Abstract
Study Objectives:
Most organisms have behavioral and physiological circadian rhythms, which are controlled by an endogenous clock. Although genetic analysis has revealed the intracellular mechanism of the circadian clock, the manner in which this clock communicates its temporal information to produce systemic regulation is still largely unknown.
Design:
Sleep behavior was measured using the Drosophila Activity Monitoring System (DAMS) monitor under a 12 h light:12 h dark cycle and constant darkness (DD), and 5 min without recorded activity were defined as a bout of sleep.
Results:
Here we show that Drosophila insulin-like peptides (DILPs) and their receptor (DInR) regulate sleep behavior. All mutants of the seven dilps and the mutant of their receptor exhibit decreases of total sleep except dilp4 mutants, whereas upregulation of DILP and DInR in the nervous system led to increased sleep. Histological analysis identified four previously unidentified neurons expressing DILP: D1, P1, L1, and L2, of which L1 and L2 belong to the LNd and LNv clock neurons that separately regulate different times of sleep. In addition, dilp2 levels significantly decrease when flies were fasted, which is consistent with a previous report that starvation inhibits sleep, further indicating that the dilp system is involved in sleep regulation.
Conclusion:
Taken together, the results indicate that the Drosophila insulin-like peptide system is a crucial regulator of sleep.
Citation:
Cong X, Wang H, Liu Z, He C, An C, Zhao Z. Regulation of sleep by insulin-like peptide system in Drosophila melanogaster. SLEEP 2015;38(7):1075–1083.
Keywords: Drosophila melanogaster, insulin-like peptide, insulin receptor, sleep
INTRODUCTION
Sleep is widespread from insects to mammals.1 Longer waking periods lead to longer and more intense sleep periods.2 The fruit fly Drosophila melanogaster exhibits all the behavioral characteristics of mammalian sleep,3 thereby establishing Drosophila as a powerful genetic model organism to identify novel genes that modulate sleep.4
Sleep does not appear to be controlled by a single locus or dedicated genes. Over the past decade, some genes and pathways that modulate Drosophila sleep have been identified, such as cyclic adenosine monophosphate response-element binding protein (CREB),5,6 epidermal growth factor receptor, Shaker,2 sleepless,7 the gamma-aminobutyric acid (GABA) receptor,8 EcR,9 cyclin A,10 and insomniac.11
Approximately 150 clock neurons in the central nervous system are divided into three lateral neuron groups—dorso-lateral neurons (LNds), PDF-positive ventrolateral neurons (LNvs) and the fifth small ventrolateral neuron (fifth small LNv)—and three dorsal neuron groups—dorsal neurons 1, 2, and 3 (DN1, DN2, DN3). The LNvs in the circadian neuronal system contribute to sleep regulation by promoting wakefulness.12,13 The LNds function in modulating sleep suppression during starvation.14 The mushroom body (MB) and pars intercerebralis (PI) have also been recently identified as centers of regulation for sleep and wakefulness in Drosophila.15,16
Neuropeptides are small polypeptide molecules that have a wide variety of effects on regulation of development, growth, homeostasis, or behavior.17 Recent studies showed that some neuropeptides in Drosophila were found to be involved in sleep regulation, such as pigment-dispersing factor (PDF),8,12 amnesiac (amn),18 neuropeptide F (NPF),19 and short neuropep-tide F (sNPF).6 Insulin is the most extensively studied peptide hormone20 and seems to serve as both a neurotransmitter and growth factor.21 It affects diverse processes in all multicellular organisms, including growth, metabolism, development, reproduction, aging, and stress resistance.22,23 Moreover, the expression profile of insulin-like peptides (ILPs) is evolutionarily conserved among organisms. The insulin-producing cells (IPCs) in invertebrates and vertebrates may be derived from a common ancestor,24 in which the signaling mechanisms, biochemical components, and target tissues all appear to be conserved.25
The D. melanogaster genome contains seven genes encoding Drosophila insulin-like peptides (DILPs) 1 through 7, of which DILPs 1 through 5 were predicted to be most closely related to mammalian insulin,26,27 whereas DILP6 and DILP7 were predicted to be more similar to insulin-like growth factor 1 and relaxin (respectively) in vertebrates.26,28 These dilps are expressed in diverse spatiotemporal patterns during development, suggesting their different and multiple functions.25 dilp2 displays the highest messenger RNA (mRNA) expression, and it can rescue various phenotypes caused by ablation of insulin producing cells (IPCs).29 The Drosophila insulin receptor (DInR), highly similar to human InR (hInR), is a membrane-spanning tetrameric protein (α2β2).30 Essential for Drosophila development, it is expressed in the fat body surrounding the adult brain and in the corpora allata (CA).31 Once insulin binds to specific regions in the α subunit of DInR, the β subunit is activated by a rapid conformational change. This in turn causes intracellular autophosphorylation on β subunits, which initiates tyrosine kinase activity of the receptor to activate the insulin signaling pathway.26
The ILPs and InR have been identified as conserved and ubiquitous in multicellular animals.21 They have been implicated in controlling a wide range of physiological activities.32 Insulin and nutrient level have been reported to be involved in regulation of sleep in Caenorhabditis elegans and D. melanogaster, in which day and night sleep are differentially regulated by nutrient level and distinct mechanisms.26,27,33 However, the role of ILPs in regulation of sleep is still largely unexplored, and therefore we undertook further analysis of this role, as presented herein.
MATERIALS AND METHODS
Fly Strains and Rearing
In this study, wild type and mutant strains of D. melanogaster were used, including: yw and w1118, w[1118]; Ilp1[1], w[1118]; Ilp2[1], w[1118]; Ilp3[1], w[1118]; Ilp4[1], w[1118]; Ilp5[1], w[1118]; Ilp6[41], w[1118]; Ilp7[1], w[1118]; InRGC25/+, GMR28E02-gal4 (insulin receptor driver), tim-gal4, UAS-InRdel (expressing a constitutively active DInR) and UAS-mCD8-GFP [expressing green fluorescent protein (GFP)]. The Insulin recepor mutant (InR/+) must be tested as a heterozygote because the homozygote is lethal. All these lines were purchased from the Bloomington stock center, and the Ilp mutants were generated by ends-out homologous recombination or (for Ilp6) overlapping deficiencies, and all mutants are loss-of-function as verified by Gronke et al.34 The pdf-gal4 was from Dr Rouyer's laboratory (INSERM, France) and dilp2-gal4 and UAS-dilp2(II) was from Dr Ping's laboratory (University of Georgia, Athens, GA). Flies were reared at 25°C and 65% relative humidity on a standard cornmeal-yeast-agar medium in a 12 h light/12 h dark cycle.
