebola viruses. FEBS Lett. 1993;322(1):41–46. doi: 10.1016/0014-5793(93)81107-b. [DOI] [PubMed] [Google Scholar]
- 40••.Licata JM, Simpson-Holley M, Wright NT, Han Z, Paragas J, Harty RN. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J Virol. 2003;77(3):1812–1819. doi: 10.1128/JVI.77.3.1812-1819.2003. Demonstrates roles for overlapping VP40 L-domain motifs in EBOV VLP assembly and budding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Timmins J, Schoehn G, Kohlhaas C, Klenk HD, Ruigrok RW, Weissenhorn W. Oligomerization and polymerization of the filovirus matrix protein VP40. Virology. 2003;312(2):359–368. doi: 10.1016/s0042-6822(03)00260-5. [DOI] [PubMed] [Google Scholar]
- 42.Jasenosky LD, Kawaoka Y. Filovirus budding. Virus Res. 2004;106(2):181–188. doi: 10.1016/j.virusres.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 43.Yamayoshi S, Kawaoka Y. Mapping of a region of Ebola virus VP40 that is important in the production of virus-like particles. J Infect Dis. 2007;196(Suppl 2):S291–S295. doi: 10.1086/520595. [DOI] [PubMed] [Google Scholar]
- 44.Liu Y, Cocka L, Okumura A, Zhang YA, Sunyer JO, Harty RN. Conserved motifs within Ebola and Marburg virus VP40 proteins are important for stability, localization, and subsequent budding of virus-like particles. J Virol. 2010;84(5):2294–2303. doi: 10.1128/JVI.02034-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Urata S, Noda T, Kawaoka Y, Morikawa S, Yokosawa H, Yasuda J. Interaction of Tsg101 with Marburg virus VP40 depends on the PPPY motif, but not the PT/SAP motif as in the case of Ebola virus, and Tsg101 plays a critical role in the budding of Marburg virus-like particles induced by VP40, NP, and GP. J Virol. 2007;81(9):4895–4899. doi: 10.1128/JVI.02829-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mittler E, Kolesnikova L, Herwig A, Dolnik O, Becker S. Assembly of the Marburg virus envelope. Cell Microbiol. 2013;15(2):270–284. doi: 10.1111/cmi.12076. [DOI] [PubMed] [Google Scholar]
- 47.Stahelin RV. Membrane binding and bending in Ebola VP40 assembly and egress. Front Microbiol. 2014;5:300. doi: 10.3389/fmicb.2014.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dessen A, Volchkov V, Dolnik O, Klenk HD, Weissenhorn W. Crystal structure of the matrix protein VP40 from Ebola virus. EMBO J. 2000;19(16):4228–4236. doi: 10.1093/emboj/19.16.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jasenosky LD, Neumann G, Lukashevich I, Kawaoka Y. Ebola virus VP40-induced particle formation and association with the lipid bilayer. J Virol. 2001;75(11):5205–5214. doi: 10.1128/JVI.75.11.5205-5214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50•.Gomis-Rüth FX, Dessen A, Timmins J, et al. The matrix protein VP40 from Ebola virus octamerizes into pore-like structures with specific RNA binding properties. Structure. 2003;11(4):423–433. doi: 10.1016/S0969-2126(03)00050-9. Comprises early efforts highlighting the importance of VP40 octamer formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCarthy SE, Johnson RF, Zhang YA, Sunyer JO, Harty RN. Role for amino acids 212KLR214 of Ebola virus VP40 in assembly and budding. J Virol. 2007;81(20):11452–11460. doi: 10.1128/JVI.00853-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Panchal RG, Ruthel G, Kenny TA, et al. In vivo oligomerization and raft localization of Ebola virus protein VP40 during vesicular budding. Proc Natl Acad Sci USA. 2003;100(26):15936–15941. doi: 10.1073/pnas.2533915100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adu-Gyamfi E, Digman MA, Gratton E, Stahelin RV. Single-particle tracking demonstrates that actin coordinates the movement of the Ebola virus matrix protein. Biophys J. 2012;103(9):L41–L43. doi: 10.1016/j.bpj.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu J, Qu Y, Liu Y, et al. Host IQGAP1 and Ebola virus VP40 interactions facilitate virus-like particle egress. J Virol. 2013;87(13):7777–7780. doi: 10.1128/JVI.00470-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soni SP, Adu-Gyamfi E, Yong SS, Jee CS, Stahelin RV. The Ebola virus matrix protein deeply penetrates the plasma membrane: an important step in viral egress. Biophys J. 2013;104(9):1940–1949. doi: 10.1016/j.bpj.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56•.Weissenhorn W. Oligomerization and polymerization of the filovirus matrix protein VP40. Virology. 2003;312(2):359–368. doi: 10.1016/s0042-6822(03)00260-5. Comprises early efforts highlighting importance of VP40 oligomerization. [DOI] [PubMed] [Google Scholar]
- 57••.Hoenen T, Volchkov V, Kolesnikova L, et al. VP40 octamers are essential for Ebola virus replication. J Virol. 2005;79(3):1898–1905. doi: 10.1128/JVI.79.3.1898-1905.2005. Demonstrates essential role for VP40 octamers in viral replication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang M, Orenstein JM, Martin MA, Freed EO. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69(11):6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Irie T, Licata JM, Harty RN. Functional characterization of Ebola virus L-domains using VSV recombinants. Virology. 2005;336(2):291–298. doi: 10.1016/j.virol.2005.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yasuda J, Nakao M, Kawaoka Y, Shida H. Nedd4 regulates egress of Ebola virus-like particles from host cells. J Virol. 2003;77(18):9987–9992. doi: 10.1128/JVI.77.18.9987-9992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Silvestri LS, Ruthel G, Kallstrom G, et al. Involvement of vacuolar protein sorting pathway in Ebola virus release independent of TSG101 interaction. J Infect Dis. 2007;196(Suppl 2):S264–S270. doi: 10.1086/520610. [DOI] [PubMed] [Google Scholar]
- 62.Adu-Gyamfi E, Soni SP, Jee CS, Digman MA, Gratton E, Stahelin RV. A Loop region in the N-terminal domain of Ebola virus VP40 is important in viral assembly, budding, and egress. Viruses. 2014;6(10):3837–3854. doi: 10.3390/v6103837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miao B, Skidan I, Yang J, et al. Small molecule inhibition of phosphatidylinositol-3,4,5-triphosphate (PIP3) binding to pleckstrin homology domains. Proc Natl Acad Sci USA. 2010;107(46):20126–20131. doi: 10.1073/pnas.1004522107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Soni SP, Stahelin RV. The Ebola virus matrix protein VP40 selectively induces vesiculation from phosphatidylserine-enriched membranes. J Biol Chem. 2014;289(48):33590–33597. doi: 10.1074/jbc.M114.586396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Radzimanowski J, Effantin G, Weissenhorn W. Conformational plasticity of the Ebola virus matrix protein. Protein Sci. 2014;23(11):1519–1527. doi: 10.1002/pro.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hoenen T, Biedenkopf N, Zielecki F, et al. Oligomerization of Ebola virus VP40 is essential for particle morphogenesis and regulation of viral transcription. J Virol. 2010;84(14):7053–7063. doi: 10.1128/JVI.00737-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alfadhli A, McNett H, Eccles J, et al. Analysis of small molecule ligands targeting the HIV-1 matrix protein-RNA binding site. J Biol Chem. 2013;288(1):666–676. doi: 10.1074/jbc.M112.399865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beth McDonald JM-S. No strings attached: the ESCRT machinery in viral budding and cytokinesis. J Cell Sci. 2009;122(Pt 13):2167–2177. doi: 10.1242/jcs.028308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Z, Tian H, Liu T. Cellular ESCRT complex and its roles in enveloped viruses budding. Sheng Wu Gong Cheng Xue Bao. 2012;28(9):1031–1037. [PubMed] [Google Scholar]