Sleep Analysis
Three- to five-day-old male flies were housed in monitor tubes (5[W] × 65[L] mm) with fly food. Experiments were performed in an incubator at a temperature of 25 ± 1°C and a relative humidity of 60% ± 5%. Lights were turned on at Zeitgeber (ZT) 0 (local time 06:30) and off at ZT12 (local time 18:30). The sleep activity was recorded using the Drosophila Activity Monitoring System (Trikinetics, Waltham, MA). A sleep bout was defined as 5 min or more of behavioral immobility. The waking activity was calculated by dividing the total activity counts during the observation period by the length of the wake period. The details for the experimental protocol and data analysis were described by Chen.6
Immunofluorescence Analysis
Adult brains from dilp2-gal4 > UAS-mCD8-GFP flies were dissected in chilled phosphate buffered saline (PBS, pH 7.4), fixed by immersion in ice-cold 4% paraformaldehyde in PBS at room temperature for 2 h, and then rinsed for 3 × 15 min in PBS with 0.5% Triton X-100 (PBST). The brain samples were analyzed with Nikon ECLIPSE TE2000-E and Nikon D-ECLIPSE (Nikon, Japan) confocal microscopes. Confocal images were obtained at an optical section thickness of 1–2 μm and finally analyzed with Image J. Staining intensity of NPF was calculated and normalized as described.6
In order to explore the relationship between DILP neurons and clock neurons, pdf-gal4 > UAS-mCD8-GFP or timgal4 > UAS-mCD8-GFP fly brains were dissected quickly in dim light. Then samples were fixed and incubated with rabbit polyclonal anti-DILP2 as a primary antibody and anti-rabbit TRITC-tagged antibody (diluted 1:300) as a secondary antibody. Then brains were viewed under a laser-scanning confocal microscope (Nikon ECLIPSE TE2000-E and Nikon D-ECLIPSE, Japan), with an optical section thickness of 1–2 μm.
Effects of Food Deprivation on dilp2 Levels
For food deprivation experiments, 4-day-old wild type (w1118) flies were loaded into tubes containing standard fly food for 1 day of acclimation. Flies were then transferred at ZT0 (start of lights on, Day 2) to a tube containing either standard fly food (ad libitum control) or 0.5% agarose. Flies were maintained at 25°C with 12:12 LD cycles. Control and treated flies after fasting for different times were analyzed by quantitative (q)RT-PCR, separately at ZT6, ZT12, ZT18, and ZT24.
For qRT-PCR, total RNA was extracted from the heads using Trizol reagents (Qiagen, Germany) and complementary DNA was synthesized using a Quantscript RT kit (Qiagen, Germany). D. melanogaster actin was used as an internal standard. The primers for amplifying dilp2 and actin were as follows: dilp2 For (5′-CTCAATCCCCTGCAGTTTGT-3′) and dilp2 Rev (5′-CTCTCCACGATTCCTT GCC-3′), and actin For (5′-CA GAGCAAG CGTGGTATCCT-3′) and actin Rev (5′-CTCATT GTAGAAGGTGTGGTGC-3′). qRT-PCR was performed using thermal cycling conditions for dilp2 as follows: 95°C for 2 min, followed by 40 cycles of 95°C for 15 sec, 57°C for 25 sec, and 68°C for 35 sec. For each time point, three independent replicates were analyzed.
Data Analysis
Statistical analysis was performed with SPSS version 11.5 (SPSS, Chicago, IL). P values were obtained with one-way analysis of variance and considered significantly different at P < 0.05 and extremely different at P < 0.001.
RESULTS
Effects of Seven DILPs and DInR Deficient Mutants on Sleep
In order to determine whether the DILP system is involved in regulation of sleep, we analyzed mutants of all seven different insulins (from insulin 1–7) and the insulin receptor (DInR; InR[GC25]). Under photoperiod 12 light/12 dark cycles, loss of any of the seven types of DILPs (except for DILP4) and the insulin receptor significantly decreased the total sleep amount during the daytime, principally caused by reduction of sleep bout duration (Figure 1A–1D), and DILP3 also caused a significant reduction during the nighttime (Figure 1A). Under constant darkness (DD) condition, loss of all of them (except for DILP4) also caused a decrease of total sleep during daytime, mainly caused by reduction of the sleep bout duration (Figure 2A–2D), and DILPs 2, 3, 5, and 7 mutants showed lower rhythmicity, with reductions of about 24%, 63%, 25%, and 34%, respectively (Table 1). However, waking activity was not significantly affected between the controls and the mutants, indicating the effects on sleep are independent of waking activity (Figure 1E and Figure 2E). All these results above showed that DILP system has an action on Drosophila sleep.
Figure 1.
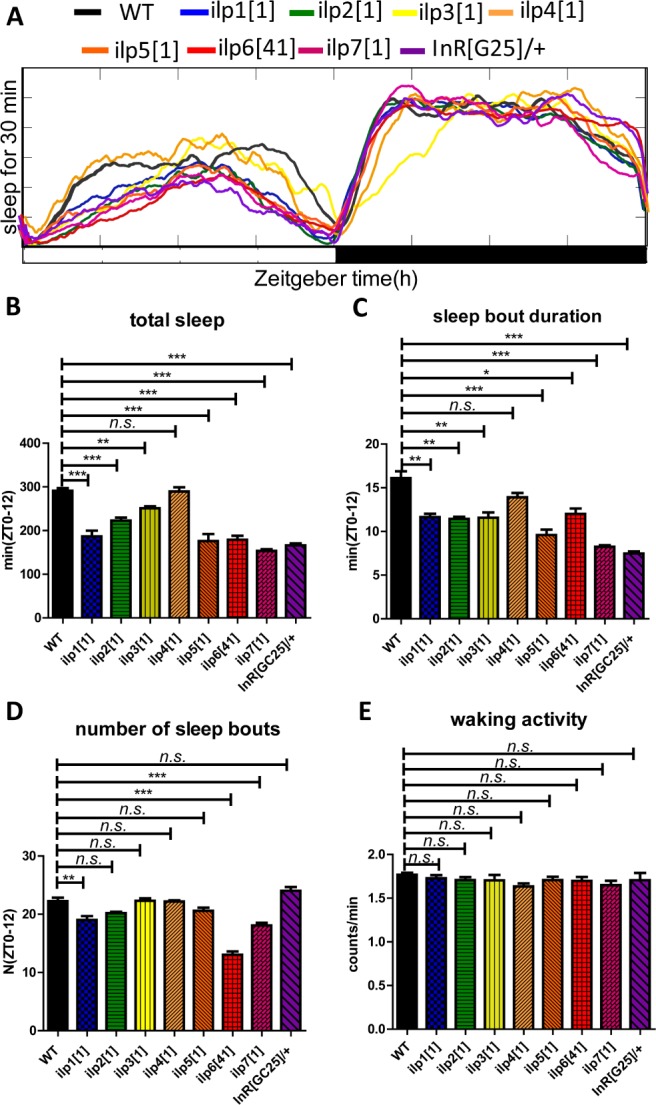
Sleep behavior of mutants of different insulin peptides and receptor under light-dark (LD) conditions. (A) Sleep profiles of the control and insulin and receptor mutant flies. A daily time course (30-min intervals) of the amount of sleep was recorded for the mutant flies and control (wild-type, WT) flies (n = 64 for each group). (B–E) The total sleep amount, sleep bout duration, number of sleep bouts and waking activity during the day (ZT 0–12) were calculated separately in male flies. The x axis indicates light periods of LD, respectively. *P < 0.05; **P < 0.01; ***P < 0.001.
Figure 2.
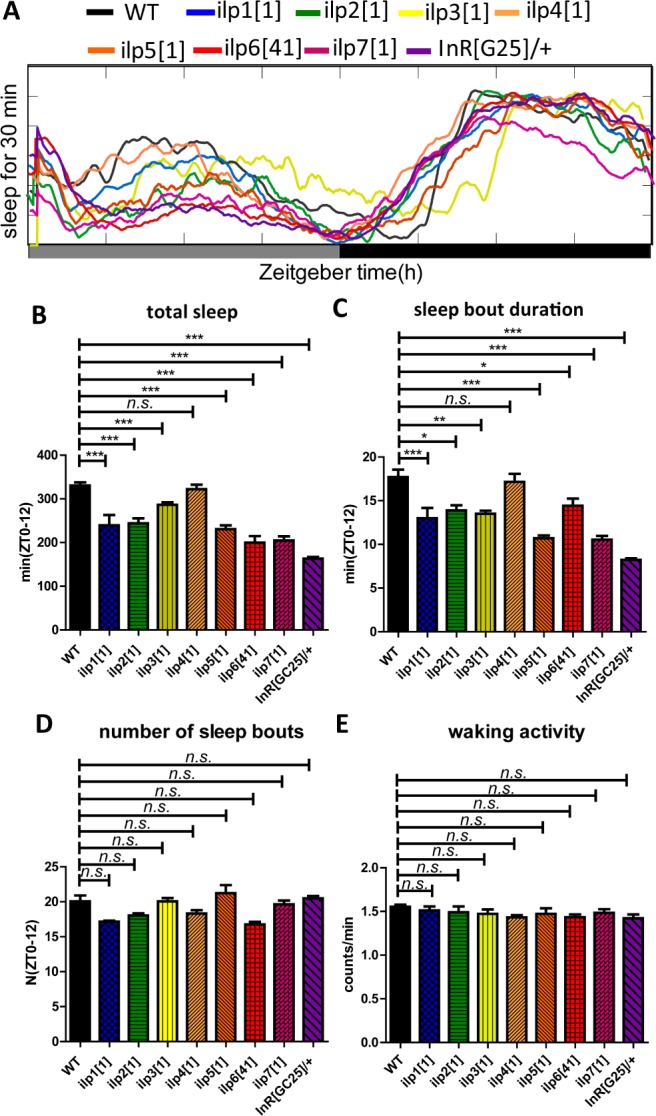
Sleep behavior of mutants of different insulin peptides and receptor under constant darkness (DD). (A) Sleep profiles of the control and insulin and receptor mutant flies. Daily time course (30-min intervals) of the amount of sleep was recorded for the mutant flies and control (wild-type, WT) flies (n = 64 for each group). (B–E) The total amounts of sleep, the sleep bout duration, number of sleep bouts, and waking activity during the day (ZT 0–12) were calculated separately in male flies. The x-axis indicates subjective day and subjective night of DD, respectively. *P < 0.05; **P < 0.01; ***P < 0.001.
Table 1.
Activity rhythms of null mutants in constant darkness.
DILP2 is Expressed in Several Neurons in the Brain, Including in Clock Neurons That Have Not Been Previously Described as Sites of Expression
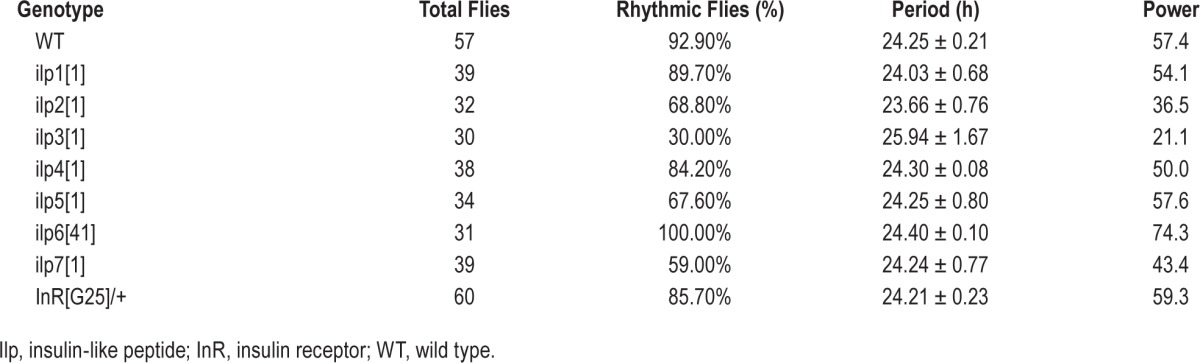
To determine the cells of the brain that could release insulin to regulate sleep, we analyzed DILP2 because it is widely expressed in the brain. We mapped the DILP2 neurons in the adult brain by specific expression of green fluorescent protein (GFP) with a dilp2-gal4 driver. We identified the known IPCs distributed in the median nerve secretory cell (MNC),35 and found four new sets of brain neurons expressing dilp2. They were classified into one posterior group (P1), one dorsal group (D1), and two lateral groups (L1 and L2) based on the positions and neurite arborization patterns in brains (Figure 3). DILP2 was detected consistently in MNC, P1, D1, and L1 neurons in 100% of 23 studied individual brains, and in L2 neurons in 56.5% of 23 studied individual brains (Table 2). Further inspection revealed that P1, D1, and L1 consisted of single neurons per brain lobe, whereas the MNC groups ranged from five to eleven cells (average of 8.33 ± 2.17) and the L2 group from one to three cells (average of 1.88 ± 0.22) (Table 2).
Figure 3.