- 70.Votteler J, Sundquist WI. Virus budding and the ESCRT pathway. Cell Host Microbe. 2013;14(3):232–241. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71••.Neumann G, Ebihara H, Takada A, et al. Ebola virus VP40 late domains are not essential for viral replication in cell culture. J Virol. 2005;79(16):10300–10307. doi: 10.1128/JVI.79.16.10300-10307.2005. Demonstrates that PTAP and PPEY VP40 L-domain motifs are not essential for viral replication in cell culture. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72•.Lu J, Han Z, Liu Y, et al. A host-oriented inhibitor of Junin Argentine hemorrhagic fever virus egress. J Virol. 2014;88(9):4736–4743. doi: 10.1128/JVI.03757-13. Demonstrates efficacy of small-molecule PTAP inhibitor for blocking arenavirus replication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73•.Han Z, Lu J, Liu Y, et al. Small-molecule probes targeting the viral PPxY-host Nedd4 interface block egress of a broad range of RNA viruses. J Virol. 2014;88(13):7294–7306. doi: 10.1128/JVI.00591-14. Demonstrates efficacy of small-molecule PPxY inhibitor for blocking egress of a broad array of RNA viruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen BJ, Lamb RA. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology. 2008;372(2):221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ehrlich LS, Carter CA. HIV assembly and budding: Ca(2+) signaling and non-ESCRT proteins set the stage. Mol Biol Int. 2012 doi: 10.1155/2012/851670851670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Teis SA. The ESCRT machinery. Science. 2012;22(4):R116–R120. doi: 10.1016/j.cub.2012.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pornillos O, Garrus JE, Sundquist WI. Mechanisms of enveloped RNA virus budding. Trends Cell Biol. 2002;12(12):569–579. doi: 10.1016/s0962-8924(02)02402-9. [DOI] [PubMed] [Google Scholar]
- 78.Zhai Q, Fisher RD, Chung HY, Myszka DG, Sundquist WI, Hill CP. Structural and functional studies of ALIX interactions with YPX(n)L late domains of HIV-1 and EIAV. Nat Struct Mol Biol. 2008;15(1):43–49. doi: 10.1038/nsmb1319. [DOI] [PubMed] [Google Scholar]
- 79.Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe. 2011;9(3):235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Strack B, Calistri A, Craig S, Popova E, Göttlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114(6):689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 81.Weiss ER, Popova E, Yamanaka H, Kim HC, Huibregtse JM, Göttlinger H. Rescue of HIV-1 release by targeting widely divergent NEDD4-type ubiquitin ligases and isolated catalytic HECT domains to Gag. PLoS Pathog. 2010;6(9):e1001107. doi: 10.1371/journal.ppat.1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82•.García M, Cooper A, Shi W, et al. Productive replication of Ebola virus is regulated by the c-Abl1 tyrosine kinase. Sci Transl Med. 2012;4(123):123ra124. doi: 10.1126/scitranslmed.3003500. Demonstrates regulation of EBOV replication via phosphorylation of VP40 by the c-Abl1 tyrosine kinase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reid SP, Leung LW, Hartman AL, et al. Ebola virus VP24 binds karyopherin alpha1 and blocks STAT1 nuclear accumulation. J Virol. 2006;80(11):5156–5167. doi: 10.1128/JVI.02349-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Halfmann P, Neumann G, Kawaoka Y. The Ebola virus VP24 protein blocks phosphorylation of p38 mitogen-activated protein kinase. J Infect Dis. 2011;204(Suppl 3):S953–S956. doi: 10.1093/infdis/jir325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Noda T, Kolesnikova L, Becker S, Kawaoka Y. The importance of the NP: VP35 ratio in Ebola virus nucleocapsid formation. J Infect Dis. 2011;204(Suppl 3):S878–S883. doi: 10.1093/infdis/jir310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lu J, Qu Y, Liu Y, et al. Host IQGAP1 and Ebola virus VP40 interactions facilitate virus-like particle egress. J Virol. 2013;87(13):7777–7780. doi: 10.1128/JVI.00470-13. [DOI] [PMC free article] [PubMed] [Google Scholar]