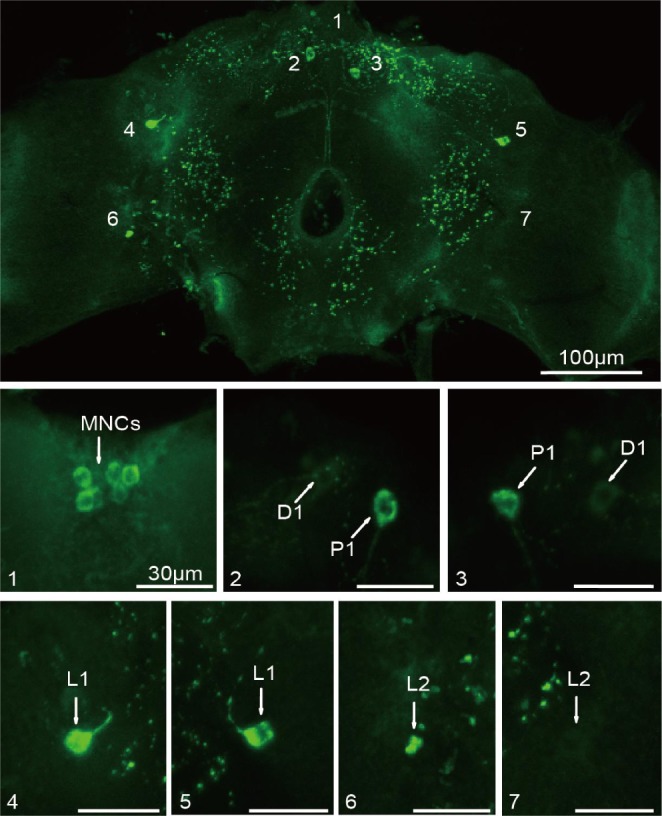
Localization of Drosophila insulin-like peptides (DILP) neurons in adult brains. DILP neurons labeled by expressing green fluorescent protein (GFP) through a dilp2-Gal4 driven GFP reporter in whole-mount brains. Panels 1 through 7 are the enlarged neurons from the whole-mount figure. Specific neurons are indicated by arrows. Scale bars show 100 μm for the whole-mount figure and 30 μm for all detailed figures.
Table 2.
Distribution of Drosophila insulin-like peptide 2 neurons in adult brains.

Furthermore, we determined the relationship between DILP2 neurons and circadian clock neurons. By using double immunofluorescence, fly brains expressing GFP (green) driven separately by pdf- and tim-gal4 as markers for the LNvs (including l-LNvs and s-LNvs) and all clock neurons (including the LNds), respectively, were constructed. Then, DILP neurons (red) in the brains were detected using an anti-DILP2 antibody. Results showed that the L1 and L2 neurons expressing DILP2 were colocalized with the dorsal lateral neurons (LNds) (Figure 4C and 4D) and ventral lateral neurons (LNvs) (Figure 4A and 4B), respectively.
Figure 4.
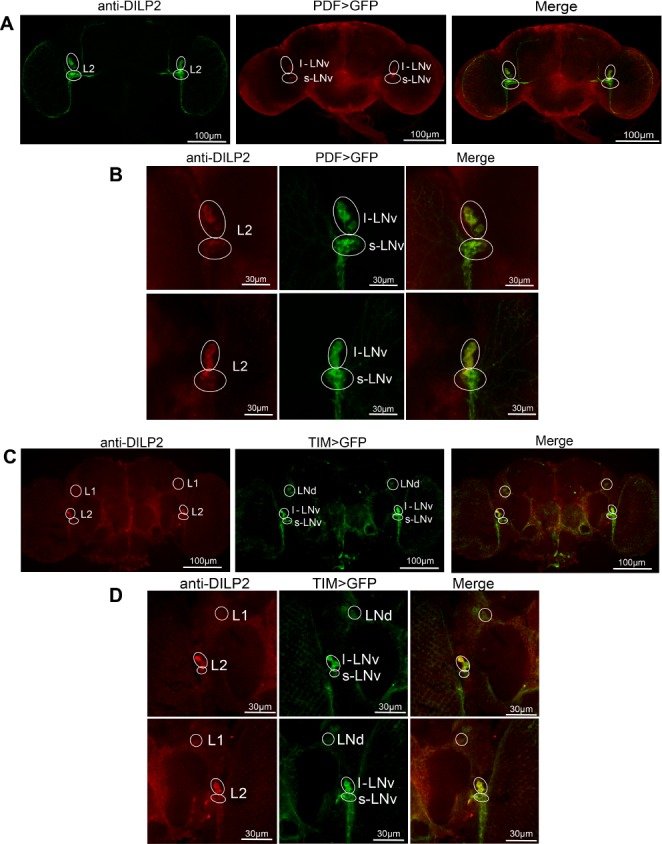
Double immunofluorescence of Drosophila insulin-like peptides (DILP) neurons and clock neurons. (A) Projections of the whole brain were observed, pdf-gal4;;UAS-mCD8-GFP brains were stained with anti-DILP2 antibody. Green cells are the PDF neurons (the LNvs), red cells are the DILP2 neurons, and yellow cells exhibit co-localization between both signals. (B) Magnified views of merged regions in A, revealing colocalization of L2 and LNvs (s-LNvs and l-LNvs) in some of the cell bodies in which the two are expressed. (C) Localization of L1 and L2 neurons (red) with the LNd and LNv (respectively) (tim-gal4;;UAS-mCD8-GFP). (D) Higher magnifications of lateral segments of the whole brain in B, revealing colocalization of L1/LNds and L2/LNvs.
Effects of DILP2 and DInR in Clock Neurons and Other Neurons on Sleep
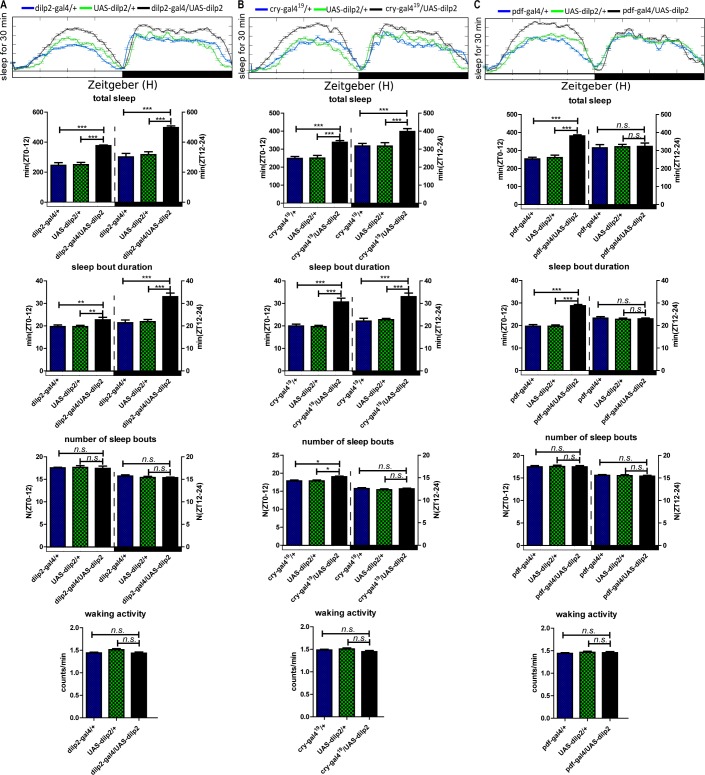
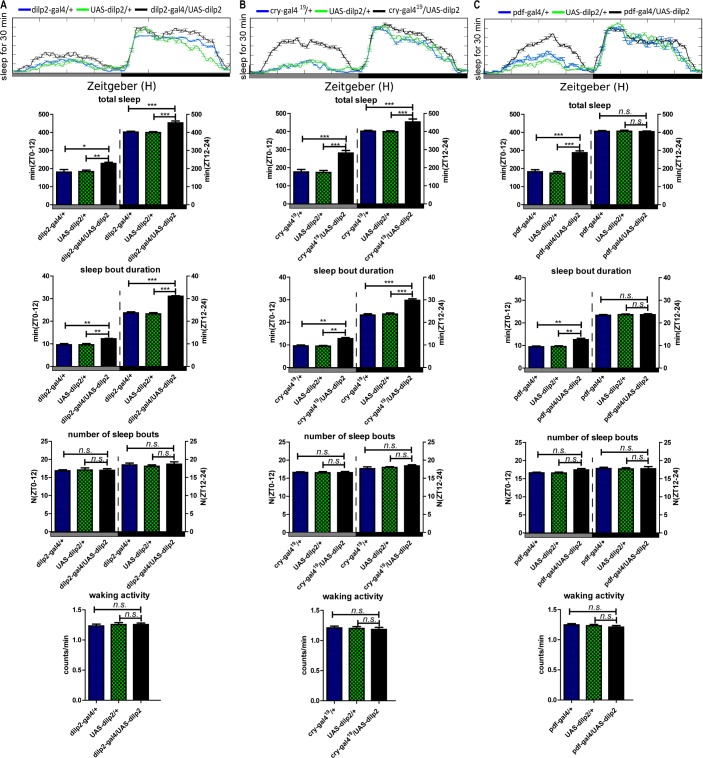
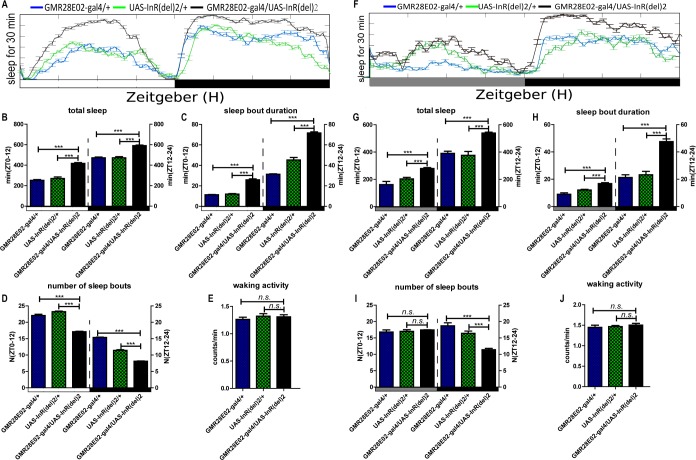
From previous reports, the LNd and LNv clock neurons are involved in sleep regulation.36 To determine the sleep actions of DILP2 in clock and all dilp-expressing neurons, we used pdfgal4, which drives gene expression in all large LNvs (l-LNvs) and 4 small LNvs (s-LNvs) neurons in the fly brain, cry-gal4, which drives gene expression in some LNds, DN1, and LNvs,37 and dilp2-gal4, which drives gene expression in the dilp2-expressing neurons. Upregulated expression of dilp2 in dilp2 and cry neurons significantly increased the total sleep both during the daytime and nighttime, caused by an increase of sleep bout duration under both LD and DD cycles (Figure 5A and 5B and Figure 6A and 6B). But upregulated dilp2 in pdf neurons significantly increased the total sleep only during the daytime under both LD and DD conditions (Figure 5C and Figure 6C). Results also showed that upregulated dilp2 flies exhibited a higher rhythmic percentage in DD (Table 3). Moreover, upregulation of the insulin receptor (DInR) in its endogenous neurons driven by the GMR28E02-gal4 (a dinr driver) had similar effects on the sleep pattern as upregulation of DILP2 (Figure 7). In contrast with the sleep bout duration, waking activity was also not significantly affected by gain of dilp2 and DInR function (Figures 5–7). All the aforementioned results show DILP2 regulates sleep, with the LNvs mainly controlling sleep during the daytime. The regulation of total sleep is mainly through the control of the sleep bout duration.
Figure 5.
Sleep behavior of the Drosophila insulin-like peptides (DILP)2 gain-of-function flies in a light-dark (LD) cycle. (A) Results for DILP2 expression driven by dilp2-gal4. (B) Results for DILP2 expression driven by cry-gal4. (C) Results for DILP2 expression driven by pdf-gal4. The total sleep amounts, sleep bout durations, number of sleep bouts during the day (ZT 0–12) and night (ZT 12–24), and waking activity were calculated separately. Horizontal white and black boxes along the x-axis indicate light and dark periods of LD, respectively. (n = 62 for each group) **P < 0.01; ***P < 0.001.
Figure 6.
Sleep behavior of the Drosophila insulin-like peptides (DILP)2 gain-of-function male flies in a constant darkness (DD) condition. (A) Results for DILP2 expression driven by dilp2-gal4. (B) Results for DILP2 expression driven by cry-gal4. (C) Results for DILP2 expression driven by pdf-gal4. The total sleep amounts, sleep bout durations, number of sleep bouts during the subjective day (CT 0–12) and subjective night (CT 12–24) and waking activity were calculated separately. Horizontal white and black boxes along the x-axis indicate subjective day and night periods of DD, respectively. (n = 62 for each group). *P < 0.05; **P < 0.01; ***P < 0.001.
Table 3.
Activity rhythms of flies with up regulated dilp and dInR in DD.
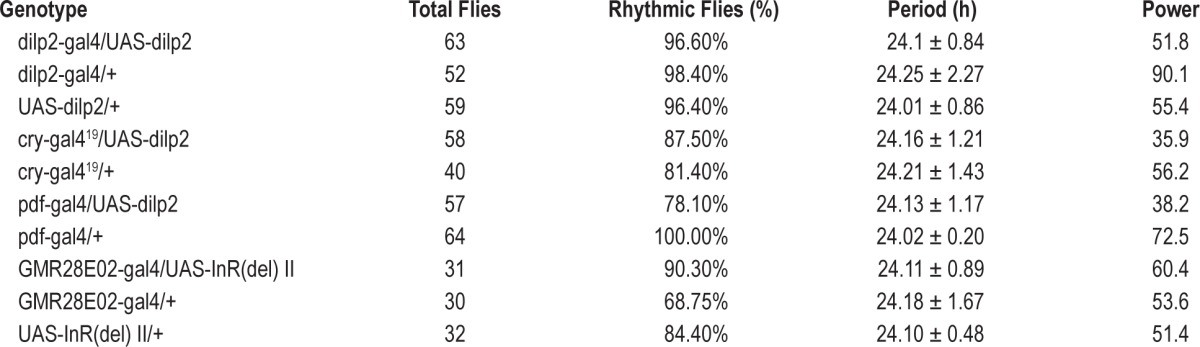
Figure 7.
Sleep behavior of the Drosophila insulin-like receptor (DInR) gain-of-function male flies in light-dark (LD) and constant darkness (DD) conditions. (A) Results for DInR endogenous expression driven by GMR28E02-gal4 in LD cycle. (F) Results for DInR endogenous expression driven by GMR28E02-gal4 in DD condition. (A–E) Total sleep amounts, sleep bout durations, number of sleep bouts during the day (ZT 0–12) and night (ZT 12–24), and waking activity were calculated separately. Horizontal white and black boxes along the x-axis indicate light and dark periods of LD (A–E), respectively (n = 58 for each group). **P < 0.01; ***P < 0.001. (F–J) Horizontal gray and black boxes along the x-axis indicate subjective day and night periods of DD, respectively (n = 58 for each group) *P < 0.05; **P < 0.01; ***P < 0.001.
To be sure that the binary expression approach (GAL4 > UAS) was overexpressing the responder gene, we measured the mRNA levels of the responder gene in each transgenic line through quantitative reverse transcription-polymerase chain reaction (RT-PCR), and thereby validated the transgenic system because the responder genes were overexpressed (see Figure S1, supplemental material).
Food Deprivation Reduces dilp2 Level
After the flies had fasted for 6 h, dilp2 levels in wild type flies started to significantly decrease compared to controls. The dilp2 levels after fasting 6, 12, 18, and 24 h decreased 36.85% (P < 0.001), 48.71% (P < 0.001), 49.25% (P < 0.001), and 44.22% (P < 0.001), respectively (Figure 8). These results are consistent with a previous report that starvation inhibits sleep,14 further indicating that the dilp system is involved in sleep regulation.
Figure 8.
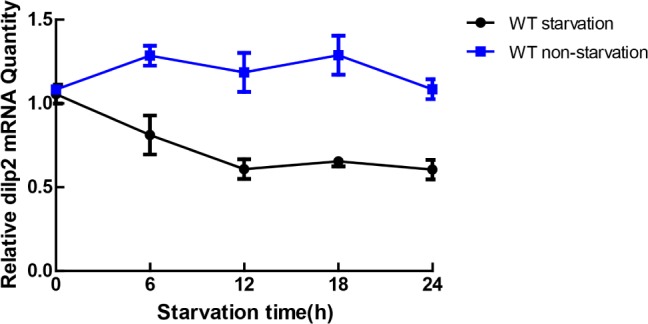
Starvation reduces Drosophila insulin-like peptide (dilp) level analyzed by quantitative polymerase chain reaction (qPCR). Profile of dilp level by qPCR in starved and unstarved flies. The 4-day-old wild-type (w118) flies were deprived for 24 h starting at Zeitbeger time (ZT) 0. The x-axis represents the ZT. RNA levels are normalized to the amount present in the flies at ZT0. Data were collected every 6 h. Error bars indicate the standard error of the mean from three independent replicates.
DISCUSSION
Here, we used genetic and molecular analysis in adult flies to show that insulin (principally dilp2) and its receptor regulate sleep, because loss-of-function mutants sleep less and gain-of-function mutants sleep more. Interestingly, Metaxakis et al.38 found that loss-of-function mutations in multiple dilp genes (dilp2-3,5, a long-lived Drosophila strain) led to decreased day sleep but also increased night sleep, and night and day sleep were regulated through distinct mechanisms.26,27 This means that dilp single gene mutants probably did not reduce total ILPs enough to initiate signaling of night sleep in this study.
Sleep circuits are intimately linked to the circadian system, thereby ensuring the appropriate times for sleep and wake. Sleep homeostatic and circadian regulation have been shown to be controlled by an intricate neuronal circuitry involving the circadian clock neurons, the MB, and the PI.6 Mice with clk and bmal mutations (especially bmal loss of function) showed impaired glucose tolerance and reduced insulin secretion,39 indicating that the clock genes regulate insulin secretion.
Previous studies have identified only one group of DILP cells, the MNCs (including PIs), in larval brains.31,40 In this study, we identified several more in adult brains. Our observation of additional cells is probably because the LNds and l-LNv are not typically seen in larvae. Here in adult brains, we found four more DILP-secreting neurons—the P1, D1, L1, and L2 neurons (Figure 3)—stably expressing DILP. By colocalizing DILP2 with pdfgal4 and tim-gal4 expression, we found that the L1 and L2 neurons expressing DILP actually are the clock neurons LNds and LNvs, respectively (Figure 4). The LNvs promote wakefulness controlled by the GABA receptor and PERIOD protein.13,41 The LNds suppress sleep during starvation.14 Our data show that the LNvs mostly regulate daytime sleep and the LNds mostly regulate nighttime sleep. We also found that upregulation of the dilp system (both dilp and dInR) increase both daytime and nighttime sleep, whereas lowered signaling through this system has some effects on nighttime sleep but the daytime effects are more prominent, indicating that day sleep is more sensitive to insulin signaling. Furthermore, a previous study showed that starvation inhibits sleep.14 In this study, we found that starvation for 6 h or more caused a significant decrease in dilp2 mRNA (Figure 8). All these data indicate DILP2 is involved in the relationship between food deprivation and sleep.
In this study, we found that loss of most dilps could reduce sleep. However, its receptor has been reported to have only one in Drosophila, which means that all dilps work probably through the same receptor. From Figures 1 and 2, we found that only the heterozygous (InR[GC25]/+) dilp receptor mutants did reduce sleep more than any single dilp mutant, especially on the sleep bout duration. In addition, different dilps are expressed in diverse spatiotemporal patterns, suggesting their actions at different time and sites.25 However, it seems to be more complicated because these dilps may partially work together or work independently at different times. The temporal and spatial requirements for dilp function will be addressed by further work in the future.
DILP (mainly DILP2 in our experiments) is transferred to the corpus cardiacum/corpus allatum (CC/CA) ring gland complex through long axons.40 In addition, the receptor DInR has also been found in the endocrine cells of the CA in adult Drosophila bodies, where important hormones, such as juvenile hormone (JH), are synthesized and released.42 Specifically knocking down DInR in the CA downregulates the expression of 3-hydroxy-3-methylglutaryl CoA reductase (HMG-COAR), a key enzyme in JH synthesis, resulting in an 80% decrease in the level of JH.43 Taking this into account, it can be concluded that the DILP/DInR system, which is regulated by the clock neurons LNds and LNvs, may serve as a key regulator of the neuroendocrine system, and may play roles in the synthesis and release of JH. Thus, sleep could be regulated by interaction of these aforementioned factors. However, DILP has also been detected in other types of brain neurons, indicating that DILP is a multieffector involved in other functions, such as feeding behavior and growth.21,28
DISCLOSURE STATEMENT
This was not an industry supported study. The authors have indicated no financial conflicts of interest.
ACKNOWLEDGMENTS
The authors thank Dr Francois Rouyer (INSERM, France) for pdf-gal4, Ping Shen (University of Georgia, Athens, GA) for dilp2-gal4 and UAS-dilp2(II), and Giorgio F. Gilestro (Imperial College London, UK) for helping with the use of sleep analysis software. We also thank J. Price (University of Missouri) for the comments and revision on an earlier draft of this manuscript. This work was supported by the National Basic Research Program from Ministry of Science and Technology of the Peoples Republic of China (“973” Program Grant number 2012CB114100) and the National Natural Science Foundation of China (Grant number 31272371) to Z. Zhao, and the National Natural Science Foundation of China (Grant number 31172090) to C. An.
SUPPLEMENTAL MATERIAL
Assays of dilp2 levels to verify the GAL4/UAS system. (A) UAS-dilp2/dilp-Gal4, UAS-dilp2/+ and dilp-Gal4/+. (B) UAS-dilp2/cry-Gal4, UAS-dilp2/+ and cry-Gal4/+. (C) UAS-dilp2/pdf-Gal4, UAS-dilp2/+ and pdf-Gal4/+. (D) UAS-InR(del)/GMR28E02-gal4, UAS-InR(del)/+ and GMR28E02-gal4/+. The quantitative polymerase chain reaction assays from adult heads were explored at two time points in all tested strains. Levels were normalized relative to actin. *P < 0.05; **P < 0.01; ***P < 0.001, and the bar heights indicate mean values ± standard error of the mean.
REFERENCES
- 1.Allada R, Siegel JM. Unearthing the phylogenetic roots of sleep. Curr Biol. 2008;18:R670–9. doi: 10.1016/j.cub.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cirelli C, Bushey D, Hill S, et al. Reduced sleep in Drosophila shaker mutants. Nature. 2005;434:1087–92. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- 3.Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–7. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 4.Harbison ST, Mackay TFC, Anholt RRH. Understanding the neurogenetics of sleep: progress from Drosophila. Trends Genet. 2009;25:262–9. doi: 10.1016/j.tig.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hendricks JC, Williams JA, Panckeri K, et al. A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat Neurosci. 2001;4:1108–15. doi: 10.1038/nn743. [DOI] [PubMed] [Google Scholar]
- 6.Chen WF, Shi W, Li LZ, et al. Regulation of sleep by the short neuropeptide F (sNPF) in Drosophila melanogaster. Insect Biochem Molec. 2013;43:809–19. doi: 10.1016/j.ibmb.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Koh K, Joiner WJ, Wu MN, Yue ZF, Smith CJ, Sehgal A. Identification of SLEEPLESS, a sleep-promoting factor. Science. 2008;321:372–6. doi: 10.1126/science.1155942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung BY, Kilman VL, Keath JR, Pitman JL, Allada R. The GABA(A) receptor RDL acts in epileptogenic PDF neurons to promote sleep in Drosophila. Curr Biol. 2009;19:386–90. doi: 10.1016/j.cub.2009.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishimoto H, Kitamoto T. The steroid molting hormone ecdysone regulates sleep in adult Drosophila melanogaster. Genetics. 2010;185:269–U403. doi: 10.1534/genetics.110.114587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rogulja D, Young MW. Control of sleep by cyclin A and its regulator. Science. 2012;335:1617–21. doi: 10.1126/science.1212476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stavropoulos N, Young MW. Insomniac and Cullin-3 regulate sleep and wakefulness in Drosophila. Neuron. 2011;72:964–76. doi: 10.1016/j.neuron.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parisky KM, Agosto J, Pulver SR, et al. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron. 2008;60:672–82. doi: 10.1016/j.neuron.2008.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shang YH, Haynes P, Pirez N, et al. Imaging analysis of clock neurons reveals light buffers the wake-promoting effect of dopamine. Nat Neurosci. 2011;14:889–95. doi: 10.1038/nn.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keene AC, Duboue ER, McDonald DM, et al. Clock and cycle limit starvation-induced sleep loss in Drosophila. Curr Biol. 2010;20:1209–15. doi: 10.1016/j.cub.2010.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crocker A, Shahidullah M, Levitan IB, Sehgal A. Identification of a neural circuit that underlies the effects of octopamine on sleep:wake Behavior. Neuron. 2010;65:670–81. doi: 10.1016/j.neuron.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foltenyi K, Greenspan RJ, Newport JW. Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat Neurosci. 2007;10:1160–7. doi: 10.1038/nn1957. [DOI] [PubMed] [Google Scholar]
- 17.Nassel DR, Winther AME. Drosophila neuropeptides in regulation of physiology and behavior. Prog Neurobiol. 2010;92:42–104. doi: 10.1016/j.pneurobio.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 18.Liu WJ, Guo F, Lu BK, Guo AK. Amnesiac regulates sleep onset and maintenance in Drosophila melanogaster. Biochem Bioph Res Co. 2008;372:798–803. doi: 10.1016/j.bbrc.2008.05.119. [DOI] [PubMed] [Google Scholar]
- 19.He CX, Yang YY, Zhang MM, Price JL, Zhao ZW. Regulation of sleep by neuropeptide y-like system in Drosophila melanogaster. Plos One. 2013;8:e74237. doi: 10.1371/journal.pone.0074237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuo WL, Gehm BD, Rosner MR. Cloning and expression of the cDNA for a Drosophila insulin-degrading enzyme. Mol Endocrinol. 1990;4:1580–91. doi: 10.1210/mend-4-10-1580. [DOI] [PubMed] [Google Scholar]
- 21.Wu Q, Brown MR. Signaling and function of insulin-like peptides in insects. Annu Rev Entomol. 2006;51:1–24. doi: 10.1146/annurev.ento.51.110104.151011. [DOI] [PubMed] [Google Scholar]
- 22.Butler AA, Le Roith D. Control of growth by the somatropic axis: growth hormone and the insulin-like growth factors have related and independent roles. Annu Rev Physiol. 2001;63:141–64. doi: 10.1146/annurev.physiol.63.1.141. [DOI] [PubMed] [Google Scholar]
- 23.Porte D, Baskin DG, Schwartz MW. Insulin signaling in the central nervous system: a critical role in metabolic homeostasis and disease from C-elegans to humans. Diabetes. 2005;54:1264–76. doi: 10.2337/diabetes.54.5.1264. [DOI] [PubMed] [Google Scholar]
- 24.Wang S, Tulina N, Carlin DL, Rulifson EJ. The origin of islet-like cells in Drosophila identifies parallels to the vertebrate endocrine axis. Proc Natl Acad Sci U S A. 2007;104:19873–8. doi: 10.1073/pnas.0707465104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. 2001;11:213–21. doi: 10.1016/s0960-9822(01)00068-9. [DOI] [PubMed] [Google Scholar]
- 26.Giannakou ME, Partridge L. Role of insulin-like signalling in Drosophila lifespan. Trends Biochem Sci. 2007;32:180–8. doi: 10.1016/j.tibs.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, Liu JN, Li CR, Momen B, Kohanski RA, Pick L. Deletion of Drosophila insulin-like peptides causes growth defects and metabolic abnormalities. Proc Natl Acad Sci U S A. 2009;106:19617–22. doi: 10.1073/pnas.0905083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slaidina M, Delanoue R, Gronke S, Partridge L, Leopold P. A Drosophila insulin-like peptide promotes growth during nonfeeding states. Dev Cell. 2009;17:874–84. doi: 10.1016/j.devcel.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296:1118–20. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- 30.Fernandez R, Tabarini D, Azpiazu N, Frasch M, Schlessinger J. The Drosophila insulin-receptor homolog - a gene essential for embryonic-development encodes 2 receptor isoforms with different signaling potential. Embo J. 1995;14:3373–84. doi: 10.1002/j.1460-2075.1995.tb07343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belgacem YH, Martin JR. Disruption of insulin pathways alters trehalose level and abolishes sexual dimorphism in locomotor activity in Drosophila. J Neurobiol. 2006;66:19–32. doi: 10.1002/neu.20193. [DOI] [PubMed] [Google Scholar]
- 32.Hafen E. Cancer, type 2 diabetes, and ageing: news from flies and worms. Swiss Med Wkly. 2004;134:711–9. doi: 10.4414/smw.2004.09885. [DOI] [PubMed] [Google Scholar]
- 33.You YJ, Kim J, Raizen DM, Avery L. Insulin, cGMP, and TGF-beta signals regulate food intake and quiescence in C-elegans: a model for satiety. Cell Metabolism. 2008;7:249–57. doi: 10.1016/j.cmet.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gronke S, Clarke DF, Broughton S, Andrews TD, Partridge L. Molecular evolution and functional characterization of Drosophila insulin-like peptides. Plos Genetics. 2010;6:e1000857. doi: 10.1371/journal.pgen.1000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haselton AT, Fridell YWC. Adult Drosophila melanogaster as a model for the study of glucose homeostasis. Aging. 2010;2:523–6. doi: 10.18632/aging.100185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardin PE. Molecular mechanisms of circadian timekeeping in Drosophila. Sleep Biol Rhythms. 2009;7:235–42. [Google Scholar]
- 37.Im SH, Li WH, Taghert PH. PDFR and CRY signaling converge in a subset of clock neurons to modulate the amplitude and phase of circadian behavior in Drosophila. Plos One. 2011;6:e18974. doi: 10.1371/journal.pone.0018974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Metaxakis A, Tain LS, Gronke S, et al. Lowered insulin signalling ameliorates age-related sleep fragmentation in Drosophila. Plos Biology. 2014;12:e1001824. doi: 10.1371/journal.pbio.1001824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.la Fleur SE, Kalsbeek A, Wortel J, Fekkes ML, Buijs RM. A daily rhythm in glucose tolerance: a role for the suprachiasmatic nucleus. Diabetes. 2001;50:1237–43. doi: 10.2337/diabetes.50.6.1237. [DOI] [PubMed] [Google Scholar]
- 40.Cao C, Brown MR. Localization of an insulin-like peptide in brains of two flies. Cell Tissue Res. 2001;304:317–21. doi: 10.1007/s004410100367. [DOI] [PubMed] [Google Scholar]
- 41.Bushey D, Tononi G, Cirelli C. Sleep and synaptic homeostasis: structural evidence in Drosophila. Science. 2011;332:1576–81. doi: 10.1126/science.1202839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tu MP, Yin CM, Tatar M. Mutations in insulin signaling pathway alter juvenile hormone synthesis in Drosophila melanogaster. Gen Comp Endocr. 2005;142:347–56. doi: 10.1016/j.ygcen.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 43.Belgacem YH, Martin JR. Hmgcr in the Corpus Allatum Controls Sexual Dimorphism of Locomotor Activity and Body Size via the Insulin Pathway in Drosophila. Plos One. 2007;2:e187. doi: 10.1371/journal.pone.0000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Assays of dilp2 levels to verify the GAL4/UAS system. (A) UAS-dilp2/dilp-Gal4, UAS-dilp2/+ and dilp-Gal4/+. (B) UAS-dilp2/cry-Gal4, UAS-dilp2/+ and cry-Gal4/+. (C) UAS-dilp2/pdf-Gal4, UAS-dilp2/+ and pdf-Gal4/+. (D) UAS-InR(del)/GMR28E02-gal4, UAS-InR(del)/+ and GMR28E02-gal4/+. The quantitative polymerase chain reaction assays from adult heads were explored at two time points in all tested strains. Levels were normalized relative to actin. *P < 0.05; **P < 0.01; ***P < 0.001, and the bar heights indicate mean values ± standard error of the mean.